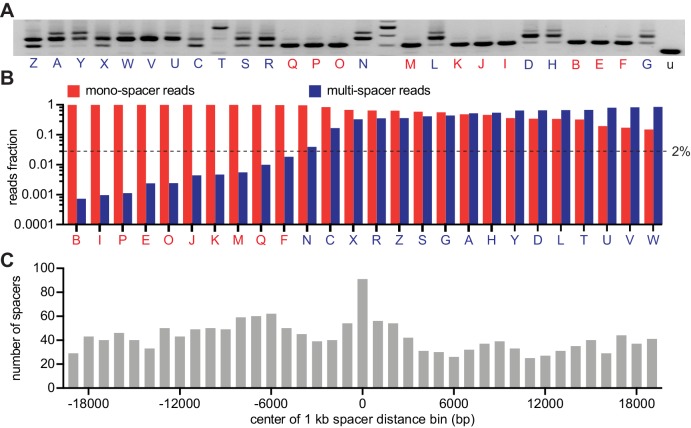
Figure 2. CRISPR expansion likely involves priming by the first acquired spacer.
(A) Agarose gel electrophoresis of the products of the amplification of the pCRISPR array present in staphylococci infected in semi-solid media with ϕNM4γ4, using plasmid DNA templates extracted from 26 different surviving colonies and the unexpanded array (u); red and blue letters: mono- and multi-spacer colony names. (B) Fraction of the reads obtained after NGS of the PCR products shown in (A) containing either a single or multiple spacers (red and blue bars, respectively). Dashed line indicates the 2% value, the minimal fraction of multi-spacer reads that leads to multiple PCR products after amplification of the pCRISPR array. (C) Distance between the targets in the ϕNM4γ4 genome specified by the first and second spacers acquired after infection of staphylococci carrying pCRISPR; obtained from analysis of NGS data. The number of different second spacers within 1 kb bins of the ϕNM4γ4 genome are shown; the position of first spacer acquired in each array is set as 0 kb.