Abstract
Understanding polymicrobial interactions involving fungi in the environment and the human mycobiome is necessary to address environmental and medically related problems such as drought or antimicrobial resistance. The diversity of these interactions highlights the complexity of fungi, considering how some interactions can be antagonistic, while others synergistic. Over the years, an increase in studies on the mycobiome have revealed similarities between the human and environmental hosts. More recently, studies have focused on microbial commensal relationships and identifying causative agents of human disease. The overlap of some of these interactions is impossible to ignore, indicating that there are areas for medical exploitation that need to be further investigated. This review provides the latest advances in polymicrobial interactions involving fungi and discusses the importance of the fungal lifestyle in the environment and in human disease.
Keywords: biofilms, disease, environment, fungi, mycobiome, polymicrobial interactions
1. INTRODUCTION
Polymicrobial interactions involving fungi are important in plant growth, prevention and exacerbation of human disease, enhanced resistance to antimicrobials (Frey-Klett et al., 2011; Peleg et al., 2010; Shirtliff et al., 2009) and vary in complexity, depending on the microbes involved. The recognition of these complex symbioses, particularly in some bacterial-fungal interactions, has led to the consideration that these microbial communities act as a single metaorganism (Deveau et al., 2018). Polymicrobial interactions involving fungi are also essential in the growth and survival of plants aiding to overcome challenges like drought, alkaline soil environments, and disease. Interactions of fungi with other microbes may enhance antimicrobial resistance likely by adaptation and co-existence. This area of study is of particular significance, as drug resistance continues to be a contemporary problem, especially among medically important microorganisms. Moreover, the human microflora, mainly the bacterial microbiome, has been widely studied to understand the relationships, both commensal and pathogenic, between microbes and the human host. The fungal microbiome, often referred to as mycobiome (Ghannoum et al., 2010), has been of recent interest, with studies now focusing on how these eukaryotic microbes’ factor into the commensal network. In the last decade, the amount of publications that show up with a “mycobiome” search on PubMed jumped from 1 to 491 and counting. Studies conducted on the human mycobiome fall within different anatomical systems of the body including the digestive, respiratory, genitourinary, and integumentary.
The diversity of the mycobiome has just begun elucidation and its impact in the human microflora can be explored in the context of disease. The recent boom in understanding the fungal diversity in the mycobiome is credited to the burden of medically important fungi, particularly in immunocompromised patients. The balance of the mycobiome plays important roles in disease prevention, onset, and progression. Understanding the symbiosis of fungal species present in the host mycobiome and other microbes would provide insight on chronic diseases such as cystic fibrosis (CF) and Crohn’s disease (Cui et al., 2013). Multiple studies have found that, similar to the microbiome, most organisms that reside in the human mycobiome are unculturable (Cui et al., 2013; Nash et al., 2017). Hence, recent advances in next generation sequencing techniques have provided novel understanding of the function of the mycobiome in human disease. This information can be utilized to assess the physical and chemical interactions fungi use to compete for nutrients and niche dominance as well as being able to cause disease. Moreover, this knowledge can be incorporated in the development of therapeutics that specifically target keeping the balance of these polymicrobial interactions, enhancing health and diminishing or preventing the exacerbation of these often-fatal illnesses. In this review, we discuss polymicrobial interactions involving fungi in the environment, highlighting the importance of further studies particularly in the role of these communities on antimicrobial resistance (Table 1). We then transition to interactions within the mycobiome (Table 2), where we discuss the larger impact fungi have in human disease, and how best to approach treatment in these patients.
Table 1.
Environmentally and medically important polymicrobial interactions involving fungi.
| Prevalent fungi | Interactions | Description and reference (s) |
|---|---|---|
| Rhizophagus intraradices | Bacillus thuringiensis Pseudomonas putida | Increased water and nutrient uptake, and decreased electrolyte leakage and stomatal conductance in Trifolium repens plants [Ortiz et al 2015; Lane et al., 1997] |
| Acaulospora laevis Glomus geosporum Glomus mosseae Scutellospora armeniaca | Rhizobium leguminosarum bv. viciae | Increased nodule formation, root length, nitrogenase activity, and decreased malondialdehyde content in faba beans [Ahmed et al., 2008; Abd-Alla et al., 2014] |
| Rhizoglomus irregularis | rhizobia strains from Lotus corniculatus | Maximal nitrogen fixation and plant diversity, and increased seedling establishment in Dutch dune grassland microcosms [van der Heijden et al.; 2016, Veer et al., 1997] |
| Fusarium oxysporum | Pseudomonas chlororaphis strain PCL1391 | Production of an antifungal factor by PCL1391 was capable of preventing root rot disease in tomatoes [Chin-A-Woeng et al., 1998] |
| Phytophthora infestans | Fusarium oxysporum strain EF119 | Interaction resulted in growth inhibition of P. infestans [Kim et al., 2007] |
| Fusarium culmorum | Piriformospora indica chlamydospores | Chlamydospore inoculation prevented root rot caused by F. culmorum [Harrach et al., 2013] |
| Puccinia triticina | Chaetomium spp. and Phoma spp. | Decreased number of P. triticina pustules in wheat [Dingle and McGee, 2003] |
| Pseudomonas | Botrytis cinerea | Inoculation of fungal isolate on Arabidopsis thaliana plants caused enhanced plant resistance to B. cinerea [Ritpitakphong et al., 2016] |
| Neotyphodium uncinatum | barley yellow dwarf virus (BYDV) | Meadow ryegrass infected with fungal endophytes had decreased susceptibility to BYDV [Lehtonen et al., 2006] |
| Cryphonectria parasitica | reovirus isolate similar to the Reoviridae family | Reovirus reduced fungal virulence and altered colony morphology [Hillman et al., 2004] |
| Cryphonectria parasitica | Cryphonectria hypovirus 1 strains | Hypovirulence of C.parasitica and alteration of cytosine methylation on fungal genome [Krstin et al., 2017] |
| Sclerotinia nivalis, S. minor, and Botrytis cinerea | S. sclerotiorum partitivirus 1 | Partitivirus causes fungal hypovirulence [Xiao et al., 2014; Xu et al., 2015] |
| Candida albicans | Escherichia coli | β−1, 3-glucans present in interaction affected E. coli ofloxacin tolerance [De Brucker et al., 2015] |
| C. albicans | Staphylococcus aureus | FKS1 gene inhibition in fungus increased bacterial susceptibility to vancomycin; dual-species biofilm resulted in increased fungal resistance to miconazole [Kong et al., 2016; Kean et al., 2017] |
| C. albicans | Pseudomonas aeruginosa | Bacterium was unable to kill fungi in its yeast form [(Hogan and Kolter, 2002] |
| Cryptococcus neoformans var. grubii (serotype A) and C. neoformans var. neoformans (serotype D) | Acinetobacter baumannii | Serotype A was more bacterial-resistant and had increased biofilm and capsule formation compared to serotype D [Abdulkareem et al., 2015] |
| C. neoformans | S. aureus | Fungal viability decreased in capsular mutants co-cultured with bacteria [Saito and Ikeda, 2005] |
| Aspergillus niger | Salmonella enterica | Absence of cellulose synthesis genes in bacteria prevented biofilm formation on fungus [Brandl et al., 2011] |
| Glomus intraradices | Azospirillum brasiliense and Rhizobium leguminosarum mutants | Bacteria with extracellular polysaccharides (EPS) attached to AM fungi roots better than EPS-deficient mutants [Bianciotto et al., 2001] |
| C. neoformans | Acanthamoeba castellanii | Encapsulated fungi can survive co-incubation with amoebae better than acapsular mutants [Steenbergen et al., 2001] Rate of phagocytosis decreases as fungal capsule size increases [Chrisman et al., 2010] |
| Aspergillus fumigatus | A. castellanii | Interaction results in increased fungal growth, although the presence of conidia decreased amoeba viability [Maisonneuve et al., 2016] |
| F. oxysporum | A. fumigatus and Vermamoeba vermiformis | Co-incubation results in increased fungal growth [Cateau et al., 2014] |
Table 2.
Impact of the mycobiome on infection and disease.
| Mycobiome | Prevalent fungi | Associated diseases and reference (s) |
|---|---|---|
| Gut |
Candida Cladosporium Malassezia Saccharomyces |
Crohn’s disease, irritable bowel syndrome, ulcerative colitis [Hager and Ghannoum, 2017; Mar Rodriguez et al., 2015] |
| Lung | Ascomycota Basidiomycota |
Asthma, chronic obstructive pulmonary disorder, cystic fibrosis, HIV [Tipton et al., 2017; Willger et al., 2014; Harrison et al., 2013; Cui et al., 2015; Krause et al., 2016] |
| Oral |
Aspergillus Candida spp. Epicoccum Malassezia |
HIV, periodontal disease [Ghannoum et al., 2010; Dupuy et al., 2014; Monteiro-da-Silva et al., 2014; Zakaria et al., 2017; Peters et al., 2017; Mukherjee et al., 2014] |
| Vaginal/Urinary tract | Candida spp. | Bacterial vaginosis, intra-uterine device associated infections, urinary tract infection [Drell et al., 2013; Srinivasan et al., 2012; Goldacre et al., 1979; Ackerman and Underhill, 2017; Bradford and Ravel, 2017] |
| Skin | Malassezia | Atopic dermatitis, atopic eczema, dandruff, folliculitis, psoriasis [Kong and Morris, 2017; Zhang et al., 2011; Gaitanis et al., 2012; Kato et al., 2007; Sugita et al., 2003; Chang et al., 2011; Park et al., 2012; McGinley et al., 1975] |
| Nails |
Trichophyton rubrum Aspergillus spp. Candida spp. |
Onychomycosis [Veer and Damle, 2007; Kotwal, 2018; Yang, et al., 2011; Foster et al., 2005] |
2. POLYMICROBIAL INTERACTIONS INVOLVING FUNGI
2.1. Fungal-bacterial interactions that enhance plant growth
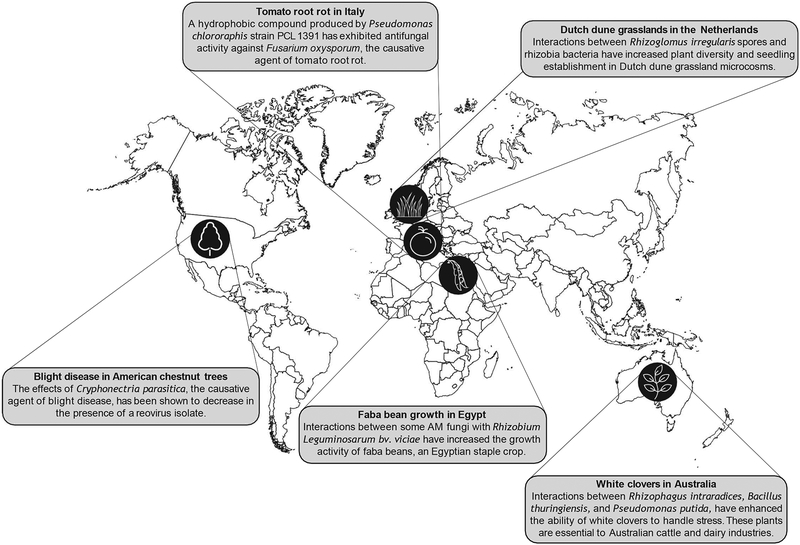
Challenging environments around the world make it difficult for essential plants to grow in abundance. Plants play pivotal roles in global and local economies, which highlights the need to implement measures that can sustain proper crop growth. Certain bacterial-fungal interactions have demonstrated synergistic abilities that can promote plant growth and survival in extreme conditions. Interactions between the plant Trifolium repens, the arbuscular mycorrhizal (AM) fungus Rhizophagus intraradices — in addition to other AM fungi collected from the plant’s rhizosphere — and bacteria such as Bacillus thuringiensis and Pseudomonas putida, provide T. repens the ability to handle stressful environments (e.g., drought) and increase the plant’s nutrient uptake (Ortiz et al., 2015). These interactions promote increased water and nutrient uptake and decrease electrolyte leakage and stomatal conductance (Ortiz et al., 2015), an application of widespread importance, especially in Australia, where white clovers are essential to the cattle and dairy industries (Lane et al., 1997). Interactions between AM fungi and the bacterium Rhizobium leguminosarum bv. viciae have shown similar results on their impact on faba beans survival. This staple crop is cultivated mostly in the Middle East due to its versatility, which includes culinary uses, crop rotation, and animal feed. In fact, this crop is one of the backbones of the Egyptian economy, which highlights the necessary efforts for its preservation (Ahmed et al., 2008). Unfortunately, the successful growth of faba beans in Egypt can be challenging due to the alkaline soil environment. Interactions between AM fungi, specifically Acaulospora laevis, Glomus geosporum, Glomus mosseae, and Scutellospora armeniaca, with R. leguminosarum bv. viciae, have shown to increase nodule formation, root length, nitrogenase activity, and a decrease in malondialdehyde content, all important for faba beans growth (Abd-Alla et al., 2014).
Other interactions between AM fungi and rhizobia bacteria have had positive effects on Dutch dune grassland microcosms. Microcosms inoculated with AM spores from Rhizoglomus irregularis, rhizobia strains from Lotus corniculatus, and T. repens causes maximal nitrogen fixation, plant diversity, and increases seedling establishment (van der Heijden et al., 2016). This observation could be applied to solve the ongoing problem of grass-encroachment and low species diversity in grasslands (Veer et al., 1997). Large-scale application of these interactions represents an agricultural alternative to produce and sustain global crops, which can positively impact other world regions with similar challenges.
2.2. Fungal-bacterial interactions that prevent plant disease
There are emerging fungal species that pose a threat to tomato crops, arguably one of the world’s most important crops, as they are a part of the staple food diet in many countries. Fusarium oxysporum and Phytophthora infestans, the causative agents of tomato root rot and tomato blight, respectively, are examples of such species (Fig. 1). Reports of these tomato plant diseases have been documented worldwide including the United States (U.S.), Malta, Japan, and Italy (Porta-Puglia et al., 2005). Interestingly, a possible solution to this threat is present in the tomato rhizosphere given that Pseudomonas chlororaphis strain PCL1391 can produce antifungal factors, including the hydrophobic compound phenazine 1-carboxamide. This molecule exhibits antifungal activity and prevents root rot disease caused by F. oxysporum, thus playing a major role in biocontrol and plant disease suppression (Chin-A-Woeng et al., 1998).
Fig. 1. Potential applications of polymicrobial interactions involving fungi.
Polymicrobial interactions involving fungi and their role in disease prevention and plant growth have been extensively documented. Examples of symbiotic associations involving fungi with significant impact around the world include the tomato rot in Italy, the Dutch dune grassland in the Netherlands, the blight disease in chestnut trees in the U.S., the faba bean growth in Egypt, and the white clovers in Australia.
Tomato late blight caused by P. infestans destroyed the majority of tomato crops in the northeastern U.S. in 2009. The effects of this plant pandemic were economically devastating, starting in small gardens and quickly expanding to commercial growers who reported losses that were indicative of total crop destruction (Fry et al., 2013). In this regard, the tomato late blight is minimized by the fermentation broths of numerous fungal species taken from numerous vegetable species, including Capsicum annuum and Cucurbita pepo. Endophytic fungi that live inside of plants such as red pepper, tomato, and cucumber were effective in controlling the tomato late blight, particularly F. oxysporum EFF119, a strain isolated from red pepper roots (Kim et al., 2007). F. oxysporum also inhibits the growth of other causative agents of plant disease including Pythium ultimum and Phytophthora spp., infestans and capsici. Similar synergistic relationships involving Fusarium spp. prevent disease in barley roots. For instance, inoculation of Piriformospora indica chlamydospores on barley roots prevented root rot caused by F. culmorum. Barley roots pre-treated with chlamydospores displayed undetectable levels of ascorbate to mock-inoculated control roots and had increased levels of antioxidant activity (Harrach et al., 2013).
Leaf rust disease caused by Puccinia triticina is known for thousands of years. The earliest references of this plant disease are described in the Bible and classical Greek/Roman literature. Even now, the devastating effects of the leaf rust continues to impact the U.S. economy and food supply. For instance, Kansas — a U.S. state leader in wheat-production — recorded a 14% loss due to leaf rust in 2007 (Bolton et al., 2008) (Fig. 1). These economic losses can easily cripple essential food supplies and emphasize the need for preventative measures. For example, in the presence of endophytes, the number of P. triticina pustules in wheat decrease significantly (Dingle and McGee, 2003), an observation that may prevent or minimize leaf rust disease.
Interactions between a Pseudomonas species and Arabidopsis thaliana have also shown increased resistance to Botrytis cinerea, a significant fungal plant pathogen (Ritpitakphong et al., 2016). Similarly, the susceptibility of meadow ryegrass to the barley yellow dwarf virus is similarly affected by the presence of endophytic fungi, indicating that endophytes can play a major role in disease prevention beyond bacterial-fungal interactions. Additionally, meadow ryegrass infected with endophytes have decreased susceptibility to barley yellow dwarf virus compared to those that are endophyte-free (Lehtonen et al., 2006). These instances demonstrate that synergistic interactions can play a role in preventing disease. Together, these findings may aid in preserving and better protecting the world’s supply of life-sustaining crops for future generations.
2.3. Attenuation of fungal disease in plants by viruses
Chestnut blight disease is another example of the destructive potential of fungi. In the 1900s, after the introduction of Cryphonectria parasitica to the U.S. — a species autochthonous of Asia — American chestnut trees were wiped out almost entirely (Rigling and Prospero, 2018) (Fig. 1). C. parasitica’s destructive ability is reduced in the presence of a reovirus isolate with similar characteristics to the Reoviridae family. The reovirus decreased fungal virulence and resulted in altered colony morphology compared to other isolates (Hillman et al., 2004). The hypovirulence of C. parasitica results during interactions with some Cryphonectria hypovirus 1 (CHV1) strains (Krstin et al., 2017). CHV1 alters the levels of cytosine methylation on the genome of C. parasitica depending on the strain of the virus (Nuskern et al., 2018), suggesting that some viral strains may be better at preventing disease than others. Viruses in Sclerotinia sclerotiorum exhibit similar instances of hypovirulence. S. sclerotiorum partitivirus 1 and S. sclerotiorum mitovirus 1 have hypovirulent effects on their host during interaction. The partitivirus can cause hypovirulence of S. nivalis, S. minor, and Botrytis cinerea (Xiao et al., 2014; Xu et al., 2015), all of which are responsible for plant diseases, such as white mold, lettuce drop, and gray mold, respectively. These examples provide evidence of the importance in understanding the beneficial interactions of fungal pathogens and viruses to prevent plant disease.
2.4. Fungal-bacterial interactions that interfere with antibiotic efficacy
Antifungal drug resistance has become a major problem. Fungi have evolved abilities that bypass the action of commonly used antifungal drugs, making fungal infections much harder to control and treat. For example, Candida albicans can form biofilms on indwelling medical devices and resist antifungal treatment (Andes et al., 2004; Chandra et al., 2001). Interactions of C. albicans and Escherichia coli in mixed species biofilms showed that the presence of β−1, 3-glucans affects the susceptibility of E. coli to antibiotics (De Brucker et al., 2015). A decrease in E. coli’s ofloxacin tolerance occurred when β−1, 3-glucans were degraded by the enzyme lyticase. In contrast, C. albicans mutants that produce high levels of β−1, 3-glucans evinced increased tolerance of E. coli to ofloxacin during these interactions (De Brucker et al., 2015), a discovery whose chances of exploitation are high, and would have widespread ramifications given the high percentage of patients with indwelling medical devices.
Interactions between Staphylococcus aureus and C. albicans in a biofilm results in the upregulation of 27 proteins, many of which are involved in metabolic processes, stress response, and cell wall synthesis (Peters et al., 2010). Inhibition of the FKS1 gene in C. albicans strains, which is involved in β−1, 3-glucan synthesis, during these interactions increased susceptibility of S. aureus to vancomycin (Kong et al., 2016). Potential future directions for these promising observations could involve elucidating the proteome of these 27 proteins and a closer examination to β−1, 3-glucans as a target for therapeutic drugs. In dual-species biofilms of S. aureus and C. albicans, an increased resistance of C. albicans to miconazole was observed (Kean et al., 2017). Increased concentrations of farnesol correlate to higher rates of fungal survival following treatment with vancomycin (Kong et al., 2017), highlighting yet another target for therapeutic development.
Dimorphism, or the ability to switch between the mycelial and yeast form at specific temperatures, is a property displayed by several medically important fungi such as Sporothrix schenckii, Histoplasma capsulatum, and C. albicans. This fungal ability is advantageous for host colonization and during polymicrobial interactions. For example, the opportunistic bacterium Pseudomonas aeruginosa is able to form a biofilm on C. albicans but can only kill the fungus in its filamentous form. P. aeruginosa cannot bind to C. albicans yeast cells (Hogan and Kolter, 2002). Nevertheless, P. aeruginosa produces the phenazine 5MPCA, and interactions between a 5MPCA analogue with C. albicans causes fungal death by altering protein synthesis (Morales et al., 2010). Fungal-bacterial competition studies are necessary to understand fundamental questions related to microbial virulence evolution, antimicrobial resistance, and may lead to the identification of novel molecules with antimicrobial potential.
2.5. Phenotypic attributes that contribute to survival in polymicrobial interactions
Interactions involving Cryptococcus neoformans and the Gram-negative bacterium Acinetobacter baumannii have revealed fungal serotype differences in biofilm formation. C. neoformans var grubii, widely distributed serotype A strains, resists killing by A. baumannii and displays increased biofilm and capsule formation compared to serotype D counterparts, C. neoformans var neoformans (Abdulkareem et al., 2015). Confocal microscopy demonstrated that even though the biofilm thickness of both serotypes was similar, morphologically they were significantly different (Abdulkareem et al., 2015). For example, C. neoformans serotype A H99 strain exhibited a uniform distribution of cells throughout the imaged field whereas serotype D B3501 strain displayed aggregates of cells scattered in the imaged field, in both cases surrounded by substantial amounts of capsular polysaccharide. These morphological differences might have important implications in the pathogenesis of these serotypes after infection of the human host.
Likewise, C. neoformans’ capsular production reduces fungal susceptibility to S. aureus (Saito and Ikeda, 2005). The viability of C. neoformans decreases in capsular mutants when co-cultured with S. aureus, and direct contact by the bacterium is essential for fungal death. Exogenous capsular polysaccharide protects C. neoformans against S. aureus, suggesting that active capsular production and release is used as a defensive mechanism by the fungus in competitive polymicrobial interactions. Similarly, certain Salmonella enterica strains have better ability to adhere and form biofilms on the hyphae of A. niger. Chitin and cellulose are important in regulating this interaction. For instance, Salmonella strains that lacked cellulose are unable to aggregate on A. niger, which subsequently prevents the formation of a dense biofilm. Only after cellulose synthesis genes are restored in these bacterial strains do they attach to and form a biofilm on A. niger (Brandl et al., 2011). Furthermore, wild-type Azospirillum brasiliense and Rhizobium leguminosarum species that produce extracellular polysaccharides attach firmly to AM fungi roots compared to strains that have impaired production, or do not produce extracellular polysaccharides (Bianciotto et al., 2001). Together, these studies demonstrate that certain fungal phenotypic characteristics can be advantageous during interactions with bacteria.
2.6. Interactions between fungi and amoebae
Amoebae are capable of engulfing large and small propagules (e.g., spores, conidia, etc.) from species such as C. neoformans, Botrytis cinerea, Cochliobolus sativus, Alternaria alternata, Aureobasidium pullulans, and Cladosporium spp., all of which play an important role in biocontrol of fungi in the environment (Casadevall et al., 2019; Delafont et al., 2018; Guimaraes et al., 2016; Novohradska et al., 2017). The dimorphic fungi Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum can also be phagocytosed by Acanthamoeba castellanii in their yeast form, although the conidia of H. capsulatum in particular, are cytotoxic to these amoeba (Steenbergen et al., 2004). C. neoformans is one of the most studied fungi interacting with amoebae (Bunting et al., 1979; Steenbergen et al., 2001). Following phagocytosis of C. neoformans serotype A and D strains by A. castellanii, the fungus replicates inside of the amoeba and can subsequently kill its host by lysis. Encapsulated C. neoformans can survive co-incubation with A. castellanii better than acapsular mutants. Melanization, an accumulation of a dark pigment surrounding the fungal cell wall, protects these cells against killing by amoebae. However, a phospholipase cryptococcal mutant showed decreased replication rate in amoebae compared with isogenic strains (Steenbergen et al., 2001). C. gattii also interacts with A. castellanii, though the rate at which phagocytosis occurs is lower than that of C. neoformans (Malliaris et al., 2004) and decreases as the capsule size of C. neoformans increases (Chrisman et al., 2010). Gene expression of 656 genes in C. neoformans, some of which encode for virulence factors, are modulated during these interactions. Some of the 322 genes that are upregulated in C. neoformans are involved in metabolism and stress response, while those that are downregulated genes are involved in ergosterol synthesis (Derengowski Lda et al., 2013). Interestingly, it was recently described that mannose-binding proteins may be involved in fungal recognition by amoebae and promotes interactions that allow the emergence and maintenance of fungal virulence in animals (Goncalves et al., 2019).
A. castellanii is also capable of phagocytosing A. fumigatus, resulting in the amoeba’s death and intracellular fungal replication (Maisonneuve et al., 2016). The interaction of A. fumigatus and Vermamoeba vermiformis results in increased fungal growth, though the presence of fungal conidia can decrease the viability of the amoeba (Maisonneuve et al., 2016). When the two free living amoebae A. castellanii and V. vermiformis are co-incubated with F. oxysporum, fungal growth increases (Cateau et al., 2014), which parallels those studies finding synergistic reactions involving F. oxysporum. Given that drinking water distribution systems can be a major reservoir for several opportunistic microorganisms, the impact of F. oxysporum-amoebae interactions is worth exploring since traces of V. vermiformis have been previously observed in hospital drinking water (Delafont et al., 2018), therefore, understanding how these interactions contribute to fungal growth can be used to anticipate or limit human exposure.
3. THE MYCOBIOME
3.1. The gut mycobiome
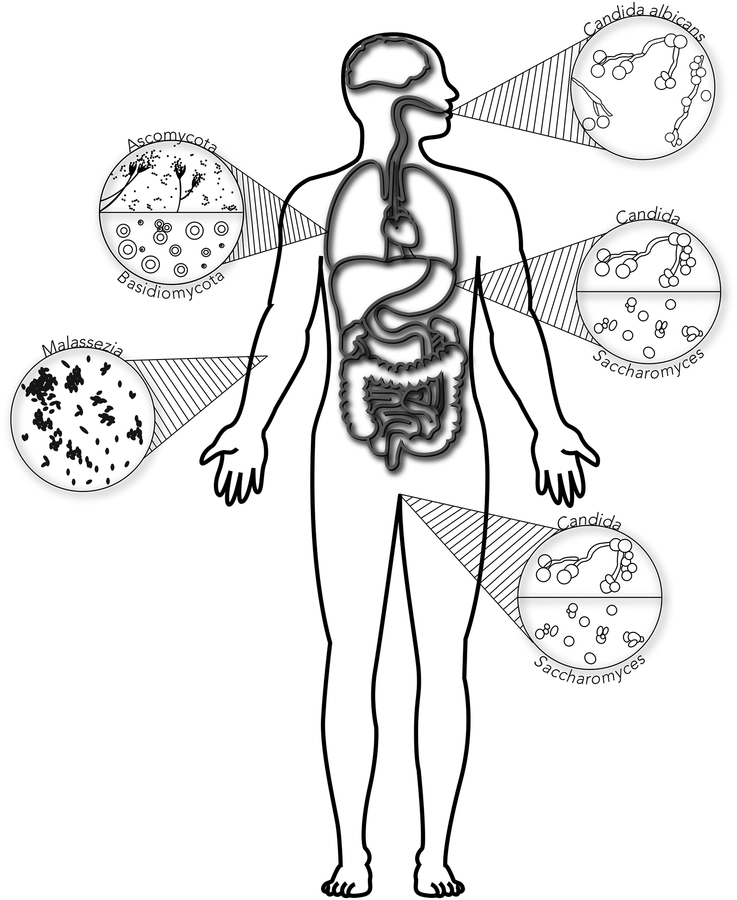
The gut mycobiome is a topic of many current studies, particularly in healthy individuals, seeking to identify the fungi present in the stomach and their roles in health determination. Many studies conducted in the gut have focused on assessing the microbial population in healthy individuals while comparing it to the population of individuals affected by gastrointestinal illnesses, particularly ulcerative colitis and Crohn’s disease. Changes in the fungal communities between healthy individuals as well as individuals affected by illness play a role in disease susceptibility and progression. Identifying the key players in gut-related diseases can lead to specialized treatment and create preventative measures for those deemed at risk (Nash et al., 2017). For instance, the gut mycobiome of the Human Microbiome Project (HMP) was assessed and conducted on stool samples of apparently healthy individuals using a combination of Internal Transcribed Spacer (ITS), 18S and 16S rRNA, and whole genome shotgun metagenomic sequencing, revealing that the most abundant fungi in the gastrointestinal tract are Malassezia, Candida, and Saccharomyces species (Nash et al., 2017) (Fig. 2). These results provide a baseline for the fungi in the healthy gut, correlation analyses across the samples, and samples collected longitudinally were able to show that the gut mycobiome is not only variable among individuals, but is also alterable with time (Nash et al., 2017).
Fig. 2. Mycobiome composition specific to different regions of the human body.
The human body of a healthy individual with microscopic images of the dominant fungi inhabiting organ-specific mycobiomes. The oral mycobiome has a dominance of Candida albicans, the lung mycobiome a shared dominance of Ascomycota and Basidiomycota, the gastrointestinal mycobiome a shared dominance of Candida and Saccharomyces, the urinary mycobiome has a dominance of Saccharomyces, the vaginal mycobiome has a split dominance of Saccharomyces and Candida, and the skin mycobiome has a dominance of Malassezia.
The mycobiome between obese and non-obese individuals was compared, aiming to find whether there were any specific fungal species associated with susceptibility to obesity (Mar Rodriguez et al., 2015). Seven cited studies found that about 184 species of fungi resided in the human gut mycobiome with Candida, Saccharomyces, and Cladosporium species the most common (Mar Rodriguez et al., 2015). However, the inability to distinguish the fungi in the mycobiome as commensals or isolates gained from diet and the environment was noted as a limitation of these studies (Nash et al., 2017). The findings reported by Nash et al. (2017) (Nash et al., 2017) and Mar Rodriguez et al. (2015) (Mar Rodriguez et al., 2015) independently identified Saccharomyces and Candida genera as the most common fungi in the gut mycobiome. However, there were disparities obtained from those studies in the third most common genera isolated in the gut mycobiome with Nash et al. (2017) reporting Malassezia whereas Mar Rodriguez et al. (2015) finding Cladosporium. This discrepancy is possibly attributed to differences in analyzed cohorts (e.g., healthy vs. obese), patients’ selection criteria, region of the world (e.g., U.S. vs. Spain), diet (e.g., Western vs. Eastern), and confounding variables associated with the inability to distinguish between commensal and transient fungi.
Disruption of the healthy mycobiome of the gut is linked to two main gastrointestinal inflammatory illnesses: ulcerative colitis and Crohn’s disease. Different studies have identified links between these illnesses and particular species in the mycobiome. Hager and Ghannoum, (2017) compared the common fungal species found in the gut of Crohn’s disease patients and their healthy relatives, with the aim of identifying genetic components to the mycobiome, as well as increased susceptibility to Crohn’s disease. E. coli, Serratia marcescens, and Candida tropicalis were elevated in the Crohn’s patients as compared to their relatives (Hager and Ghannoum, 2017). C. tropicalis, in particular, was significantly increased in Crohn’s patients, especially in its ability to interact with anti-Saccharomyces cerevisiae antibodies (ASCA), an important biomarker associated with Crohn’s disease (Hager and Ghannoum, 2017; Hoarau et al., 2016). C. tropicalis was also in a close relationship with pathogenic bacteria in Crohn’s patients, while S. cerevisiae was not correlated with bacteria (Hager and Ghannoum, 2017; Hoarau et al., 2016). Similarly, the association between C. tropicalis, E. coli, and S. marcescens on biofilm formation was investigated. This triple-species biofilm is several layers thicker compared to monospecies biofilms and these interkingdom interactions cause intestinal inflammation and activation of the host immune response (Hager and Ghannoum, 2017). This observation might be important in understanding the basis of Crohn’s disease in patients.
A similar study conducted in the mycobiome of Crohn’s patients, comparing it to that of their relatives and nonrelatives, showed that Ascomycota and Basidiomycota were responsible for more than 1% of the mycobiome composition (Hoarau et al., 2016). Ascomycota, in particular, was the most abundant, responsible for more than 74% of the mycobiome composition in individuals with Crohn’s and their healthy family members (Hoarau et al., 2016). Specifically, S. cerevisiae and C. tropicalis were the most common fungal species in Crohn’s patients at abundance levels of 24% and 10%, respectively. S. cerevisiae and Galactomyces geotrichum were the most abundant fungal species discovered in the gut of healthy family members of Crohn’s patients (Hoarau et al., 2016). Higher levels of ASCA were associated with Crohn’s patients; C. tropicalis was the only fungi positively associated with ASCA (Hoarau et al., 2016). Candida had five positive intrakingdom correlations with Fusarium, Haematonectria, Nectria, Thanatephorus, and Trichosporon; and one negative correlation with Saccharomyces (Hoarau et al., 2016). ASCA data in Crohn’s patients demonstrate the risk factors associating Crohn’s disease and susceptibility to C. tropicalis proliferation within the mycobiome, which can lead to Crohn’s onset (Hager and Ghannoum, 2017; Hoarau et al., 2016). The further identification of the species residing in each type of individual can be looked into deeper, particularly the presence of high ASCA levels. Perhaps, this observation in Crohn’s disease patients have therapeutic applications and might be useful in preventative care and treatment of those susceptible or affected.
Conversely, in Irritable Bowel Syndrome (IBS), previous studies conducted on the gut microbiota were unable to find consistent differences in the microbiome of IBS patients versus healthy patients, nor were they able to find results showing a relationship between microbiota composition and IBS manifestations (Frost et al., 2019; Gu et al., 2019; Maharshak et al., 2018). One study conducted revealed S. cerevisiae and C. albicans as the prominent fungal species in healthy and IBS patients, although the number of these fungal species was relatively larger in IBS patients (Botschuijver et al., 2017; Gu et al., 2019). There are limited number of studies conducted on the mycobiome of IBS patients, with none able to definitively associate fungal and bacterial interactions in IBS (Gu et al., 2019).
The association between diet and physiology on the composition of the gut mycobiome, including the effect of cholesterol levels, has been investigated (Chacon et al., 2018; Mar Rodriguez et al., 2015). These studies assessed whether modifications to the mycobiome are related to human nutrition, and how these different fungal species may contribute to obesity (Mar Rodriguez et al., 2015), which in turn, could lead to the development of treatments to fight obesity. Through random forest-type classification, higher levels of Aspergillus and lower levels of Mucor, Penicillium, Saccharomyces, and Eupenicillium were associated with obesity (Mar Rodriguez et al., 2015). In contrast, Agaricomycetes were significantly more abundant in non-obese subjects than obese subjects, with Mucor being the most prevalent genus in non-obese subjects, its abundance was associated with weight loss (Mar Rodriguez et al., 2015). This observation can be applied in obese subjects to change the diversity of their mycobiomes and, in turn, help treat or reduce their obesity (Mar Rodriguez et al., 2015). Notably, Penicillium, Aspergillaceae, and Eurotiomycetes positively correlated with HDL-cholesterol levels whereas Saccharomycetes, Tremellomycetes, Cystobasidiomycetes and Erythrobasidiaceae were negatively correlated (Mar Rodriguez et al., 2015). Overall, diversity in the mycobiome decreases with obesity where obese subjects had higher levels of Ascomycota, Saccharomycetes, Dipodascaceae, and Tremellomycetes in comparison to non-obese control subjects (Mar Rodriguez et al., 2015). The growth of particular fungal genus was associated with body mass index (BMI), fat mass, hip circumference, HDL cholesterol, and fasting triglycerides (Mar Rodriguez et al., 2015). A few studies have looked into the correlation between diet and the mycobiome of different individuals, comparing a plant-based diet with a typical western or high fat diet. Nevertheless, Fusarium was frequent among vegetarians, present in 88% of vegetarians and only present in 3% of those with western diets (Hallen-Adams and Suhr, 2017). Likewise, other fungi abundantly present in vegetarians were Malassezia, Penicillium, and Aspergillus (Hallen-Adams and Suhr, 2017). Future studies should aim to distinguish isolates from diet/environment and those naturally residing in the gut in order to properly identify the resident species and further determine correlations between gut health, illness, and fungal communities.
3.2. The lung mycobiome
Ascomycota and Basidiomycota predominantly colonize the healthy respiratory tract (Tipton et al., 2017) (Fig. 2). Healthy individuals have Penicllium, Aspergillus, Candida, Malassezia, Pneumocystis, Cladosporium, and other genera in minor quantities (Tipton et al., 2017). The variability of respiratory fungi have been found to be a case-by-case basis, as studies on individuals with similar illnesses possess different populations of fungi in their lungs (Tipton et al., 2017). Additional factors contributing to the diversity in samples found among patients are associated with the different environmental exposures patients face since samples are thought to be acquired through inhalation, and are dependent on location, weather, and subsequent influences. Although the lower the diversity of the mycobiome, the higher the risk of lung illnesses, such as CF and chronic obstructive pulmonary disease (COPD) (Tipton et al., 2017). Data analyses from patients with CF demonstrated that Candida was the most prevalent genus with C. albicans, C. dubliniensis, and C. parapsilosis identified as the main species (Willger et al., 2014). The only other fungus detected across all samples was Malassezia spp., but in much smaller quantities, while three species of Aspergillus were detected in low levels in a few samples. Interestingly, none of which was A. fumigatus, a fungus commonly associated with CF (Willger et al., 2014). When comparing lung function as a result of CF with fungal growth in the airway, an inverse relationship between Malassezia spp. and lung function was observed whereas a positive relationship between Eleutheromyces spp. and lung function was identified (Harrison et al., 2013). Furthermore, Candida spp. and Aspergillus spp. were observed in the airways of 25% of the CF patients (Harrison et al., 2013).
Patients immunocompromised with HIV infection have the highest rates of fungal associated respiratory illnesses. A common respiratory disease afflicting HIV patients is COPD, with high rates of infection regardless of antiretroviral therapy (Cui et al., 2015). The lung mycobiome of healthy and HIV individuals with and without COPD was compared (Cui et al., 2015). Candida is the overpoweringly present fungi in about 90% of the samples analyzed with ITS sequencing, with a dominance in oral wash samples versus bronchoalveolar lavage (Cui et al., 2015). Ceriporia lacerate, S. cerevisiae, and Penicillium brevicompactum were more abundant in the bronchoalveolar lavage than in the oral wash samples (Cui et al., 2015). These particular organisms cause opportunistic infection, suggesting that they possibly are commensals within the lungs (Cui et al., 2015). It is important to note that these studies were conducted in non-smoker patients (both the control and HIV infected) (Cui et al., 2015). These parameters were able to determine whether the incidence of particular fungi in the COPD patients was due to a direct association with HIV infection (Cui et al., 2015). Additionally, Pneumocystis jirovecii and Ceriporia lacerata, known organisms associated with respiratory infections in immunocompromised patients, were overrepresented in the lungs of HIV patients (Cui et al., 2015). An experimental group identified as HIV infected with CD4 counts lower than 500 cells/microliter had an abundance of Zasmidium nocoxi and Teratosphaeria jonkershoekensis (Cui et al., 2015). The investigators described multiple limitations of this study. They were unable to determine causation between the mycobiome variability in HIV infected and COPD patients. The smoking status was self-reported and it is possible that the subjects’ accounts were not 100% accurate. Finally, limited number of microorganisms were identified using sequencing techniques, which are unable to distinguish the viability of the microbes among the species detected (Cui et al., 2015).
There are limited studies on other respiratory illnesses such as patients with asthma. Of the studies conducted on the lungs of individuals with asthma, Psathyrella and Malassezia spp. were identified in 25% of the samples, while Eremothecium spp. was reported in 40% of the lung samples extracted from the healthy patients (Krause et al., 2016; Tipton et al., 2017). Lung transplant individuals with healthy participants demonstrated the presence of Candida in the oral wash but not in the bronchoalveolar lavage. Environmental fungi such as Davidiellaceae and Cladosporium were mainly identified in bronchoalveolar lavage of these patients (Charlson et al., 2012). Conversely, in the lung transplant patients, there were high levels of Candida in oral washes and bronchoalveolar lavages, while Aspergillus was typically identified either strictly in bronchoalveolar lavages or minimally in oral washes (Krause et al., 2016).
3.3. The oral mycobiome
The oral mycobiome is one of the most studied areas of the mycobiomes due to its increasing rates of infection in immunosuppressed individuals, most notably HIV patients. A pioneering study analyzed the oral mycobiome in healthy individuals utilizing panfungal ITS probes with 454 pyrosquencing (Ghannoum et al., 2010). Candida was the most abundant genus in the oral cavity, present in 75% of the participants, with C. albicans the most common species found in 40% of the participants (Ghannoum et al., 2010) (Fig. 2). Other genera commonly present, in descending order were Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium, and Cryptococcus. The genera with the lowest abundance may be associated to contamination acquired from the environment through inhalation or food ingestion (Ghannoum et al., 2010). When testing for variance in gender and ethnicity, Caucasian and Asian males differed from one another in their oral mycobiome, but females did not (Ghannoum et al., 2010). This was not conclusive as the sample size was too small but warrants further study (Ghannoum et al., 2010).
In contrast, the analysis of saliva samples from healthy individuals utilizing 18S ITS1 amplification, pyrosequencing, and curation showed that Malassezia and Epicoccum were among the highest colonizers in the oral mycobiome. Malassezia is most often regarded for its role in the skin mycobiome (Dupuy et al., 2014). Another study conducted in 40 healthy individuals showed that species of Candida, Rhodotorula, Penicillium, Aspergillus, and Cladosporium were the most prevalent in their oral mycobiome (Monteiro-da-Silva et al., 2014). Despite the variability of fungi assessed in the participants, the abundance and quantity of fungi present in the oral cavity were consistent after a 30-week follow-up (Monteiro-da-Silva et al., 2014). Saliva samples from community dwelling elderly subjects (~75 to 99 years old) showed prevalence of C. albicans, C. glabrata, and C. dubliniensis (Zakaria et al., 2017).
Analysis of the oral mycobiome between individuals with periodontal disease and those with good oral health showed no difference between the groups (Peters et al., 2017). All the participants had Candida and Aspergillus as the prevalent genera (Peters et al., 2017). Individuals with periodontal disease evinced an increasing trend in the abundance of Candida, although this finding was not significant (Peters et al., 2017). The limitations of the study stem from the small sample size of 30 total participants, 15 from each experimental group (Peters et al., 2017).
Similarities and differences in the oral mycobiome of HIV and non-HIV individuals were also observed (Mukherjee et al., 2014). HIV patients were abundant in Candida (92%), Epicoccum (33%), and Alternaria (25%); while Candida (58%), Pichia (33%), and Fusarium (33%) were the most common genera in non-HIV individuals (Mukherjee et al., 2014). Candida and Penicillium were the only two genera that resided in both HIV and non-HIV individuals, with C. albicans mostly associated to non-HIV (58%) and HIV (83%) patients (Mukherjee et al., 2014). Interestingly, there was an inversely proportional relationship between Pichia and Candida. A decrease in the presence of Pichia was associated with an increase in Candida (Mukherjee et al., 2014). Pichia had a growth inhibitory effect on Cryptococcus, Aspergillus, and Fusarium when grown on selective Pichia spent medium (Mukherjee et al., 2014). These studies were crucial in identifying polymicrobial interactions among the fungi in the oral mycobiome. The discovery of the Pichia’s inhibitory effects on the Candida species among the other fungi is the first identified interaction among the oral mycobiome. This observation is predicted to be a novel and potential target of antifungal treatment production and development (Mukherjee et al., 2014).
3.4. Onychomycosis
Fungi that frequently cause nail infection, or onychomycosis, include dermatophytes (e.g., Trichophyton rubrum), non-dermatophyte molds (e.g., Aspergillus spp.), and Candida spp. (Veer et al., 2007). Fungal disease of nails may be associated with poor circulation, heart disease, and diabetes. Recently, two species not previously considered onychomycotic, Trichosporon asteroides and Trichosporon faecale, were identified as causative agents of onychomycosis (Kotwal et al., 2018). In a recent study, samples from 7,733 patients with suspected fungal infection were analyzed and 6% of superficial fungal infections were mixed infections, mostly regarding foot lesions, especially the toenails (Gawaz and Weisel, 2018). Co-infections of toenails consisted of dermatophyte-dermatophyte (16.1%), dermatophyte-yeast (45.6%), yeast-yeast (12.1%), and other (8.7%). Concomitant fungal skin infection is observed 25% of patients with toenail onychomycosis (Lipner and Scher, 2015; Maraki and Mavromanolaki, 2016). The susceptibility of acquiring a mixed co-infection of toenails in higher in male patients between 60 and 80 years old (Gawaz and Weisel, 2018).
Polymicrobial interactions involving Fusarium solani onychomycosis co-infection with the bacterium P. aeruginosa have also been documented in Korea (Yang et al., 2011). Immunocompetent patients presenting discoloration and thickening of the thumbnail showed F. solani onychomycosis co-infected with P. aeruginosa (Yang et al., 2011). These findings support an earlier study that found increased P. aeruginosa growth in potassium hydroxide (KOH)-positive specimens compared to KOH-negative samples, suggesting that, at least in this case, interaction of the bacterium with F. solani promotes its replication (Foster et al., 2005). In contrast, P. aeruginosa inhibits fungal growth and this antifungal activity has been observed against F. solani (Yang et al., 2011), Trichophyton spp. (Treat et al., 2007), A. fumigatus (Kerr et al., 1999), and C. albicans (Hogan and Kolter, 2002). Pyocyanin and 1-hydroxyphenazine have been identified as responsible for the antifungal activity of P. aeruginosa in mixed cultures (Kerr et al., 1999) and co-infections (Yang et al., 2011). Although the mechanism behind these interactions have been superficially defined, more understanding of these co-infection may be important for diagnosis and treatment.
3.5. The vaginal/urinary tract mycobiome
Urinary tract infections (UTI) are caused by a disruption in a woman’s microflora, allowing the overgrowth of Candida. Due to this relationship, there was interest in identifying other commensal fungi within the vaginal mycobiome and possible candidates for opportunistic infection or associated illnesses. The vaginal mycobiome of healthy women was determined showing high colonization rates for Candida spp., particularly, C. parapsilosis, C. dubliniensis, C. krusei, and C. albicans (Drell et al., 2013) (Fig. 2). These fungal species were observed to interact with lactobacilli, a consistent resident in women without medical history of bacterial vaginosis (Drell et al., 2013; Srinivasan et al., 2012). Vaginal mycobiome studies are difficult due to the presence of air contaminants and limited availability of database on vaginal fungi for analysis (Drell et al., 2013).
Vaginal swabs were performed in aged women attending a family planning clinic and the composition of their microbial flora and its associations with normal vaginal discharge and vulval itching were assessed (Goldacre et al., 1979). C. albicans was significantly more abundant in women younger than 25 years of age, than in those older. Fungal diversity outside of C. albicans was more frequent in women older than 35 years, than in those younger (Goldacre et al., 1979). When comparing contraceptive methods, there was no change in C. albicans abundance levels (Goldacre et al., 1979). Comparisons of asymptomatic and symptomatic vaginal yeast colonization in young females demonstrated that asymptomatic adolescent females had high vaginal fungal burden with >500 cfu per vaginal swab culture in ~50% of the females tested (Barousse et al., 2004). A different study conducted in the urinary tract, utilized next generation sequencing was able to amass a wide number of fungi in urine samples, although the only class of fungi detected belonged to Saccharomycetes (Ackerman and Underhill, 2017) (Fig. 2). Issues in detecting fungi from urine samples stem from the difficulty in culturing all types of fungi (Ackerman and Underhill, 2017).
The vaginal and urinary tract mycobiomes are difficult to assess due to the large amount of fungal species that are unculturable, lack of known sequences in the database, air contaminants, and ITS primer bias (Drell et al., 2013). There are relatively a few articles documenting the vaginal and urinary mycobiome composition, particularly among healthy individuals in order to create a baseline. Most of the literature currently published on the female genitourinary mycobiome revolves around C. albicans and its interactions (Bradford and Ravel, 2017). Future studies should incorporate the data in these studies and analyze the mycobiome implications in different diseases associated with these anatomical areas, especially in UTI and intrauterine devices (IUD) associated infections. Studies conducted in the urinary tract can help determine treatment and upkeep of bladder health and related diseases (Ackerman and Underhill, 2017).
3.6. The skin mycobiome
Fungi are associated with infections of the skin, such as ringworms caused by dermatophytes. The skin is the most susceptible organ to fungal infection due to its exposure to the environment. This aspect makes skin mycobiome studies difficult to distinguish between resident and transient species. Malassezia furfur’s need for lipids finds them in areas with available sebum, causing them to be the dominant fungi at all sites of the skin including the scalp, back, face, neck, and limbs (Kong and Morris, 2017; Zhang et al., 2011) (Fig. 2). Additionally, Malassezia spp. interact with other components of skin such as keratinocytes and immunological cells such as dendritic cells and macrophages (Gaitanis et al., 2012). They are found predominantly on healthy skin but have been implicated in disease such as in cases of atopic dermatitis, folliculitis, psoriasis, and atopic eczema (Zhang et al., 2011). Mast cell expression of dectin-1, and their response to M. sympodialis, is modified in atopic eczema patients in comparison to mast cells in healthy individuals, leading to exacerbated atopic eczema (Gaitanis et al., 2012).
Malassezia is present in 79.2% of healthy individuals compared to 68.7% in atopic dermatitis patients (Zhang et al., 2011). Other fungi abundantly found in the skin of atopic dermatitis patients were C. albicans, Cryptococcus diffluens, Wickerhamomyces anomalus, Cryptococcus liquefaciens, and Trichosporum asahii (Zhang et al., 2011). C. diffluens and C. liquefaciens colonized the skin of atopic dermatitis patients at a higher rate than in normal skin (Kato et al., 2007; Sugita et al., 2003). C. diffluens was isolated in 42% of atopic dermatitis patients in samples collected from Sabouraud dextrose agar colonies grown after contact with patient dressings, and in 97% of patients in samples extracted directly from the patient dressings; C. liquefaciens was isolated in 33% and 86% of patients in these collection methods, respectively (Sugita et al., 2003). Utilizing logistic regression analysis, Chang and colleagues demonstrated a relationship between skin sensitization to C. albicans and development of atopic dermatitis (Chang et al., 2011).
Samples from individuals with dandruff scalps were compared to individuals with no dandruff aiming to distinguish the fungi associated with dandruff through GS-FLX Titanium sequencing (Park et al., 2012). Basidomycota was isolated from a dandruff scalp two-fold higher than in healthy scalps (Park et al., 2012). Other fungi in dandruff scalps were Eupencillium, Filobasidium, Malassezia, and Penicllium (Park et al., 2012). In healthy individuals, samples isolated include Acremonium, Coniochaeta, Cryptococcus, Didymella, Rhodotorula, and Ascomycete (Park et al., 2012). Both dandruff and healthy individuals had Acremonium spp. as the dominant growing fungi, although in dandruff individuals, Malassezia was present twice as abundant as the healthy scalp (McGinley et al., 1975; Park et al., 2012).
4. MEDICAL CONDITIONS: FUNGAL OPPORTUNISM OR SOCIALIZATION?
Fungi causes opportunistic infections mainly in immunocompromised individuals. Fungi also contribute to the microflora of the human host, and are typically commensals, but when there are vulnerabilities in the host immune system, they interact with other fungi/bacteria/viruses to cause polymicrobial infections. This section discusses examples of interactions in which the presence of fungi is important in the development of disease.
4.1. Polymicrobial infections involving Candida spp.
C. albicans related hospital acquired infections often involve polymicrobial interactions (Harriott and Noverr, 2011). The most common polymicrobial interactions involving C. albicans infection includes those with Staphylococcus epidermidis, Enterococcus spp., and S. aureus (Harriott and Noverr, 2011). Together, S. aureus/C. albicans or S. epidermidis/C. albicans biofilms enhance antimicrobial resistance of the bacterium and fungus against vancomycin and fluconazole, respectively (Harriott and Noverr, 2011). The antimicrobial resistance associated with these interactions are one of the leading causes behind the difficulty of properly treating these infections in immunocompromised patients. Other studies have investigated the relationship between C. albicans and streptococci. In the presence of C. albicans, streptococci have robust biofilm formation on biotic surfaces such as the oral, esophageal, and vaginal mucosa (Xu et al., 2014). However, binding of streptococci reduced colonization of Candida species and S. aureus, inhibiting their ability to cause disease (Xu et al., 2014).
IUD harbor biofilms made up of C. albicans and various bacterial strains (Harriott and Noverr, 2011). These IUD related polymicrobial interactions were associated with higher rates of bacterial vaginosis, higher risk of pelvic inflammatory disease, and other recurrent vaginal infections in IUD users versus non-users (Harriott and Noverr, 2011). Likewise, E. coli and C. albicans interactions in the urinary tract increase susceptibility to infection, whereas C. albicans colonization alone did not cause infection (Dhamgaye et al., 2016). C. albicans is also associated with skin wounds. Cutaneous lesions demonstrated polymicrobial biofilms often formed by Candida, Malassezia, Cladosporium, Trichtophyton, and other yeasts, with >50% made up of fungi (Harriott and Noverr, 2011). Bacterial species in association with C. albicans biofilms include isolates of Lactobacillus/Enterococcus spp., and Staphylococcus spp., although their contribution to disease is still being investigated (Harriott and Noverr, 2011). Due to the common occurrence of polymicrobial infections, a potential treatment option should focus on identifying areas that different microorganisms have in common, such as the binding of streptococci or the presence of a biofilm that can target both fungal and bacterial species.
Fungal peritonitis, an inflammatory disease affecting the abdominal wall in dialysis patients, has been linked to extractions of Candida, Staphylococci, Streptococci, Enterococci, Pseudomonas, E. coli, Klebsiella, and other bacterial isolates (Miles et al., 2009). Intraperitoneal co-infection with E. coli and C. albicans was lethal in mice compared to single species infections (Dhamgaye et al., 2016; Klaerner et al., 1997). Emphysematous cystitis is a common infection in the bladder wall affecting diabetes patients, occurring more often in women, and linked to infection with E. coli, Enterobacter, Proteus, Klebsiella, and Candida (Casqueiro et al., 2012). There is currently limited literature and investigations on fungal peritonitis, but the relationship between Klebsiella and Candida should be looked into further considering their association in infections of the gut and urinary tract.
Graft-versus-host is a medical complication in which a patient who recently underwent organ transplantation mounts an immune response attacking the donor organ. About 35% of graft-versus-host disease (GVHD) and gastrointestinal graft-versus-host disease (GI-GVHD) patients are commonly colonized with the Candida spp.: C. albicans, C. glabrata, C. krusei, C. parapsilosis, and C. tropicalis (van der Velden et al., 2013). Half of Candida colonized individuals are reported to have grades II-IV acute GVHD compared to 32% in non-colonized individuals (van der Velden et al., 2013). There was no difference observed between Candida colonized skin GVHD patients and non-colonized (van der Velden et al., 2013). The mechanism for this phenotypic observation is currently unknown, although it is hypothesized to be similar to those identified by bacterial interactions in GVHD (van der Velden et al., 2013). Current preventative treatments of fungal associated GVHD are antifungal therapies, which is beneficial in reducing the severity of GI-GVHD (van der Velden et al., 2013). Discovery of GVHD associated infections would dramatically change the field of medical intervention, as organ transplant recipients are considerably susceptible to fungal infections.
4.2. Interactions between P. aeruginosa and fungi in CF
CF, a respiratory disease characteristic of increased mucus production in the lungs, is a breeding ground for microbial infections, including those caused by fungi (Delhaes et al., 2012; Peters et al., 2012). In CF patients, isolates collected from sputum samples contained C. albicans, Aspergillus spp., Malassezia restricta, and Pneumocystis jirovecii (Delhaes et al., 2012). P. aeruginosa co-colonizes with A. fumigatus, an interaction that is becoming a common and an increasing problem in CF patients (Costa-Orlandi et al., 2017). Aspergillus interacts with multiple Gram-negative bacteria aside from P. aeruginosa, most notably Achromobacter spp., Burkholderia cepacia, and Stenotrophomonas maltophilia (Dhamgaye et al., 2016). P. aeruginosa inhibited Candida germination and lyses C. albicans hyphae but has no lethal effect on its yeast cells (Peters et al., 2012). A reduction in the fungal community in CF patients correlated with poor clinical status and lung function compared to healthy individuals (Delhaes et al., 2012).
Advanced molecular techniques were able to identify fungi, bacteria, and viruses together in the lungs of CF patients (Filkins and O’Toole, 2015). The current polymicrobial interactions of interest in the lungs of CF patients includes Streptococcus spp., Trichosporon spp., and rhinovirus (Filkins and O’Toole, 2015). All other organisms that reside in the lungs of CF patients include Streptococcus pneumoniae, Achromobacter spp., and others, which were once thought to contribute to pathogenesis, were commensals (Filkins and O’Toole, 2015). Although the best approach at determining pathogenic organisms is to do so individually, it has been noted that pathogenicity needs to be assessed within polymicrobial interactions as these infections differ mechanistically from singular infection (Filkins and O’Toole, 2015). Elucidating these polymicrobial interactions can then be incorporated into treatment assessment in CF patients (Filkins and O’Toole, 2015).
4.3. C. albicans interactions with other microbes in oral infections
The study of oral infections are of current interest due to their diverse polymicrobial environment, such as the one present in dental caries, a common oral infection that results when carbohydrates are fermented into lactic acid by Streptococcus mutans, Lactobacillus acidophilus, or C. albicans (Peters et al., 2012). These microbes harvest nutrients and demineralize the tooth surface causing a reduction in pH (Peters et al., 2012). C. albicans interactions with streptococci have been associated with an increase in patients with dental caries (Arvanitis and Mylonakis, 2015; Diaz et al., 2014; Koo et al., 2018). Another common oral infection is denture stomatitis, which typically affects the elderly and is characterized by redness and swelling of the soft palate and tissues. In addition to C. albicans, 82 species of bacteria were present in biofilms of healthy and infected patients, a third of which were present in both (Peters et al., 2012). C. glabrata, S. cerevisiae, C. krusei, and C. parapsilosis have also been isolated from patients affected with denture stomatitis. The mixture of C. albicans and C. glabrata was the most common combination, present in 25% of the patients (Coco et al., 2008). Of all oral infections involving fungi, candidiasis, caused by C. albicans, is perhaps the predominant oral infection in immunocompromised patients, particularly those with HIV (Harriott and Noverr, 2011; Rouabhia and Chmielewski, 2012). Streptococcus gordonii and C. albicans interactions promote hyphal development and biofilm formation, although it is unclear if this interaction leads to oral colonization in vivo (Harriott and Noverr, 2011).
C. albicans interactions involving the mitis group of streptococci (MGS) results in a mutually beneficial relationship that promote colonization of the oral cavity, as well as, in exacerbating the host inflammatory response (Rapala-Kozik et al., 2018). In contrast, C. albicans biofilm production in periodontal disease is inhibited by the Gram-negative, Aggregatibacter actinomycetemcomitans (Rapala-Kozik et al., 2018). Additionally, Streptococcus mutans inhibits the filamentous formation of C. albicans in early biofilm formation (Rapala-Kozik et al., 2018). Similarly, Streptococcus gordonii also inhibits C. albicans biofilm formation through similar CSP competence peptide, although it has no effect on C. albicans hyphae (Rapala-Kozik et al., 2018). Further studies involving polymicrobial interactions in the oral cavity are currently being conducted to elucidate the particular role of fungi in oral disease (Peters et al., 2012).
4.4. Involvement of P. aeruginosa and other fungi in malignant external otitis
Malignant external otitis (MEO) is an infection of the temporal bone mainly caused by P. aeruginosa, although there are rare cases associated with other fungi, such as in the case of immunocompromised patients (Bovo et al., 2012). One example was that of a 21-year old man with AIDS, hepatitis C, and a history of opportunistic infections, including Pneumocystis carinii pneumonia and invasive pulmonary aspergillosis. Cultures revealed the presence of the fungus Scedosporium apiospermum and S. aureus (Yao and Messner, 2001). Another example is that of a diabetic, 69-year old male with P. aeruginosa MEO symptoms. However, the treatment did not resolve the symptoms and the patient suffered of cranial nerves paralysis due to a co-infection with A. fumigatus (Bovo et al., 2012). Although these cases are rare, fungal infection should always be considered when presumptive bacterial infection symptoms do not improve after antibiotic therapy.
4.5. Association of fungi and Alzheimer’s disease
Alzheimer’s disease is a neurodegenerative disease caused by neuroinflammation and neuronal death (Pisa et al., 2017). Tissue sections from the external frontal cortex, cerebellar hemisphere, entorhinal cortex/hippocampus, and choroid plexus regions in the central nervous system (CNS) of an Alzheimer’s patient were analyzed and compared to a healthy subject (Pisa et al., 2015). Using anti-C. glabrata antibodies, fungal cells were present in each section analyzed in the Alzheimer’s patient, some even located at intranuclear regions and detected within neurons, but no fungal cells were present in the control participant (Pisa et al., 2015). Specific antibodies against C. albicans, Candida famata, Phoma betae, and Syncephalastrum racemosum showed the presence of fungal cells in three of the regions examined from an Alzheimer’s patient, but were absent in the control participant samples (Pisa et al., 2015). Entorhinal cortex/hippocampus samples from Alzheimer’s and healthy subjects identified the presence of fungi in all Alzheimer’s patients, but in none of the control participants (Pisa et al., 2015). Samples taken from the entorhinal cortex of Alzheimer’s patients were positive for figures resembling punctate, hyphal, and yeast-like structures (Pisa et al., 2016). Some of these structures were located surrounding or within the nuclei of brain cells (Pisa et al., 2016). A fungal enolase and β-tubulin were also identified in brain tissue samples from Alzheimer’s patients and β-tubulin staining indicated several punctate bodies within the cytoplasm (Pisa et al., 2016). Furthermore, macromolecules from diverse fungi were identified in the bloodstream, cerebrospinal fluid, and brain tissue of Alzheimer’s patients (Pisa et al., 2017). Among the common fungi isolated from brain tissue of Alzheimer’s patients were Alternaria, Botrytis, Candida, Cladosporium, Cryptococcus, Fusarium, Malassezia, and Penicillium (Pisa et al., 2017). These fungi coexist in brain tissue alongside bacteria, predominantly Burkholderia spp. (Pisa et al., 2017). These findings are a good starting point for future studies on the contribution of these polymicrobial interactions in the etiology of Alzheimer’s disease and other neurological diseases (Pisa et al., 2017).
5. CONCLUSION
Polymicrobial interactions in the environment are diverse, ranging from synergistic to antagonistic. These interactions are important because they are capable of enhancing life-sustaining crops. Other interactions are not as beneficial, such as those that decrease antimicrobial efficacy. Understanding environmental polymicrobial interactions may be helpful in the clinical setting, as many of the documented environmental interactions involving fungi are similar to causative agents of human disease including but not limited to C. neoformans, C. albicans, and Fusarium spp. For instance, candidemia is the fourth cause of bloodstream infections in the U.S., with the presence of a central venous catheter as a known risk factor, exacerbating patients’ morbidity and mortality (Barter et al., 2019). Estimations of C. albicans biofilm in individuals with indwelling medical devices continue to climb, emphasizing the need for better therapeutic interventions. Studying the mycobiome is important for understanding the role of the fungal population in the human microflora. However, it is necessary to realize the involvement of the mycobiome in human disease since the presence of certain fungal species in any given part of the body doesn’t necessarily indicate that these eukaryotic microorganisms are the etiological agents of disease (Ackerman and Underhill, 2017). Therefore, future studies would need to focus on differentiating fungi involved in disease or their role as transient or resident microbiota. In addition, current challenges in understanding the mycobiome rise from the inability to isolate samples from different regions of the body and to distinguish those that naturally reside in that region from those that are contaminated by environmental sources. Optimization of fungal isolation techniques is a crucial element to identification of the components of the mycobiome. Once studies are able to incorporate these into their findings, this information can be utilized to assess the molecular and structural factors fungi use to compete and co-exist in polymicrobial interactions. We will be able to determine the implications of these interactions in the evolution of fungal virulence and pathogenesis in plants, animals, and humans. Finally, studies focused on polymicrobial interactions involving fungi may lead to the development of treatments for the common types of fungal infections, thus, preventing infection in individuals at risk or reducing the population that are affected by fungi.
HIGHLIGHTS.
Polymicrobial interactions involving fungi are important in the environment and medicine.
Fungi facilitate plant growth and adaptations to stressful conditions.
Microbial symbiosis in the environment highlights the complexity and diversity of fungi.
The mycobiome has an essential role in health and disease that has begun to be elucidated.
Knowledge acquired from interkingdom interactions can be used in therapeutic development.
ACKNOWLEDGEMENTS
S. Z. A. was supported by the National Science Foundation (NSF)’s ACSScellence program (COURI) grant number DUE-1565063. G. J. C. was supported by the NSF’s Louis Stokes Alliance for Minority Participation (LSAMP) grant number HRD-1202008. L.R.M. was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health (NIH) under award number R01AI145559. L.R.M. is funded and has an appointment in the Infectious Diseases and Immunology cluster of the Border Biomedical Research Center (BBRC; National Institute on Minority Health and Health Disparities [NIMHHD] award number 5U54MD007592), UTEP’s Research Centers in Minority Institutions Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Abd-Alla MH, El-Enany AW, Nafady NA, Khalaf DM, Morsy FM, 2014. Synergistic interaction of Rhizobium leguminosarum bv. viciae and arbuscular mycorrhizal fungi as a plant growth promoting biofertilizers for faba bean (Vicia faba L.) in alkaline soil. Microbiol Res 169, 49–58. [DOI] [PubMed] [Google Scholar]
- Abdulkareem AF, Lee HH, Ahmadi M, Martinez LR, 2015. Fungal serotype-specific differences in bacterial-yeast interactions. Virulence 6, 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman AL, Underhill DM, 2017. The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann Transl Med 5, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed AK, Tawfik KM, Zinab A, El-Gawad A, 2008. Tolerance of seven faba bean varieties to drought and salt stresses. Res J Agr Biol Sci 4, 175–186. [Google Scholar]
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A, 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun 72, 6023–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis M, Mylonakis E, 2015. Fungal-bacterial interactions and their relevance in health. Cell Microbiol 17, 1442–1446. [DOI] [PubMed] [Google Scholar]
- Barousse MM, Van Der Pol BJ, Fortenberry D, Orr D, Fidel PL Jr., 2004. Vaginal yeast colonisation, prevalence of vaginitis, and associated local immunity in adolescents. Sex Transm Infect 80, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter DM, Johnston HL, Williams SR, Tsay SV, Vallabhaneni S, Bamberg WM, 2019. Candida Bloodstream Infections Among Persons Who Inject Drugs - Denver Metropolitan Area, Colorado, 2017–2018. MMWR Morb Mortal Wkly Rep 68, 285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianciotto V, Andreotti S, Balestrini R, Bonfante P, Perotto S, 2001. Extracellular polysaccharides are involved in the attachment of Azospirillum brasilense and Rhizobium leguminosarum to arbuscular mycorrhizal structures. Eur J Histochem 45, 39–49. [DOI] [PubMed] [Google Scholar]
- Bolton MD, Kolmer JA, Garvin DF, 2008. Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol 9, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botschuijver S, Roeselers G, Levin E, Jonkers DM, Welting O, Heinsbroek SEM, de Weerd HH, Boekhout T, Fornai M, Masclee AA, Schuren FHJ, de Jonge WJ, Seppen J, van den Wijngaard RM, 2017. Intestinal Fungal Dysbiosis Is Associated With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology 153, 1026–1039. [DOI] [PubMed] [Google Scholar]
- Bovo R, Benatti A, Ciorba A, Libanore M, Borrelli M, Martini A, 2012. Pseudomonas and Aspergillus interaction in malignant external otitis: risk of treatment failure. Acta Otorhinolaryngol Ital 32, 416–419. [PMC free article] [PubMed] [Google Scholar]
- Bradford LL, Ravel J, 2017. The vaginal mycobiome: A contemporary perspective on fungi in women’s health and diseases. Virulence 8, 342–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl MT, Carter MQ, Parker CT, Chapman MR, Huynh S, Zhou Y, 2011. Salmonella biofilm formation on Aspergillus niger involves cellulose--chitin interactions. PLoS One 6, e25553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting LA, Neilson JB, Bulmer GS, 1979. Cryptococcus neoformans: gastronomic delight of a soil ameba. Sabouraudia 17, 225–232. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Fu MS, Guimaraes AJ, Albuquerque P, 2019. The ‘Amoeboid Predator-Fungal Animal Virulence’ Hypothesis. J Fungi (Basel) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casqueiro J, Casqueiro J, Alves C, 2012. Infections in patients with diabetes mellitus: A review of pathogenesis. Indian J Endocrinol Metab 16 Suppl 1, S27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cateau E, Hechard Y, Fernandez B, Rodier MH, 2014. Free living amoebae could enhance Fusarium oxysporum growth. Fungal Ecol 8. [Google Scholar]
- Chacon MR, Lozano-Bartolome J, Portero-Otin M, Rodriguez MM, Xifra G, Puig J, Blasco G, Ricart W, Chaves FJ, Fernandez-Real JM, 2018. The gut mycobiome composition is linked to carotid atherosclerosis. Benef Microbes 9, 185–198. [DOI] [PubMed] [Google Scholar]
- Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA, 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183, 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang FY, Lee JH, Yang YH, Yu HH, Wang LC, Lin YT, Chiang BL, 2011. Analysis of the serum levels of fungi-specific immunoglobulin E in patients with allergic diseases. Int Arch Allergy Immunol 154, 49–56. [DOI] [PubMed] [Google Scholar]
- Charlson ES, Diamond JM, Bittinger K, Fitzgerald AS, Yadav A, Haas AR, Bushman FD, Collman RG, 2012. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am J Respir Crit Care Med 186, 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin-A-Woeng TF, Bloemberg GV, van der Bij AJ, van der Drift KM, Schripsema J, Kroon B, Scheffer RJ, Keel C, Bakker PA, Tichy HV, de Bruijn FJ, Thomas-Oates JE, Lugtenberg BJ, 1998. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant Microbe Interact 11, 1069–1077. [Google Scholar]
- Chrisman CJ, Alvarez M, Casadevall A, 2010. Phagocytosis of Cryptococcus neoformans by, and nonlytic exocytosis from, Acanthamoeba castellanii. Appl Environ Microbiol 76, 6056–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coco BJ, Bagg J, Cross LJ, Jose A, Cross J, Ramage G, 2008. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol Immunol 23, 377–383. [DOI] [PubMed] [Google Scholar]
- Costa-Orlandi CB, Sardi JCO, Pitangui NS, de Oliveira HC, Scorzoni L, Galeane MC, Medina-Alarcon KP, Melo W, Marcelino MY, Braz JD, Fusco-Almeida AM, Mendes-Giannini MJS, 2017. Fungal Biofilms and Polymicrobial Diseases. J Fungi (Basel) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Lucht L, Tipton L, Rogers MB, Fitch A, Kessinger C, Camp D, Kingsley L, Leo N, Greenblatt RM, Fong S, Stone S, Dermand JC, Kleerup EC, Huang L, Morris A, Ghedin E, 2015. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am J Respir Crit Care Med 191, 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L, Morris A, Ghedin E, 2013. The human mycobiome in health and disease. Genome Med 5, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brucker K, Tan Y, Vints K, De Cremer K, Braem A, Verstraeten N, Michiels J, Vleugels J, Cammue BP, Thevissen K, 2015. Fungal beta-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob Agents Chemother 59, 3052–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delafont V, Rodier MH, Maisonneuve E, Cateau E, 2018. Vermamoeba vermiformis: a Free-Living Amoeba of Interest. Microb Ecol 76, 991–1001. [DOI] [PubMed] [Google Scholar]
- Delhaes L, Monchy S, Frealle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabe M, Viscogliosi E, 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PLoS One 7, e36313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derengowski Lda S, Paes HC, Albuquerque P, Tavares AH, Fernandes L, Silva-Pereira I, Casadevall A, 2013. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell 12, 761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, Hacquard S, Herve V, Labbe J, Lastovetsky OA, Mieszkin S, Millet LJ, Vajna B, Junier P, Bonfante P, Krom BP, Olsson S, van Elsas JD, Wick LY, 2018. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev 42, 335–352. [DOI] [PubMed] [Google Scholar]
- Dhamgaye S, Qu Y, Peleg AY, 2016. Polymicrobial infections involving clinically relevant Gram-negative bacteria and fungi. Cell Microbiol 18, 1716–1722. [DOI] [PubMed] [Google Scholar]
- Diaz PI, Strausbaugh LD, Dongari-Bagtzoglou A, 2014. Fungal-bacterial interactions and their relevance to oral health: linking the clinic and the bench. Front Cell Infect Microbiol 4, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle J, McGee PA, 2003. Some endophytic fungi reduce the density of pustules of Puccinia recondita f. sp. tritici in wheat. Mycol Res 107, 310–316. [DOI] [PubMed] [Google Scholar]
- Drell T, Lillsaar T, Tummeleht L, Simm J, Aaspollu A, Vain E, Saarma I, Salumets A, Donders GG, Metsis M, 2013. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One 8, e54379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy AK, David MS, Li L, Heider TN, Peterson JD, Montano EA, Dongari-Bagtzoglou A, Diaz PI, Strausbaugh LD, 2014. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9, e90899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkins LM, O’Toole GA, 2015. Cystic Fibrosis Lung Infections: Polymicrobial, Complex, and Hard to Treat. PLoS Pathog 11, e1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KW, Thomas L, Warner J, Desmond R, Elewski BE, 2005. A bipartite interaction between Pseudomonas aeruginosa and fungi in onychomycosis. Arch Dermatol 141, 1467–1468. [DOI] [PubMed] [Google Scholar]
- Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A, 2011. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev 75, 583–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost F, Kacprowski T, Ruhlemann MC, Franke A, Heinsen FA, Volker U, Volzke H, Aghdassi AA, Mayerle J, Weiss FU, Homuth G, Lerch MM, 2019. Functional abdominal pain and discomfort (IBS) is not associated with faecal microbiota composition in the general population. Gut 68, 1131–1133. [DOI] [PubMed] [Google Scholar]
- Fry WE, McGrath MT, Seaman A, Zitter TA, McLeod A, Danies G, Small IM, Myers K, Everts K, Gevens AJ, Gugino BK, Johnson SB, Judelson H, Ristaino J, Roberts P, Secor G, Seebold K Jr., Snover-Clift K, Wyenandt A, Grunwald NJ, Smart CD, 2013. The 2009 Late Blight Pandemic in the Eastern United States - Causes and Results. Plant Dis 97, 296–306. [DOI] [PubMed] [Google Scholar]
- Gaitanis G, Magiatis P, Hantschke M, Bassukas ID, Velegraki A, 2012. The Malassezia genus in skin and systemic diseases. Clin Microbiol Rev 25, 106–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawaz A, Weisel G, 2018. Mixed infections are a critical factor in the treatment of superficial mycoses. Mycoses 61, 731–735. [DOI] [PubMed] [Google Scholar]
- Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM, 2010. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6, e1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldacre MJ, Watt B, Loudon N, Milne LJ, Loudon JD, Vessey MP, 1979. Vaginal microbial flora in normal young women. Br Med J 1, 1450–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves DS, Ferreira MDS, Gomes KX, Rodriguez-de La Noval C, Liedke SC, da Costa GCV, Albuquerque P, Cortines JR, Saramago Peralta RH, Peralta JM, Casadevall A, Guimaraes AJ, 2019. Unravelling the interactions of the environmental host Acanthamoeba castellanii with fungi through the recognition by mannose-binding proteins. Cell Microbiol, e13066. [DOI] [PubMed] [Google Scholar]
- Gu Y, Zhou G, Qin X, Huang S, Wang B, Cao H, 2019. The Potential Role of Gut Mycobiome in Irritable Bowel Syndrome. Front Microbiol 10, 1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes AJ, Gomes KX, Cortines JR, Peralta JM, Peralta RH, 2016. Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiol Res 193, 30–38. [DOI] [PubMed] [Google Scholar]
- Hager CL, Ghannoum MA, 2017. The mycobiome: Role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig Liver Dis 49, 1171–1176. [DOI] [PubMed] [Google Scholar]
- Hallen-Adams HE, Suhr MJ, 2017. Fungi in the healthy human gastrointestinal tract. Virulence 8, 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrach BD, Baltruschat H, Barna B, Fodor J, Kogel KH, 2013. The mutualistic fungus Piriformospora indica protects barley roots from a loss of antioxidant capacity caused by the necrotrophic pathogen Fusarium culmorum. Mol Plant Microbe Interact 26, 599–605. [DOI] [PubMed] [Google Scholar]
- Harriott MM, Noverr MC, 2011. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol 19, 557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M, Twomey K, McCarthy Y, O’Connell O, Febrer M, Alston M, Ryan R, Plant B, 2013. LSC 2013 abstract - The role of second-generation sequencing to characterize the fungal microbiota in the adult cystic fibrosis airway, and its correlation with standard culture-based methods and clinical phenotype. European Respiratory Journal 42, OP02. [Google Scholar]
- Hillman BI, Supyani S, Kondo H, Suzuki N, 2004. A reovirus of the fungus Cryphonectria parasitica that is infectious as particles and related to the coltivirus genus of animal pathogens. J Virol 78, 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA, 2016. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DA, Kolter R, 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296, 2229–2232. [DOI] [PubMed] [Google Scholar]
- Kato H, Sugita T, Ishibashi Y, Nishikawa A, 2007. Evaluation of the levels of specific IgE against Cryptococcus diffluens and Cryptococcus liquefaciens in patients with atopic dermatitis. Microbiol Immunol 51, 945–950. [DOI] [PubMed] [Google Scholar]
- Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, Burgess KE, Lang S, Millington O, Mackay WG, Williams C, Ramage G, 2017. Candida albicans Mycofilms Support Staphylococcus aureus Colonization and Enhances Miconazole Resistance in Dual-Species Interactions. Front Microbiol 8, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JR, Taylor GW, Rutman A, Hoiby N, Cole PJ, Wilson R, 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clin Pathol 52, 385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Choi GJ, Lee HB, Lee SW, Lim HK, Jang KS, Son SW, Lee SO, Cho KY, Sung ND, Kim JC, 2007. Some fungal endophytes from vegetable crops and their anti-oomycete activities against tomato late blight. Lett Appl Microbiol 44, 332–337. [DOI] [PubMed] [Google Scholar]
- Klaerner HG, Uknis ME, Acton RD, Dahlberg PS, Carlone-Jambor C, Dunn DL, 1997. Candida albicans and Escherichia coli are synergistic pathogens during experimental microbial peritonitis. J Surg Res 70, 161–165. [DOI] [PubMed] [Google Scholar]
- Kong EF, Tsui C, Kucharikova S, Andes D, Van Dijck P, Jabra-Rizk MA, 2016. Commensal Protection of Staphylococcus aureus against Antimicrobials by Candida albicans Biofilm Matrix. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EF, Tsui C, Kucharikova S, Van Dijck P, Jabra-Rizk MA, 2017. Modulation of Staphylococcus aureus Response to Antimicrobials by the Candida albicans Quorum Sensing Molecule Farnesol. Antimicrob Agents Chemother 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Morris A, 2017. The emerging importance and challenges of the human mycobiome. Virulence 8, 310–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Andes DR, Krysan DJ, 2018. Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog 14, e1007342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal S, Sumbali G, Sharma S, Kaul S, 2018. Detection of some new Trichosporon species from the dystrophied nails of three female members of a family from North Indian State of Jammu and Kashmir. Mycoses 61, 534–542. [DOI] [PubMed] [Google Scholar]
- Krause R, Moissl-Eichinger C, Halwachs B, Gorkiewicz G, Berg G, Valentin T, Prattes J, Hogenauer C, Zollner-Schwetz I, 2016. Mycobiome in the Lower Respiratory Tract - A Clinical Perspective. Front Microbiol 7, 2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstin L, Katanic Z, Jezic M, Poljak I, Nuskern L, Matkovic I, Idzojtic M, Curkovic-Perica M, 2017. Biological control of chestnut blight in Croatia: an interaction between host sweet chestnut, its pathogen Cryphonectria parasitica and the biocontrol agent Cryphonectria hypovirus 1. Pest Manag Sci 73, 582–589. [DOI] [PubMed] [Google Scholar]
- Lane LA, Ayres JF, Lovett JV, 1997. A review of the introduction and use of white clover (Trifolium repens L.) in Australia—significance for breeding objectives. Aust J Exp Agr 37, 831–839. [Google Scholar]
- Lehtonen PT, Helander M, Siddiqui SA, Lehto K, Saikkonen K, 2006. Endophytic fungus decreases plant virus infections in meadow ryegrass (Lolium pratense). Biol Lett 2, 620–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipner SR, Scher RK, 2015. Management of onychomycosis and co-existing tinea pedis. J Drugs Dermatol 14, 492–494. [PubMed] [Google Scholar]
- Maharshak N, Ringel Y, Katibian D, Lundqvist A, Sartor RB, Carroll IM, Ringel-Kulka T, 2018. Fecal and Mucosa-Associated Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Dig Dis Sci 63, 1890–1899. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Cateau E, Kaaki S, Rodier MH, 2016. Vermamoeba vermiformis-Aspergillus fumigatus relationships and comparison with other phagocytic cells. Parasitol Res 115, 4097–4105. [DOI] [PubMed] [Google Scholar]
- Malliaris SD, Steenbergen JN, Casadevall A, 2004. Cryptococcus neoformans var. gattii can exploit Acanthamoeba castellanii for growth. Med Mycol 42, 149–158. [DOI] [PubMed] [Google Scholar]
- Mar Rodriguez M, Perez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jove M, Pamplona R, Ricart W, Portero-Otin M, Chacon MR, Fernandez Real JM, 2015. Obesity changes the human gut mycobiome. Sci Rep 5, 14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraki S, Mavromanolaki VE, 2016. Epidemiology of Dermatophytoses in Crete, Greece. Med Mycol J 57, E69–E75. [DOI] [PubMed] [Google Scholar]
- McGinley KJ, Leyden JJ, Marples RR, Kligman AM, 1975. Quantitative microbiology of the scalp in non-dandruff, dandruff, and seborrheic dermatitis. J Invest Dermatol 64, 401–405. [DOI] [PubMed] [Google Scholar]
- Miles R, Hawley CM, McDonald SP, Brown FG, Rosman JB, Wiggins KJ, Bannister KM, Johnson DW, 2009. Predictors and outcomes of fungal peritonitis in peritoneal dialysis patients. Kidney Int 76, 622–628. [DOI] [PubMed] [Google Scholar]
- Monteiro-da-Silva F, Araujo R, Sampaio-Maia B, 2014. Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med Mycol 52, 498–505. [DOI] [PubMed] [Google Scholar]
- Morales DK, Jacobs NJ, Rajamani S, Krishnamurthy M, Cubillos-Ruiz JR, Hogan DA, 2010. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol 78, 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Chandra J, Retuerto M, Sikaroodi M, Brown RE, Jurevic R, Salata RA, Lederman MM, Gillevet PM, Ghannoum MA, 2014. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog 10, e1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF, 2017. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novohradska S, Ferling I, Hillmann F, 2017. Exploring Virulence Determinants of Filamentous Fungal Pathogens through Interactions with Soil Amoebae. Front Cell Infect Microbiol 7, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuskern L, Jezic M, Liber Z, Mlinarec J, Curkovic-Perica M, 2018. Cryphonectria hypovirus 1-Induced Epigenetic Changes in Infected Phytopathogenic Fungus Cryphonectria parasitica. Microb Ecol 75, 790–798. [DOI] [PubMed] [Google Scholar]
- Ortiz N, Armada E, Duque E, Roldan A, Azcon R, 2015. Contribution of arbuscular mycorrhizal fungi and/or bacteria to enhancing plant drought tolerance under natural soil conditions: effectiveness of autochthonous or allochthonous strains. J Plant Physiol 174, 87–96. [DOI] [PubMed] [Google Scholar]
- Park HK, Ha MH, Park SG, Kim MN, Kim BJ, Kim W, 2012. Characterization of the fungal microbiota (mycobiome) in healthy and dandruff-afflicted human scalps. PLoS One 7, e32847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Hogan DA, Mylonakis E, 2010. Medically important bacterial-fungal interactions. Nat Rev Microbiol 8, 340–349. [DOI] [PubMed] [Google Scholar]
- Peters BA, Wu J, Hayes RB, Ahn J, 2017. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol 17, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME, 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 25, 193–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, Scheper MA, Leid JG, Costerton JW, Shirtliff ME, 2010. Microbial interactions and differential protein expression in Staphylococcus aureus - Candida albicans dual-species biofilms. FEMS Immunol Med Microbiol 59, 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa D, Alonso R, Fernandez-Fernandez AM, Rabano A, Carrasco L, 2017. Polymicrobial Infections In Brain Tissue From Alzheimer’s Disease Patients. Sci Rep 7, 5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa D, Alonso R, Rabano A, Horst MN, Carrasco L, 2016. Fungal Enolase, beta-Tubulin, and Chitin Are Detected in Brain Tissue from Alzheimer’s Disease Patients. Front Microbiol 7, 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisa D, Alonso R, Rabano A, Rodal I, Carrasco L, 2015. Different Brain Regions are Infected with Fungi in Alzheimer’s Disease. Sci Rep 5, 15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Puglia A, Tanti R, Mifsud D, 2005. A severe outbreak of crown and root rot of tomato caused by Fusarium oxysporum f. sp. radicis-lycopersici in Malta. Phytopathol Mediterr 44, 319–321. [Google Scholar]
- Rapala-Kozik M, Bochenska O, Zajac D, Karkowska-Kuleta J, Gogol M, Zawrotniak M, Kozik A, 2018. Extracellular proteinases of Candida species pathogenic yeasts. Mol Oral Microbiol 33, 113–124. [DOI] [PubMed] [Google Scholar]
- Rigling D, Prospero S, 2018. Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol 19, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Metraux JP, L’Haridon F, 2016. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol 210, 1033–1043. [DOI] [PubMed] [Google Scholar]
- Rouabhia M, Chmielewski W, 2012. Diseases associated with oral polymicrobial biofilms. Open Mycol 6, 27–32. [Google Scholar]
- Saito F, Ikeda R, 2005. Killing of cryptococcus neoformans by Staphylococcus aureus: the role of cryptococcal capsular polysaccharide in the fungal-bacteria interaction. Med Mycol 43, 603–612. [DOI] [PubMed] [Google Scholar]
- Shirtliff ME, Peters BM, Jabra-Rizk MA, 2009. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett 299, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN, 2012. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 7, e37818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Nosanchuk JD, Malliaris SD, Casadevall A, 2004. Interaction of Blastomyces dermatitidis, Sporothrix schenckii, and Histoplasma capsulatum with Acanthamoeba castellanii. Infect Immun 72, 3478–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen JN, Shuman HA, Casadevall A, 2001. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. Proc Natl Acad Sci U S A 98, 15245–15250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T, Saito M, Ito T, Kato Y, Tsuboi R, Takeuchi S, Nishikawa A, 2003. The basidiomycetous yeasts Cryptococcus diffluens and C. liquefaciens colonize the skin of patients with atopic dermatitis. Microbiol Immunol 47, 945–950. [DOI] [PubMed] [Google Scholar]
- Tipton L, Ghedin E, Morris A, 2017. The lung mycobiome in the next-generation sequencing era. Virulence 8, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treat J, James WD, Nachamkin I, Seykora JT, 2007. Growth inhibition of Trichophyton species by Pseudomonas aeruginosa. Arch Dermatol 143, 61–64. [DOI] [PubMed] [Google Scholar]
- van der Heijden MG, de Bruin S, Luckerhoff L, van Logtestijn RS, Schlaeppi K, 2016. A widespread plant-fungal-bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J 10, 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden WJ, Netea MG, de Haan AF, Huls GA, Donnelly JP, Blijlevens NM, 2013. Role of the mycobiome in human acute graft-versus-host disease. Biol Blood Marrow Transplant 19, 329–332. [DOI] [PubMed] [Google Scholar]
- Veer P, Patwardhan NS, Damle AS, 2007. Study of onychomycosis: prevailing fungi and pattern of infection. Indian J Med Microbiol 25, 53–56. [DOI] [PubMed] [Google Scholar]
- Willger SD, Grim SL, Dolben EL, Shipunova A, Hampton TH, Morrison HG, Filkins LM, O’Toole GA, Moulton LA, Ashare A, Sogin ML., Hogan DA, 2014. Characterization and quantification of the fungal microbiome in serial samples from individuals with cystic fibrosis. Microbiome 2, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Cheng J, Tang J, Fu Y, Jiang D, Baker TS, Ghabrial SA, Xie J, 2014. A novel partitivirus that confers hypovirulence on plant pathogenic fungi. J Virol 88, 10120–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou A, 2014. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol 29, 99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wu S, Liu L, Cheng J, Fu Y, Jiang D, Xie J, 2015. A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res 197, 127–136. [DOI] [PubMed] [Google Scholar]
- Yang YS, Ahn JJ, Shin MK, Lee MH, 2011. Fusarium solani onychomycosis of the thumbnail coinfected with Pseudomonas aeruginosa: report of two cases. Mycoses 54, 168–171. [DOI] [PubMed] [Google Scholar]
- Yao M, Messner AH, 2001. Fungal malignant otitis externa due to Scedosporium apiospermum. Ann Otol Rhinol Laryngol 110, 377–380. [DOI] [PubMed] [Google Scholar]
- Zakaria MN, Furuta M, Takeshita T, Shibata Y, Sundari R, Eshima N, Ninomiya T, Yamashita Y, 2017. Oral mycobiome in community-dwelling elderly and its relation to oral and general health conditions. Oral Dis 23, 973–982. [DOI] [PubMed] [Google Scholar]
- Zhang E, Tanaka T, Tajima M, Tsuboi R, Nishikawa A, Sugita T, 2011. Characterization of the skin fungal microbiota in patients with atopic dermatitis and in healthy subjects. Microbiol Immunol 55, 625–632. [DOI] [PubMed] [Google Scholar]