Abstract
A 20-day trial was conducted to reveal bacterial community dynamics in a commercial nursery of larval Litopenaeus vannamei larvae. The bacterial communities in the ambient water were profiled by high-throughput sequencing of the V4–V5 hypervariable region of the 16S rRNA gene. The results indicated that the dominant bacterial phyla between the metamorphosis stage and postlarval stage were Bacteroidetes, Proteobacteria, Cyanobacteria, and Firmicutes, representing more than 80.09% of the bacterial operational taxonomic units. The relative abundance among bacterial phyla notably differed between the two stages. The relative abundance of Cyanobacteria was higher in the metamorphosis stage, while that of Bacteroidetes was higher and more stable in the postlarval stage. At the class level, the relative abundance of Sphingobacteriia and Alphaproteobacteria increased markedly in the postlarval stage, while that of Flavobacteriia decreased. Redundancy analysis showed that bacterial composition in the metamorphosis stage was positively correlated with salinity, alkalinity, and pH, while in the postlarval stage, it was positively correlated with ammonium nitrogen and nitrite nitrogen. Thus, microbial community diversity in the nursery phase varies per rearing stage.
Electronic supplementary material
The online version of this article (10.1007/s12088-019-00853-7) contains supplementary material, which is available to authorized users.
Keywords: Pacific white shrimp, Nursery phase, Microbial community, High-throughput sequencing, 16S rRNA
Introduction
Pacific white shrimp (Litopenaeus vannamei) is an important shrimp species. Its share in seafood has been rapidly increasing for more than two decades, with an annual percentage of 8.1%, compared with 0.62% for capture fishery and 2.6% for meat production [1]. The L. vannamei nursery phase (approximately 20 days after hatching) is a key step in the shrimp production cycle. Many infectious diseases preferentially affect L. vannamei larvae, resulting in high mortality [2]. The mass mortality of larval L. vannamei is a major constraint in its aquaculture production. It is generally attributed to opportunistic pathogenic bacteria in the ambient water. These bacteria are common in the seawater and feed that are used in hatcheries. For instance, during metamorphosis, shrimp are susceptible to pathogenic Vibrio [3].
In nursery rearing of L. vannamei, shrimp larvae encounter the ambient bacterioplankton immediately after hatching. Generally, the nursery phase can be artificially divided into two stages: metamorphosis (including nauplius, zoea, and mysis) and the postlarval stage [4]. The morphological and physiological characteristics of L. vannamei change dramatically in early development, leading to high variability in nursery rearing [5]. The dietary patterns of shrimp larvae also change with the different developmental stages [6], further influencing the aquaculture environment.
The microbiota associated with L. vannamei, such as gut and environmental microbiota, has been studied extensively [7–11]. Previous studies have also supplied further understanding of the gut microbiome in postlarvae of the nursery period [12–14]. However, most of the shrimp in these studies were postlarvae (age from Day 42 to Day 115) or older shrimp. The metamorphosis period (Day 0 to Day 20) has not received much research attention, and there is limited information about the bacterial community associated with L. vannamei larvae during the metamorphosis stage. Since the ambient bacterial community influences aquatic animals [15], the characterization of environmental microbiota would be an initial step for improving shrimp production.
Given that the aquaculture environment is closely associated with the different stages of metamorphosis of larval L. vannamei, we hypothesized that these stages may have their own pattern of bacterial community composition in the nursery rearing environment. To test this hypothesis, we explored the dynamics of bacterial communities during nursery rearing of L. vannamei larvae. In this study, we characterized the ambient microbial communities associated with the nursery phase of larval L. vannamei through high-throughput sequencing of the V4–V5 region of the bacterial 16S rRNA gene to characterize bacterial diversity and community structure.
Materials and Methods
Larval Rearing
The experiment was carried out at a commercial shrimp hatchery in Wenzhou, China. Males and females were maintained in broodstock pools with seawater until spawning. After hatching, larvae were transferred to rearing tanks with the same salinity. In this study, larvae from the same spawning pool were transferred into two rearing pools with sand-filtered seawater (30 ppt salinity) and food algae. The L. vannamei larvae were reared at approximately equal densities (2.5 × 105 individuals/m3) for a 20-day nursery stage.
Sample Collection and Water Quality Testing
Two adjacent cement pools of larval L. vannamei with similar genetic backgrounds were selected for sampling. Water samples were collected from the pools every 2 days during the rearing period. All water samples were collected in the morning prior to feeding and then immediately transferred to a nearby laboratory. Eighteen water samples were collected: L1–L9 (left pool) and R1–R9 (right pool). According to morphological observations, the samples were divided into four stages: nauplius (L1, R1), zoea (L2, L3, R2, R3), mysis (L4, L5, R4, R5), and postlarval (L6–L9, R6–R9) (Table S1).
Water samples were filtered through a 0.22 µm polycarbonate membrane filter (Millipore) to collect bacterial cells. The membrane filters were transferred to a 50 ml sterile centrifuge tube and stored at − 70 °C until DNA extraction. Alkalinity, pH, and salinity were determined in situ using a portable multiple parameter water detector (Ponsel, France). Ammonium nitrogen (NH4–N) was determined by indophenol blue spectrophotometry following the principle that ammonia reacting with phenol and hypochlorite can form indophenol blue in weak basic solutions, catalyzed by sodium nitrosyl ferricyanide, which can be measured by the absorbance value at 640 nm [16]. Nitrite nitrogen (NO2–N) was determined by naphthalene ethylenediamine spectrophotometry following the principle that nitrite can be diazotized with sulfonamide in acidic solutions, after which the product is coupled with N-(1-naphthyl (ethylenediamine) dihydrochloride to form a red azo dye that can be measured by the absorbance value at 543 nm [16].
DNA Extraction and High-Throughput Sequencing
Genomic DNA of the water samples was extracted from the membrane filters with a Water DNA Kit (Omega-Biotek) following the standard protocols. The bacterial communities in the rearing water were profiled by high-throughput sequencing of the V4–V5 hypervariable region of the 16S rRNA gene. The 16S rRNA gene was amplified using a universal primer pair, 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 909R (5′-CCCCGYCAATTCMTTTRAGT-3′), with a unique 12-digit barcode [17, 18]. Phusion® High-Fidelity Polymerase Chain Reaction (PCR) Master Mix with GC Buffer (New England Biolabs) was used for PCR as follows: 3 min at 94 °C; 30 cycles of 60 s at 94 °C, 60 s at 56 °C, and 60 s at 72 °C; and an extension of 10 min at 72 °C. Replicate PCRs of each sample were conducted, and the PCR products were combined and subjected to 1.0% agarose gel electrophoresis. The DNA band with the target size was excised, and then purified using a GeneJET DNA Gel Extraction Kit (Thermo Scientific). The sequencing samples were prepared with NEB Next® Ultra™ DNA Library Prep Kit for Illumina (New England Biolabs) per the manufacturer’s instructions. High-throughput sequencing was performed with an Illumina MiSeq platform using the Reagent Kit v2 2 × 250 bp.
Data Processing and Bioinformatics Analysis
The raw data from the high-throughput sequencing were processed with QIIME Version 1.7.0 [19]. Based on the barcodes, all the reads were trimmed and then assigned to each sample. The high-quality reads (length > 300 bp, without ambiguous base “N”, and average base quality score > 30) were chosen for further study. The aligned sequences were tested by chimera check with Gold database (Uchime algorithm), and then chimera sequences were excluded to obtain effective data. The effective sequences were clustered into operational taxonomic units (OTUs) with the software Uparse (v7.0.1001, http://drive5.com/uparse/) with a 97% identity threshold. Before downstream analysis, all the samples were re-sampled randomly at the minimum depth of all samples to minimize the biases of unequal sequencing depth. Taxonomic information of each OTU was assigned using the Ribosomal Database Project classifier (Version 2.2) and Green Gene [20].
Redundancy analysis (RDA) was further performed to determine which environmental variables were most strongly related to community composition. In addition, the 16S rRNA gene amplicon sequence data were also analyzed based on weighted UniFrac distance, alpha diversity (i.e., Shannon index), and principal coordinates analysis (PCoA). To predict the gut bacteria mediated function, the OTU table was normalized by dividing the abundance of each OTU by its predicted 16S copy number, producing the Kyoto Encyclopedia of Genes and Genomes (KEGG) classified functions of the community by the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt). Functional changes of the gut microbiome in different stages were compared using the t test.
Results
High-Throughput Sequencing Profile
High-throughput sequencing of 16S rRNA sequences was employed to determine bacterial community composition during the nursery phase of 18 water samples from two adjacent pools in the same facility. All raw sequence data were deposited under NCBI with accession numbers SRR9992992–SRR9993009 (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP218671).
In total, 409,455 effective sequences were obtained, with 8218–40,332 sequences per sample (mean = 22,747.5); 23,641 OTUs were detected by clustering at a 97% identity threshold. The sequences were randomly re-sampled at the minimum depth (8200 sequences per sample) among samples. To estimate the bacterial diversity in each sample, alpha diversity indices were calculated based on the OTUs. Bacterial diversity, estimated by the Shannon index, varied from 3.432 to 9.029 in the nursery phase. There was no significant difference in the Shannon index between the two pools (p > 0.05, t test).
Bacterial Community Composition and Beta Diversity
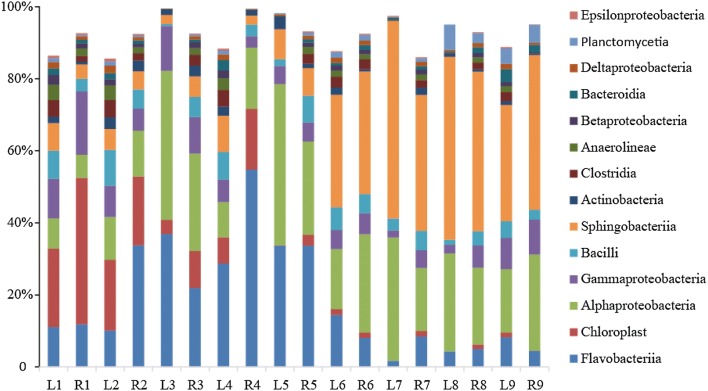
Bacteroidetes (17–58%), Proteobacteria (20–54%), Cyanobacteria (0.01–41%), and Firmicutes (1–15%) were the dominant bacterial phyla (relative abundance > 5%), accounting for more than 80.09% of the bacterial OTUs (Fig. S1). At the class taxonomic level, we found that the abundance of Sphingobacteriia and Alphaproteobacteria increased markedly in the late nursery phase, while the abundance of Flavobacteriia and Chloroplast decreased (Fig. 1).
Fig. 1.
Bacterial community structural dynamics at the class taxonomic level during the nursery phase of larval L. vannamei. L1–L9 and R1–R9 represent water samples collected successively every 2 days, and the classes with less than 1% relative abundance are not shown
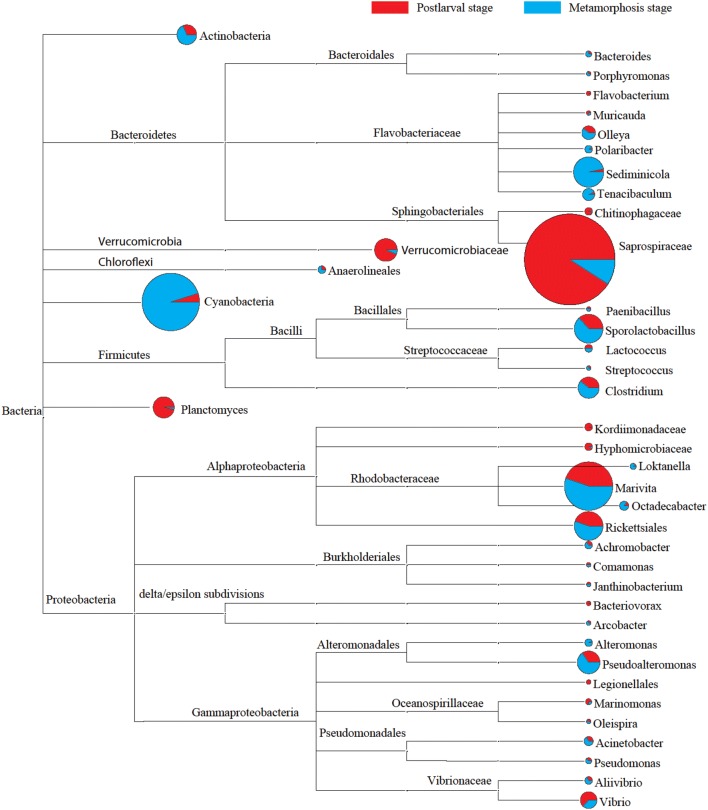
An unweighted pair group method with arithmetic mean (UPGMA) tree, using weighted UniFrac distances, revealed the similarity of water samples from the early development of L. vannamei larvae. The results indicated a noticeable difference in the bacterial community between the metamorphosis and postlarval stages (Fig. S1). The abundance of Cyanobacteria decreased significantly in the postlarval stage, while that of Bacteroidetes increased. According to the PCoA (Fig. S2), the very early parallel samples (L1 and R1) and the samples from the postlarval stage (L6–L9 and R6–R9) were clustered together, respectively, which implied a high similarity among bacterial communities from different samples. Compared to the metamorphosis stage (Day 1–Day 10), the composition of the dominant bacterial populations (relative abundance > 1%) changed in the postlarval stage of L. vannamei (Day 11–Day 20). Flavobacteriaceae, Bacillales, Rhodobacteraceae, and Alteromonadales were dominant in the metamorphosis stage. Saprosporaceae was the most dominant bacterial population among all samples (Fig. 2).
Fig. 2.
Comparison of the dominant bacterial populations at genus level in the aquaculture environment during the nursery phase of L. vannamei larvae. The OTUs without genus-level information were clustered based on the lowest taxon information. Pie charts reflect the relative abundance of each dominant operational taxonomic unit (OTU) (relative abundance > 1%) at two stages (blue represents the metamorphosis stage, Day 1 to Day 10; red represents the postlarval stage, Day 11 to Day 20); and the size of the pie chart reflects the total number of dominant OTUs in all samples
Relationship Between Bacterial Community Composition and Environmental Factors
Several environmental factors were measured during the nursery phase: NH4–N (0.04–0.59 mg/l) and NO2–N (0.03–0.18 mg/l) increased markedly, while pH (8.25–7.91), salinity (30–26.3), and total alkalinity (156–121) decreased slowly over time.
RDA showed that planktonic bacterial community compositions were influenced by different environmental factors during nursery rearing. In the metamorphosis stage, bacterial composition was mainly correlated with salinity, alkalinity, and pH, whereas that in the postlarval stage was correlated with NH4–N and NO2–N (Fig. S3). The RDA for the water quality indicators suggested that the bacterial community at different stages (metamorphosis versus postlarval) was positively correlated with different environmental factors.
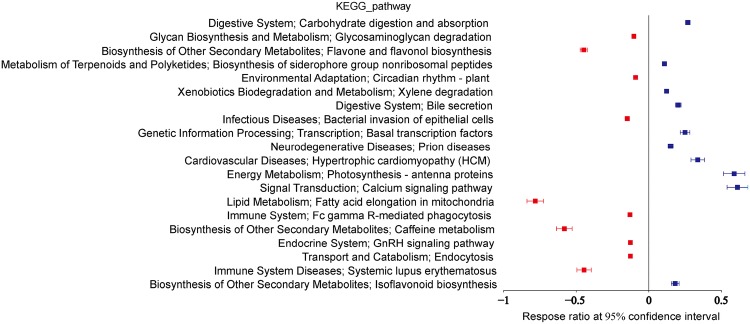
To analyze the functional changes at different stages (metamorphosis versus postlarval), KEGG Orthologies (KOs) with significant change (p < 0.05) between the two stages were selected. In total, 1, 923 differential expression KOs were found, which belong to different KEGG pathways, such as the two-component system, bacterial secretion system, and biosynthesis of secondary metabolites. Top 20 KEGG pathways were selected to depict the significant functional changes (Fig. 3).
Fig. 3.
Microbial KEGG pathways that exhibited significant changes in the postlarval stage compared to the metamorphosis stage. The top 20 KEGG pathways with significant change (p < 0.05) are shown. Red dots indicate upregulated KEGG pathways, and blue dots indicate downregulated KEGG pathways in the postlarval stage
Discussion
Despite diverse bacterial populations associated with the rearing environment of L. vannamei larvae, the dominant bacteria were Bacteroidetes, Proteobacteria, Cyanobacteria, and Firmicutes, representing more than 80.09% of the bacterial OTUs. Planktonic bacteria could be used as the indicator of shrimp health status [21, 22]. Meanwhile, as a source of intestinal microbiota, it has an important effect on the shrimp microbiome [23]. In addition, previous studies on the intestinal bacteria of shrimp, postlarvae, juveniles, and adults showed that the dominant bacteria were Proteobacteria, followed by Firmicutes and Bacteroidetes [24–27]. These phyla were also dominant in Fenneropenaeus chinensis [28], Penaeus merguiensis [29], and Nephrops norvegicus [30]. The results implied that there were numerous common bacterial populations in the intestines and the surrounding water, while the differences may be ascribed to changes in relative abundance.
The nursery phase is the key step for farming L. vannamei. During the nursery phase, the larval diet changes over time. At the nauplius stage, the nutrient needs are provided by the yolk, resulting in similar bacterial flora between the eggs and ambient water [31]. Since no plankton feeding occurs at this stage, there is little impact on bacterial community structure (Fig. S1, L1 and R1). When the shrimp larvae become zoeae on Day 3, they begin to eat unicellular algae and/or plant debris. Then, they begin to eat zooplankton or animal debris when they become myses. Thus, this shift influences the microbial community, as shown by the temporal dynamics of the bacterioplankton community (Fig. 1). The abundance of Cyanobacteria decreased noticeably after Day 3, while Flavobacteria abundance increased during the metamorphosis stage. At the start of the experiment, because of similar biotic backgrounds (same algae species and shrimp eggs from the same broodstock) and environmental conditions (same seawater, facility, and management), the results showed that bacterial communities in the two parallel pools were very similar at the beginning. Thereafter, bacterial community structure diverged in the subsequent days, leading to a high level of dissimilarity between the two parallel pools. Therefore, the bacterioplankton communities were strongly affected by the larval shrimp stage. In contrast, postlarval shrimp eat benthic or sedimentary organisms. However, there were no marked changes in microbiota observed during the postlarval stage, suggesting relatively stable bacterioplankton communities (Fig. S2). This is likely because the postlarval diet does not change as it does during metamorphosis.
Bacteroidetes and Flavobacteria may degrade various kinds of organic matter, thus providing available nutrients to Proteobacteria [32]. Compared to the healthy samples, the relative abundance of Gammaproteobacteria was higher in the diseased samples, which showed better adaptability to oligotrophic seawater environments [33, 34], indicating that the diseased shrimp ponds with low nutrient level could be more adaptable for opportunistic pathogens. In this study, Chloroplast may be prokaryotic or eukaryotic microalgae according to the taxonomic information of each OTU assigned. Therefore, Chloroplast could be taken as a symbol of the relative abundance of microalgae in a microbial community, which suggest the importance of microalgae in environmental regulation in the postlarval stage.
Zheng et al. [35] found that healthy shrimp were colonized widely by Proteobacteria and Bacteroidetes at all growth stages, which varied in relative abundance. Consistent with that study, Proteobacteria and Bacteroidetes, as the dominant phyla in the water environment, occurred at all stages of the nursery phase. The two phyla were also commonly found in other growth stages of L. vannamei [7, 8] and other aquatic animals, such as black tiger shrimp (Penaeus monodon) [22], turbot (Scophthalmus maximus), and sole (Solea senegalensis) [36]. Bacteroidetes may play an important role in dissolved organic matter uptake [37] and degradation of diverse biopolymers (cellulose, chitin, etc.) [32]. Similarly, Proteobacteria are distributed extensively in seawater environments and might participate in the degradation of complex substances [38]. Although the two dominant phyla were present throughout the nursery, the main bacterial groups shifted at different growth stages.
In mariculture of shrimp postlarvae, the developmental stage is a crucial biological factor affecting the gut microbial composition [39]. Development can be divided into three stages depicting varied gut microbial community [40]. Compared to the microbiome in the water environment, the gut microbiomes of adult shrimp among different locations were more similar, indicating the stability of the gut microbiome in adults [12, 13]. Compared to the postlarval stage, the environmental microbiome in the metamorphosis stage was highly variable. Dietary differences and digestive system development might be responsible for the changes in bacterial composition.
Several studies have predicted the functional capacity of shrimp microbiota by PICRUSt and have revealed that shrimp microbial functions varied in different organs and at different developmental stages and lifestyle [14], and even among wild, aquacultured and acute hepatopancreatic necrosis syndrome (AHPND)/early mortality syndrome (EMS) outbreak conditions [36]. In addition, in the present study, we also performed functional analysis of the water microbiome in the rearing environment of metamorphosis and postlarval stages of shrimp. Function prediction results showed that the KEGG pathways in lipid metabolism, biosynthesis of other secondary metabolites, and infectious diseases and immune system diseases were significantly upregulated in the postlarval stage, which suggests divergent functions of water environment microbiota between both stages. The results implied that environmental microorganisms may have an impact on the health of shrimp through changes in microbial function, which are caused by the high abundance of opportunistic pathogens. Meanwhile, the increase in bacterial metabolites could elevate the environmental stress level, thus leading to a low survival rate of shrimp postlarvae.
In conclusion, this study provides basic knowledge of the relationship between environmental microbiota and the nursery stage of shrimp larvae in aquaculture using high-throughput sequencing. In our study, Bacteroidetes, Proteobacteria, Cyanobacteria, and Firmicutes were the dominant phyla in the water environment during nursery rearing. The microbial community in the metamorphosis stage was unstable. However, in the postlarval stage, the microbiota varied from that of the metamorphosis stage and was more stable. The microbiota composition in the ambient water was notably affected by growth stage. Finally, our work fills the knowledge gap on the dynamics of bacterioplankton communities during the metamorphosis stage.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The project was supported by the Program for the Natural Science Foundation of China (31972821), the Zhejiang Provincial Natural Science Foundation of China (LY18C190003; LZ18C190001; LY14C190006), Science and Technology Project of Zhejiang Province (LGN18C180002; 2016C02055-5; 2015C03005; 2016F10021), the Scientific Innovation Team Project of Ningbo (2015C110018), Wenzhou science and technology project (S20150002), the Program for the Natural Science Foundation of Ningbo (2018A610342), Zhejiang Provincial Key Laboratory of Preventive Veterinary Medicine (ZPKLPVM2017KFKT005), and the KC Wong Magna Fund in Ningbo University.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Maocang Yan and Xiang Zhang have been contributed equally to this work.
References
- 1.FAO . The state of world fisheries and aquaculture. Rome: Food and Agriculture Organization of the United Nations; 2014. [Google Scholar]
- 2.Aguirre G, Ascencio V. Infectious disease in shrimp species with aquaculture potential. Recent Res Dev Microbiol. 2000;4:333–348. [Google Scholar]
- 3.Vandenberghe J, Verdonck L, Robles-Arozarena R, Rivera G, Bolland A, Balladares M, Gomez-Gil B, Calderon J, Sorgeloos P, Swings J. Vibrios associated with Litopenaeus vannamei larvae, postlarvae, broodstock, and hatchery probionts. Appl Environ Microbiol. 1999;65:2592–2597. doi: 10.1128/AEM.65.6.2592-2597.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertzler PL, Freas WR. Pleonal muscle development in the shrimp Penaeus (Litopenaeus) vannamei (Crustacea: Malacostraca: Decapoda: Dendrobranchiata) Arthropod Struct Dev. 2009;38:235–246. doi: 10.1016/j.asd.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Zacharia S, Kakati VS. Optimal salinity and temperature for early developmental stages of Penaeus merguiensis De man. Aquaculture. 2004;232:373–382. doi: 10.1016/S0044-8486(03)00538-6. [DOI] [Google Scholar]
- 6.Wei JK, Zhang XJ, Yu Y, Huang H, Li FH, Xiang JH. Comparative transcriptomic characterization of the early development in Pacific white shrimp Litopenaeus vannamei. PLoS One. 2014;9:e106201. doi: 10.1371/journal.pone.0106201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang ZB, Li XY, Wang LP, Shao ZZ. Changes in the intestinal bacterial community during the growth of white shrimp, Litopenaeus vannamei. Aquac Res. 2014;47:1737–1746. doi: 10.1111/are.12628. [DOI] [Google Scholar]
- 8.Johnson CN, Barnes S, Ogle J, Grimes DJ, Chang YJ, Peacock AD, Kline L. Microbial community analysis of water, foregut, and hindgut during growth of Pacific white shrimp, Litopenaeus vannamei, in closed-system aquaculture. J World Aquacult Soc. 2008;39:251–258. doi: 10.1111/j.1749-7345.2008.00155.x. [DOI] [Google Scholar]
- 9.Pangastuti A, Suwanto A, Lestari Y, Suhartono MT. Bacterial communities associated with white shrimp (Litopenaeus vannamei) larvae at early developmental stages. Biodiversitas. 2010;11:65–68. doi: 10.13057/biodiv/d110203. [DOI] [Google Scholar]
- 10.Zhang DM, Wang X, Xiong JB, Zhu JL, Wang YN, Zhao QF, Chen HP, Guo AN, Wu JF, Dai HP. Bacterioplankton assemblages as biological indicators of shrimp health status. Ecol Indic. 2014;38:218–224. doi: 10.1016/j.ecolind.2013.11.002. [DOI] [Google Scholar]
- 11.Zhang ML, Sun YH, Chen K, Yu N, Zhou ZG, Chen LQ, Du ZY, Li EC. Characterization of the intestinal microbiota in Pacific white shrimp, Litopenaeus vannamei, fed diets with different lipid sources. Aquaculture. 2014;434:449–455. doi: 10.1016/j.aquaculture.2014.09.008. [DOI] [Google Scholar]
- 12.Fan JQ, Chen LM, Mai GQ, Zhang HR, Yang JF, Deng D, Ma YF. Dynamics of the gut microbiota in developmental stages of Litopenaeus vannamei reveal its association with body weight. Sci Rep. 2019;9:734. doi: 10.1038/s41598-018-37042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J, Xuan L, Yu W, Zhu J, Qiu Q, Chen J. Spatiotemporal successions of shrimp gut microbial colonization: high consistency despite distinct species pool. Environ Microbiol. 2019;21:1383–1394. doi: 10.1111/1462-2920.14578. [DOI] [PubMed] [Google Scholar]
- 14.Cornejo-Granados F, Lopez-Zavala AA, Gallardo-Becerra L, Mendoza-Vargas A, Sánchez F, Vichido R, Brieba LG, Viana MT, Sotelo-Mundo RR, Ochoa-Leyva A. Microbiome of Pacific Whiteleg shrimp reveals differential bacterial community composition between Wild, Aquacultured and AHPND/EMS outbreak conditions. Sci Rep. 2017;7:11783. doi: 10.1038/s41598-017-11805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21:3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang XF. Survey, observation and analysis of lake ecology. Beijing: China Standard Press; 2000. [Google Scholar]
- 17.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiong J, Zhu J, Wang K, Wang X, Ye X, Liu L, Zhao Q, Hou M, Qiuqian L, Zhang D. The temporal scaling of bacterioplankton composition: high turnover and predictability during shrimp cultivation. Microb Ecol. 2014;67:256–264. doi: 10.1007/s00248-013-0336-7. [DOI] [PubMed] [Google Scholar]
- 22.Xiong J, Zhu J, Zhang D. The application of bacterial indicator phylotypes to predict shrimp health status. Appl Microbiol Biotechnol. 2014;98:8291–8299. doi: 10.1007/s00253-014-5941-y. [DOI] [PubMed] [Google Scholar]
- 23.Xiong J, Dai W, Qiu Q, Zhu J, Yang W, Li C. Response of host–bacterial colonization in shrimp to developmental stage, environment and disease. Mol Ecol. 2018;27:3686–3699. doi: 10.1111/mec.14822. [DOI] [PubMed] [Google Scholar]
- 24.Chaiyapechara S, Rungrassamee W, Suriyachay I, Kuncharin Y, Klanchui A, Karoonuthaisiri N, Jiravanichpaisal P. Bacterial community associated with the intestinal tract of P. monodonin in commercial farms. Microb Ecol. 2011;63:938–953. doi: 10.1007/s00248-011-9936-2. [DOI] [PubMed] [Google Scholar]
- 25.Rungrassamee W, Klanchui A, Chaiyapechara S, Maibunkaew S, Tangphatsornruang S, Jiravanichpaisal P, Karoonuthaisiri N. Bacterial population in intestines of the black tiger shrimp (Penaeus monodon) under different growth stages. PLoS One. 2013;8:e60802. doi: 10.1371/journal.pone.0060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rungrassamee W, Klanchui A, Maibunkaew S, Chaiyapechara S, Jiravanichpaisal P, Karoonuthaisiri N. Characterization of intestinal bacteria in wild and domesticated adult black tiger shrimp (Penaeus monodon) PLoS One. 2014;9:e91853. doi: 10.1371/journal.pone.0091853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong J, Zhu J, Dai W, Dong C, Qiu Q, Li C. Integrating gut microbiota immaturity and disease-discriminatory taxa to diagnose the initiation and severity of shrimp disease. Environ Microbiol. 2017;19:1490–1501. doi: 10.1111/1462-2920.13701. [DOI] [PubMed] [Google Scholar]
- 28.Liu H, Wang L, Liu M, Wang B, Jiang K, Ma S, Li Q. The intestinal microbial diversity in Chinese shrimp (Fenneropenaeus chinensis) as determined by PCR–DGGE and clone library analyses. Aquaculture. 2011;317:32–36. doi: 10.1016/j.aquaculture.2011.04.008. [DOI] [Google Scholar]
- 29.Oxley AP, Shipton W, Owens L, McKay D. Bacterial flora from the gut of the wild and cultured banana prawn, Penaeus merguiensis. J Appl Microbiol. 2002;93:214–223. doi: 10.1046/j.1365-2672.2002.01673.x. [DOI] [PubMed] [Google Scholar]
- 30.Meziti A, Mente E, Kormas KA. Gut bacteria associated with different diets in reared Nephrops norvegicus. Syst Appl Microbiol. 2012;35:473–482. doi: 10.1016/j.syapm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Phatarpekar PV, Kenkre VD, Sreepada RA, Desai UM, Achuthankutty CT. Bacterial flora associated with larval rearing of the giant freshwater prawn, Macrobrachium rosenbergii. Aquaculture. 2002;203:279–291. doi: 10.1016/S0044-8486(01)00705-0. [DOI] [Google Scholar]
- 32.Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, Cavicchioli R. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol. 2013;15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- 33.Bowman JP, McCuaig RD. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol. 2003;69:2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urakawa H, Kita-Tsukamoto K, Ohwada K. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology. 1999;145:3305–3315. doi: 10.1099/00221287-145-11-3305. [DOI] [PubMed] [Google Scholar]
- 35.Zheng YF, Yua M, Liu Y, Sua Y, Xu T, Yu MC, Zhang XH. Comparison of cultivable bacterial communities associated with Pacific white shrimp (Litopenaeus vannamei) larvae at different health statuses and growth stages. Aquaculture. 2016;451:163–169. doi: 10.1016/j.aquaculture.2015.09.020. [DOI] [Google Scholar]
- 36.Martins P, Cleary DFR, Pires ACC, Rodrigues AM, Quintino V, Calado R, Gomes NCM. Molecular analysis of bacterial communities and detection of potential pathogens in a recirculating aquaculture system for Scophthalmus maximus and Solea senegalensis. PLoS One. 2013;8:e80847. doi: 10.1371/journal.pone.0080847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 38.Cottrell MT, Kirchman DL. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol. 2000;66:1692–1697. doi: 10.1128/AEM.66.4.1692-1697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornejo-Granados F, Gallardo-Becerra L, Leonardo-Reza M, Ochoa-Romo JP, Ochoa-Leyva A. A meta-analysis reveals the environmental and host factors shaping the structure and function of the shrimp microbiota. PeerJ. 2018;6:e5382. doi: 10.7717/peerj.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muhammad ZHMZ, Wilhelm WHE, Binh TT, Christopher MA, Han MG. Microbiome analysis of Pacific white shrimp gut and rearing water from Malaysia and Vietnam: implications for aquaculture research and management. PeerJ. 2018;6:e5826. doi: 10.7717/peerj.5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.