Abstract
Bacillus spp. have been widely described for their potentials to protect plants against pathogens. Here, we reported the whole genome sequence of Bacillus velezensis ZF2, which was isolated from the stem of a healthy cucumber plant. Strain ZF2 showed a broad spectrum of antagonistic activities against many plant bacterial and fungal pathogens, including the cucumber leaf spot fungus Corynespora cassiicola. The complete genome of B. velezensis ZF2 contained a 3,931,418-bp circular chromosome, with an average G + C content of 46.50%. Genome comparison revealed closest similarity between ZF2 and other B. velezensis strains. Genes homologous to 14 gene clusters for biosynthesis of secondary metabolites were identified in the ZF2 genome. Also identified were a number of genes involved in bacterial colonization, including the genes for motility, biofilm formation, flagella biosynthesis, and capsular biosynthesis. Numerous genes associated with plant–bacteria interactions, including cellulase or protease biosynthesis, and plant growth promotion were also identified in the ZF2 genome. Overall, our data will aid future studies of the biocontrol mechanisms of B. velezensis ZF2 and promote its application in vegetable disease control.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-2165-y) contains supplementary material, which is available to authorized users.
Keywords: Antagonistic activity, Bacillus velezensis, Biological control, Comparative genomic analysis, Secondary metabolites
Introduction
Bacillus velezensis is a gram-positive, rod-shaped, motile, spore-forming, and aerobic bacterium, and forms creamy white, rough colonies with slightly irregular edges on agar-solidified culture medium (Cristina et al. 2005). B. velezensis was first isolated and described in southern Spain in 2005 (Cristina et al. 2005), and later reported as a heterotypic synonym of B. amyloliquefaciens based on DNA–DNA relatedness values (Wang et al. 2008). B. velezensis was distinguished from B. amyloliquefaciens based on core genome sequences (Dunlap et al. 2015), and these two species were separated from the “original Bacillus member” Bacillus subtilis in a recent study (Fan et al. 2017a, b). To date, many studies have investigated B. velezensis for the potential for controlling plant diseases. For example, B. velezensis CC09, which was isolated from Cinnamomum camphora, has been reported as a biocontrol agent for controlling wheat powdery mildew disease (Cai et al. 2017). B. velezensis 2A-2B, isolated from the rhizosphere of Sporobolus airoides, has been shown to exhibit a strong inhibition toward root rot (Martínez-Raudales et al. 2017). Furthermore, B. velezensis LDO2 (a peanut endophyte), B. velezensis BAC03 (isolated from potato common scab suppressive soil), and B. velezensis RC 218 (isolated from wheat anthers) have been explored for biological control of diverse plant diseases (Chen et al. 2019; Meng et al.2016; Chulze 2016).
Cucumber Corynespora leaf spot disease caused by C. cassiicola is one of the most important foliar diseases of cucumber. C. cassiicola is a major plant fungal pathogen that causes significant economic losses for many plant species, including cucumber. (Déon et al. 2014). Currently, chemical control is the primary method to control Corynespora leaf spot disease; however, C. cassiicola has developed resistance to many commonly used fungicides, including benzimidazoles, dicarboximides and N-phenylcarbamates (Miyamoto et al. 2010). Recently, biological control has been explored as an alternative solution for the prevention and suppression of plant diseases. For example, the strain B. megaterium was reported to control Aspergillus flavus on peanut kernels (Kong et al. 2010); B. amyloliquefaciens L-1 could be used to control pear ring rot (Sun et al. 2017); B. subtilis BSCBE4, Pseudomonas chlororaphis PA23 and P. fluorescens ENPF1 were reported to inhibit the growth of C. cassiicola mycelia in vitro (Mathiyazhagan et al. 2004). Nevertheless, few studies have explored the biocontrol potential of B. velezensis for Corynespora leaf spot disease, and the biocontrol mechanisms of B. velezensis against the disease still need to be elucidated.
Many strains of Bacillus spp. have been used in biocontrol strategies as antagonists of plant pathogens and/or plant growth promoters. These claimed activities mostly rely on the production of various secondary metabolites, including lipopeptides, polypeptides, macrolactones, fatty acids, polyketides, and isocoumarins, most of which exhibit a wide range of antimicrobial activities (Omura et al. 2001). These structurally diverse compounds can also affect the microflora in the rhizosphere or trigger host defense responses (Velusamy and Gnanamanickam 2008). B. velezensis was reported to harbor a large number of gene clusters involved in the biosynthesis of secondary metabolites mentioned above. For instance, the gene clusters srf, bmy, and fen were shown to direct the synthesis of the cyclic lipopeptides surfactin, bacillomycin, and fengycin; while, the gene clusters mln, bae, and dfn have been linked with the synthesis of the polyketides macrolactin, bacillaene, and difficidin, respectively (Chen et al. 2008). Furthermore, most of these secondary metabolites have been reported to facilitate the colonization in rhizosphere soil of B. velezensis strains (Abdellaziz et al. 2018). For instance, B. velezensis FZB42T showed strong colonizing ability and antimicrobial activities, one of the major factors was owing to the production of many kinds of lipopeptides including surfactin, iturin, bacillomycin, fengycin, mycosubtilin, bacillaene, bacilysin (Bochow et al. 2001; Hua et al. 2007; Alvarez et al. 2015). B. velezensis QST713, an antagonist against green mold disease, contained 15 gene clusters for the synthesis of secondary metabolites, all of which showed antimicrobial or antibacterial activities (Pandin et al. 2018). B. velezensis M75, which was isolated from cotton waste, had a number of genes associated with the synthesis of various secondary metabolites with antimicrobial activities or suppressing plant pathogens (Hua et al. 2007; Sang et al. 2017). Two antifungal compounds isolated from B. velezensis G341, which had been identified as bacillomycin and fengycin, have also been observed to inhibit the mycelial growth of various phytopathogenic fungi (Lim et al. 2017). Importantly, colonization and biofilm formation are closely associated with biological control by Bacillus spp. (Fan et al. 2017a, b). Based on previous studies, a number of genes associated with biofilm formation and colonization have been shown to play an important role in biological control, including the flagellar motility-associated genes, motA, motB, and flgM (Domka et al. 2007) and the Bacillus-specific biofilm formation pathway genes kinB, spo0A, spo0F, degU, and degS (Grossman et al. 1992). Besides these activities, plant growth promotion by Bacillus spp. was reported using growth-promoting substances, including auxins, cytokinins and gibberellins (Santner et al. 2009). For example, B. velezensis FZB42T can produce indole-3-acetic acid (IAA) and cytokinin, both of which are associated with plant growth promotion (Idris et al. 2007).
In this study, B. velezensis strain ZF2 with antagonistic activities against a broad range of plant bacterial and fungal pathogens was isolated from the stem of healthy cucumber. The entire genome of ZF2 was sequenced, annotated and compared with representative genomes of other Bacillus species. Phylogenetic analysis was performed to determine the taxonomic position of ZF2 and its relationship with other representative Bacillus spp. Genome comparison revealed that various genes involved in the biosynthesis of secondary metabolites and functional genes associated with beneficial plant–bacteria interactions occurred in the ZF2 genome. Taken together, these results provided novel insights into the biocontrol mechanisms of B. velezensis and could benefit the practical application of strain ZF2 in biocontrol of plant diseases.
Materials and methods
Bacterial isolation, antagonism assays and biocontrol assays
Bacillus velezensis ZF2 was isolated from the stem of a healthy cucumber plant in a greenhouse at Beijing, China, in November 2017, as described by Caulier et al (2018). Briefly, the stem tissues were surface sterilized in 70% ethanol for 1 min, and washed with sterile distilled water for three times. 1 mL of the fragmentized sample was subjected to heat treatment (80 °C, 15 min), 1000-fold diluted, plated on LB plate, and incubated at 28 °C for 24 h. After selective isolation, Bacillus-like strains were retained for further investigations. The antagonistic activity of B. velezensis ZF2 against plant pathogenic bacteria and fungi was assessed through plate bioassays, and the control efficacy of B. velezensis ZF2 against Corynespora leaf spot disease was tested through bioassays experiment with potted cucumbers. Plate assays were performed to assess the inhibition of C. cassiicola colony growth by B. velezensis ZF2, which was cultured for different fermentation time (48 h, 96 h, or 120 h) and in seven different fermentation media (NB: 10 g peptone, 3 g beef powder, 5 g NaCl per liter, pH 7.0; LB: 10 g tryptone, 5 g yeast extract, 10 g NaCl per liter, pH 7.0; BPY: 5 g beef extract, 10 g peptone, 5 g yeast extract, 5 g NaCl, 10 g glucose per liter; CP: 500 g corn powder per liter; SP3: 30 g soybean powder, 10 g starch, 10 g glucose, 1 g KH2PO4, 1 g K2HPO4, 0.02 g FeSO4·7H2O, 1 g CaCO3 per liter; SP8: 30 g soybean powder, 2 g peptone, 0.2 g KH2PO4, 1 g CaCO3, 5 g glucose, 3 g yeast extract per liter; and SP + CP: 30 g corn powder, 30 g soybean powder, 0.3 g (NH4)2SO4, 0.3 g MgSO4, 1 g CaCO3 per liter) (Yu et al. 2007). In addition, plate assays to assess cellulase (Cel) and protease (Prt) activity were conducted according to previously reported methods with modifications (Murata et al. 1991; Chatterjee et al. 1995). Bacterial cells were grown in LB medium overnight at 28 °C and adjusted to an OD600 of 0.8. Samples were applied to the cells, and the plates were incubated for 24 h at 28 °C to evaluate Cel and Prt activity. The Cel plates were stained with a 0.1% (w/v) Congo red solution for 30 min and then washed with a 1 M NaCl solution for 15 min. Haloes in the Prt plates became visible without any further treatment. Each treatment was repeated three times, and all of the above experiments were repeated three times.
Culture conditions, and genomic DNA extraction
The strain ZF2 was cultivated in LB (Luria broth) or NB (Nutrient broth) media at 28 °C with shaking for 24 h. The strain morphologies were observed by scanning electron microscope (SEM) and transmission electron microscopy (TEM 1230 microscope, JEOL). Genomic DNA was extracted from cultured ZF2 cells (OD600 = 0.8) using a TIANamp Bacteria DNA kit (Tiangen Biotech (Beijing) Co., Ltd.).
Genome sequencing, assembly and annotation
The genome of B. velezensis ZF2 was sequenced by Allwegene Technologies Corporation, Beijing, China. Whole-genome sequencing was performed using a Pacific Biosciences (PacBio) RS II platform, and a 10-kb SMRT Bell was used for template library construction. The sequences were assembled de novo using SMRT Link v.5.1.0 programs (https://soap.genomics.org.cn/soapdenovo.html) (Table S1). The graphical views of genome alignments were generated using CGView (Tatusova et al. 2016). Identification and annotation of the functional genes was performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP, https://www.ncbi.nlm.nih.gov/genome/annotation_prok/) (Tatusova et al. 2016). Transfer RNA (tRNA) and ribosome RNA (rRNA) genes were identified using tRNAscan-SE version 2.0 and RNAmmer version 1.2, respectively (Lagesen et al. 2007; Lowe and Chan 2016). Small nuclear RNAs (snRNAs) were predicted by BLAST searching against the R fam database (https://rfam.xfam.org) (Stanke et al. 2008). The functions of the predicted proteins were assigned through comparisons against multiple databases, including NR (nonredundant) protein databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Li et al. 2002), the RAST (Rapid Annotation using Subsystem Technology) analysis platform (Aziz et al. 2008), Pfam (https://pfam.xfam.org/), SwissProt and the enhanced COG (clusters of orthologous groups of proteins) database (https://www.ncbi.nlm.nih.gov/COG/) (Tatusov et al. 2000). In addition, SignalP 4.0 (Bendtsen et al. 2004) and TMHMM 2.0 (Krogh et al. 2001) were used to predict putative signal peptides and transmembrane helices, respectively. PHAST was used for prophage prediction (Arndt et al. 2016), and clustered regularly interspaced short palindromic repeats (CRISPR) were identified using CRISPR finder (Ibtissem et al. 2008).
Phylogenetic analysis and genome comparisons
The taxonomic position of B. velezensis ZF2 was determined by multilocus gene sequence analysis (MLSA) based on four housekeeping genes (16S rRNA, rpoD, pgk, and gyrB). The gene sequences were aligned using MUSCLE and trimmed to remove ambiguously aligned regions. Subsequently, the phylogenetic tree was constructed using the maximum likelihood method in MEGA 6.0 (Tamura et al. 2013). Other available gene sequences of closely related species for phylogenetic tree construction were downloaded from the NCBI database (Table S2). According to the phylogenetic analysis, four closely related Bacillus species with released complete genomes, including B. velezensis LS69 (GenBank: CP015911.1), B. velezensis FZB42T (GenBank: CP000560.1), B. amyloliquefaciens DSM 7 T (GenBank: FN597644.1) (Rückert et al. 2011) and B. subtilis 168 T (GenBank: AL009126.3) (Nakamura et al. 1999) were selected for genome comparison. Average nucleotide identities (ANI) (Michael and Ramon 2009) and in silico DNA–DNA hybridization (DDH) (Meier-Kolthoff et al. 2013) were calculated using the OrthoANIu algorithm (https://www.ezbiocloud.net/tools/orthoaniu) and the Genome-to-Genome Distance Calculator (GGDC) (https://ggdc.dsmz.de/ggdc.php), respectively. Furthermore, complete genome comparisons were conducted with the progressive alignment option of the Mauve 2.3.1 comparison software using the ZF2 genome as the reference genome (Michael and Ramon 2009). Venn diagrams were generated using R package (Michael and Ramon 2009).
Analysis of gene clusters for biosynthesis of secondary metabolites
Gene clusters for biosynthesis of secondary metabolites were predicted using the antiSMASH 2.0 program. The analysis was performed via the authors’ Web servers using the default parameters (https://antismash.secondarymetabolites.org). Comparative analysis of gene clusters identified in B. velezensis ZF2, B. velezensis LS69, B. velezensis FZB42T, B. amyloliquefaciens DSM 7 T, and B. subtilis 168 T was performed based on the Kyoto Encyclopedia of Genes and Genomes database (KEGG, https://www.genome.jp/kegg/) and the GenBank database.
Genome mining for genes encoding plant beneficial traits
Functional genes involved in plant–bacterial interactions such as biofilm formation, flagellar biosynthesis, motility, capsular biosynthesis were searched by NCBI database. The analysis of sequence homology of different functional genes in B. velezensis ZF2, B. velezensis FZB42T, B. velezensis LS69, B. amyloliquefaciens DSM 7 T, and B. subtilis 168 T were conducted using the KEGG database at amino acid level.
Results and discussion
Biocontrol effect of B. velezensis ZF2 against the Corynespora cassiicola
B. velezensis ZF2 showed strong antagonistic activities against the plant pathogens Pectobacterium carotovorum subsp. brasiliense, Pseudomonas syringae pv. lachrymans, P. syringae pv. tomato, Ralstonia solanacearum, Xanthomonas campestris pv. campestris, Clavibacter michiganensis subsp. michiganensis, Corynespora cassiicola, Colletotrichum sp., Phytophthora capsici, Botrytis cinerea, Fusarium oxysporum, and Alternaria solani (Fig. S1). Importantly, strain ZF2 exhibited a strong inhibitory activity against C. cassiicola. Notable inhibition zones were observed surrounding the strain ZF2 in plate assays, with an inhibition ratio of up to 60.10% (Fig. S1 G). Interestingly, microscopic observations showed that the mycelia of C. cassiicola on the antagonistic plate gathered into clusters and were significantly enlarged; whereas, the mycelia in the control plate appeared a normal state of growth (Fig. S2 A and B). Extracellular enzyme assays showed that strain ZF2 produced protease (Prt) and cellulase (Cel) (Fig. S2 C and D), which might result in mycelial expansion of C. cassiicola and suppression of cucumber leaf spot disease. Moreover, strain ZF2 showed a significant effect in controlling Corynespora leaf spot disease on potted cucumbers (Fig. S3). The control efficiency was 90.81 ± 1.62, close to the efficacy of chlorothalonil (96.09 ± 1.72), a fungicide with strong sterilizing activity (Sigler and Turco 2002) (Table S3).
Furthermore, the inhibitory effect of B. velezensis ZF2 toward C. cassiicola was evaluated following fermentation for different times or in different mediums. The results showed that strain ZF2 displayed a maximal inhibitory effect after 48 h of fermentation with a 10% of seed culture addition (1, 5, 10%) (Fig. S4A). The inhibitory effect became enhanced with the increase of the ratio of seed culture addition. Moreover, the inhibitory effect appeared most obvious (63.98%) when strain ZF2 was incubated in corn meal medium, among seven different media, with a 10% seed culture addition (Fig. S4 B).
Organism information
B. velezensis ZF2 was determined to be a motile, gram-positive, endospore-forming, and aerobic bacterium belonging to the Bacillaceae family. The strain grew readily on LB plate at 28 °C and produced creamy white colonies with irregular margins after 24 h of incubation (Fig. S5 A), showing colony morphology and cultural characteristics similar to other Bacillus spp. (Das et al. 2018; Lim et al. 2017; Martínez-Raudales et al. 2017). Strain ZF2 displayed rod-shaped cells with a length of 3–5 μm and a diameter of 0.8–1.2 μm (Fig. S5 B, C). The strain could grow in 2.0–10.0% NaCl (w/v) and over a wide temperature range (15–37 °C), with an optimal pH of 7.0. Biolog assays showed that strain ZF2 could utilize diverse carbon sources, including α-d-glucose, d-mannose, d-mannitol, d-sorbitol, l-alanine, l-glutamic acid, myo-inositol and sodium butyrate (Table S4). Pathogenicity tests showed that strain ZF2 was nonpathogenic toward cucumber (Fig. S6). Minimum information about the genome sequence (MIGS) of B.velezensis ZF2 is summarized in Table S1 (Field et al. 2008).
General genomic features of Bacillus velezensis ZF2
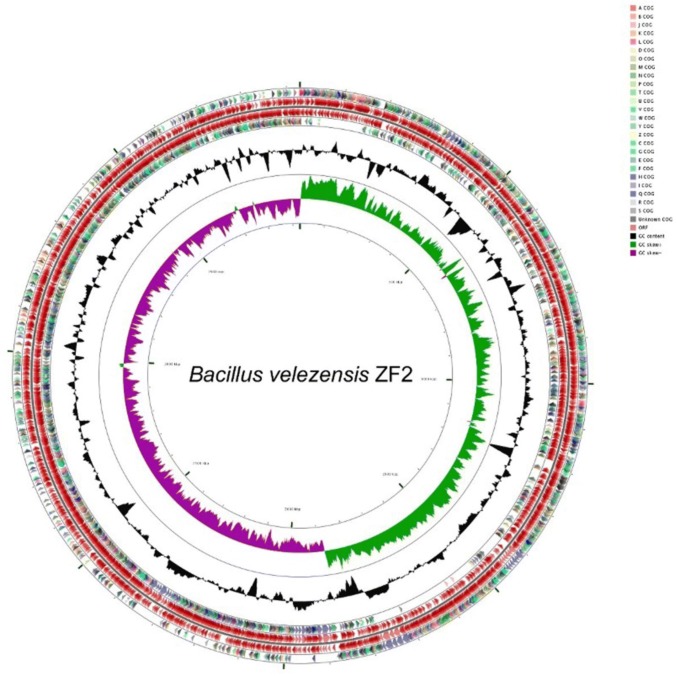
The complete genome of B. velezensis ZF2 comprised a circular 3,931,418-bp chromosome with an average G + C content of 46.50%, without plasmid. A graphical circular genomic map showing the genome structure and functions of strain ZF2 is presented in Fig. 1. A total of 4,058 open reading frames (ORFs) were predicted in the genome of ZF2. The ZF2 genome contained 3,808 protein-coding genes (CDS), 86 tRNA genes, 27 rRNA genes, 5 ncRNA genes and 123 pseudogenes (Table S5). Using the Pfam, SignalP, and TMHMM databases, 2,617 (64.49%), 150 (3.70%) and 1,054 (25.97%) of the ORFs could be classified into different groups, respectively. In addition, the ZF2 genome encoded 88 secreted proteins, 9 genomic islands, 2 prophage regions and 2 CRISPR loci (Table S5).
Fig. 1.
Graphical circular map of the B. velezensis ZF2 genome generated using the CGview server. From outside to center, rings 1 and 4 show protein-coding genes oriented in the forward (colored by COG categories) and reverse (colored by COG categories) directions, respectively. Rings 2 and 3 show genes on the forward and reverse strands. Ring 5 shows the G + C% content plot (black), and the inner most ring shows the GC skews, where green indicates positive values and purple indicates negative values
The functional categories of the 3,808 CDS in the ZF2 genome were further analyzed using the Cluster of Orthologous Groups of proteins (COG) database. The results showed that 3,335 CDS were assigned to different COG categories (Table S6), among which 7.10% were associated with transcription, 5.45% with ribosomal structure and biogenesis, 6.55% with carbohydrate transport and metabolism, and 7.47% with amino acid transport and metabolism. In addition, 723 genes were not assigned to COG categories, and their features and functions need to be determined.
Comparative genomics analysis of Bacillus velezensis ZF2 with other Bacillus strains
To understand the genetic relationships between B. velezensis ZF2 and other Bacillus strains, a phylogenetic tree was constructed based on four housekeeping genes (16S rRNA, rpoD, pgk, and gyrB) (Table S2). As expected, three primary monophyletic clades including B. velezensis, B. amyloliquefaciens, and B. subtilis, respectively, were corroborated by bootstrap values (Fig. 2) B. velezensis was closest to B. amyloliquefaciens, and both species were separated from B. subtilis. These results were consistent with the former notion that B. velezensis and B. amyloliquefaciens were classified as B. subtilis (Palazzini et al. 2016). In the study, strain ZF2 was classified as B. velezensis, and closest to JYYP2, followed by LS69 and SQR9, among 17 B. velezensis strains (Fig. 2).
Fig. 2.
Phylogenetic tree highlighting the relative position of B. velezensis ZF2 among other Bacillus species. The phylogenetic tree was constructed based on four housekeeping genes (16S rRNA, rpoD, pgk, and gyrB) according to the aligned gene sequences using the maximum likelihoods method in MEGA 6.0. Bootstrap values (1,000 replicates) are shown at the branch points. The scale bar indicates 0.01 nucleotide substitution per nucleotide position. GenBank accession numbers associated with the housekeeping loci for all strains are presented in Table S2
Genome comparison was conducted between B. velezensis ZF2 and other four available complete genomes of Bacillus strains (B. velezensis LS69, the strain closely related to ZF2; B. velezensis FZB42T, the type strain of B. velezensis; B. amyloliquefaciens DSM 7 T, the type strain of B. amyloliquefaciens; and B. subtilis 168 T, the type strain of B. subtilis) (Table 1). The results indicated that the genome size of strain ZF2 (3,929,773 bp) was larger than FZB42T and LS69 (3,918,589 and 3,917,761 bp, respectively) (Chen et al. 2009; Liu et al. 2017), but smaller than B. amyloliquefaciens DSM 7 T (3,980,199 bp) (Christian et al. 2011) and B. subtilis 168 T (4,215,606 bp) (Harwood and Wipat 1996) (Table 1). The average G + C content of ZF2 was similar to that of B. velezensis FZB42T (46.50%), B. velezensis LS69 (46.50%) and B. amyloliquefaciens DSM 7 T (46.10%), and was higher than that of B. subtilis 168 T (43.50%) (Table 1). Furthermore, all the five Bacillus genomes contained a single chromosome, without plasmid.
Table 1.
Genomic features of Bacillus velezensis ZF2 and other Bacillus spp
| Features | Bacillus velezensis ZF2 | Bacillus velezensis FZB42T | Bacillus velezensis LS69 | Bacillus amyloliquefaciens DSM 7 T | Bacillus subtilis 168 T |
|---|---|---|---|---|---|
| Size (bp) | 3,929,773 | 3,918,589 | 3,917,761 | 3,980,199 | 4,215,606 |
| G + C content (%) | 46.50 | 46.50 | 46.50 | 46.10 | 43.50 |
| Replicons | One chromosome | One chromosome | One chromosome | One chromosome | One chromosome |
| Total genes | 3927 | 3.892 | 3870 | 4120 | 4536 |
| Predicted no. of CDS | 3685 | 3687 | 3678 | 3870 | 4237 |
| Ribosomal RNA | 27 | 29 | 21 | 30 | 30 |
| Transfer RNA | 86 | 88 | 72 | 94 | 86 |
| Other RNA | 5 | 4 | 5 | 5 | 95 |
| Pseudogene | 124 | 84 | 94 | 121 | 88 |
| GenBank sequence | CP032154.1 | CP000560.1 | CP015911.1 | FN597644.1 | AL009126.3 |
ANI and DDH analyses are widely used to evaluate the similarities between bacterial strains based on whole-genome sequence, and the compared strains with ANI values ≥ 96% and DDH values ≥ 70% are typically regarded as being the same species (Fujikawa and Sawada 2016; Zhang et al. 2016). In this study, the ANI and DDH values between strains ZF2 and LS69 were 99.95 and 100.00%, respectively. Similarly, the ANI and DDH values between strains ZF2 and FZB42T were 98.30 and 95.70%, respectively. The ANI value between ZF2 and B. amyloliquefaciens DSM 7 T was 93.98%, although the DDH values was 82.50%. These results indicated that B. velezensis and B. amyloliquefaciens had a close relationship, but they were different species. Moreover, lower ANI and DDH values were obtained when the B. subtilis 168 T genome was used as reference. Altogether, these findings revealed that strains ZF2, LS69 and FZB42 were closely related to each other, and could be assigned to the same taxonomic position (Table 2).
Table 2.
Percentage of average nucleotide identities (ANI) and in silico DNA–DNA hybridization (DDH)among the selected Bacillus genomes
| Bacillus velezensis ZF2 | Bacillus velezensis LS69 | Bacillus velezensis FZB42T | Bacillus amyloliquefaciens DSM 7 T | Bacillus subtilis 168 T | |
|---|---|---|---|---|---|
| Bacillus velezensis ZF2 | 99.95 | 98.30 | 93.98 | 77.23 | |
| 100.00 | 95.70 | 82.50 | 33.70 | ||
| Bacillus velezensis LS69 | 99.95 | 98.37 | 94.07 | 77.14 | |
| 100.00 | 95.60 | 82.40 | 33.70 | ||
| Bacillus velezensis FZB42 | 98.30 | 98.37 | 94.20 | 77.12 | |
| 95.70 | 95.60 | 80.70 | 33.40 | ||
| Bacillus amyloliquefaciens DSM 7 | 93.98 | 94.07 | 94.20 | 77.10 | |
| 82.50 | 82.40 | 80.70 | 31.30 | ||
| Bacillus subtilis 168 | 77.23 | 77.14 | 77.12 | 77.10 | |
| 33.70 | 33.70 | 33.40 | 31.30 |
ANI values were computed for pairwise genome comparison with using the OrthoANIu algorithm. The percentage of ANI was shown on the top and bolded
In silico DNA–DNA hybridization was calculated using Genome-to-Genome Distance Calculator (GGDC). The percentage of DDH was shown on the bottom
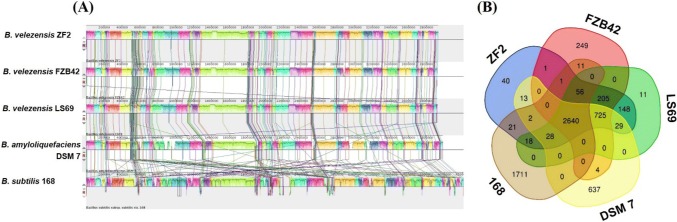
The evolutionary distance among these five Bacillus strains was further evaluated using their whole-genome sequences with the Mauve program. Compared to FZB42T and LS69, no gene insertions or deletions were detected in B. velezensis ZF2. The synteny plot of the pairwise alignments from the Mauve analysis strengthened the classification of the three strains as the same species. At the species level, the genome sequence of strain ZF2 was aligned to B. amyloliquefaciens DSM 7 T and B. subtilis 168 T. The results showed that large local collinear block (LCB) inversion occurred among the three species, especially between DSM 7 T and 168 T, and the ZF2 genome was much more similar to DSM 7 T than to 168 T, supporting the relationship described above (Fig. 3a).
Fig. 3.
Comparison of B. velezensis ZF2 genome sequences against other four Bacillus genome sequences. a Synteny analysis of the B. velezensis ZF2 and B. velezensis FZB42T, B. velezensis LS69, B. amyloliquefaciens DSM 7 T and B. subtilis 168 T genomes. Pairwise alignments of genomes were generated using Mauve. ZF2 genome as the reference genome. Boxes with same color indicate syntenic regions. Boxes below the horizontal strain line indicate inverted regions. Rearrangements are shown with colored lines. The scale is in nucleotides. b Venn diagram showing the number of shared and unique clusters of orthologous genes
To identify specific genes in B. velezensis ZF2, its genome sequence was compared to the complete genome sequences of the four Bacillus strains. As shown in Fig. 2b, 6,550 protein clusters and 2,640 core genes present in ZF2 were shared with strains FZB42T, LS69, DSM 7 T and 168 T. In addition, 3,849 orthologous genes were shared between ZF2 and LS69, while 3,628 orthologous genes were shared between ZF2 and FZB42T. Additionally, 3,437 orthologous genes were shared between ZF2 and DSM 7 T, and 2,766 orthologous genes were shared between ZF2 and 168 T. These findings indicated that strain ZF2 showed a higher level of similarity with strain LS69. Furthermore, 40 unique genes were identified in the genome of B. velezensis ZF2, and the functions of most of these genes need further confirmation.
Gene clusters involved in the synthesis of secondary metabolites
Secondary metabolites produced by Bacillus spp. exhibited broad-spectrum biological activities, including antimicrobial activity against various phytopathogens (Karlovsky et al. 2008; Mondol et al. 2013). For example, surfactin, a cyclic lipopeptide antibiotic and biosurfactant produced by Bacillus strains, showed antimicrobial and antiviral activity by altering membrane integrity (Vollenbroich et al. 1997) and could protect plants against infection by the pathogen Pseudomonas syringae (Peypoux et al. 1999). The surfactin-encoding locus comprised of four open reading frames (srfA-D). SrfA and B were three amino acid activating modules, while SrfC was a one-module enzyme and SrfD mediated the transfer of the β-hydroxy fatty acid substrate (Jaruchoktaweechai et al. 2000). Fengycin, a lipopeptide complex, was first discovered in B. subtilis strain F-29–3 and could effectively inhibit the growth of filamentous fungi (Vanittanakom et al. 1986). A five-gene cluster (fenA–E) has been shown to be responsible for the biosynthesis of fengycin. Bacilysin was reported as a dipeptide antibiotic containing a L-Ala residue at the N-terminus and a nonproteinogenic amino acid, L-anticapsin, at the C-terminus (Jaruchoktaweechai et al. 2000), and exhibited activity against a wide range of bacteria and the yeast Candida albicans due to an anticapsin moiety (Zimmerman et al. 1987). The bacABCDE operon was responsible for the biosynthesis of bacilysin (Chen et al. 2009).
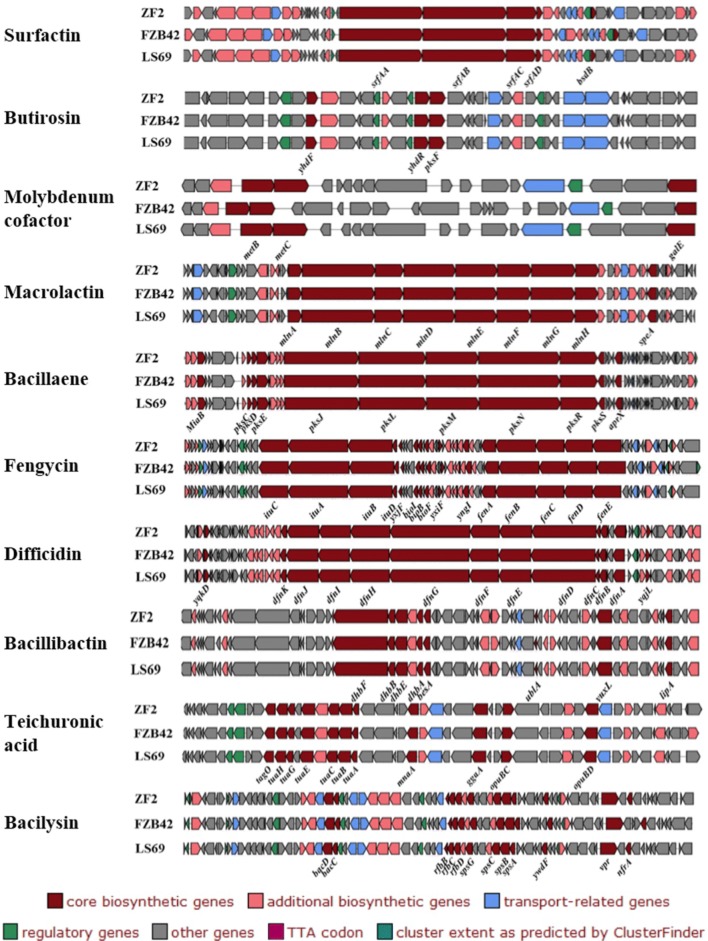
In this study, 14 gene clusters involved in the production of secondary metabolites were detected in the genome of strain ZF2, including two encoding NRPSs (nonribosomal peptide synthetases), two transATPKSs (trans-Acyl transferase polyketide synthetases), three transATPKS-NRPSs, and two bacteriocin-NRPSs, a saccharide, a lantipeptide, a putative NRPS, a fatty acid and an unknown type. In addition, gene clusters were identified that were specifically involved in the synthesis of surfactin, butirosin, macrolactin, bacillaene, fengycin, difficidin, bacillibactin, teichuronic acid, bacilysin, citrulline, molybdenum cofactor, iturin, amylolysin and amylocyclicin (Table 3). These compounds have been reported to have antimicrobial activities (Adnan et al. 2014; Dion et al. 1972b). Moreover, each of the 14 gene clusters was searched for homologues in the other four Bacillus strains (B. velezensis LS69, B. velezensis FZB42T, B. amyloliquefaciens DSM 7 T, and B. subtilis 168 T) through an antiSMASH genome analysis and the KEGG database. The results showed that the 14 gene clusters associated with the biosynthesis of secondary metabolites in B. velezensis ZF2 were also present in FZB42T and LS69 strains, and these strains possessed similar core biosynthetic genes with very high identity (approximately 100%) at the amino acid level (Table 3 and Fig. 4). These three strains had the primary gene clusters srfAA, srfAB, srfAC, srfAD and bsdB involved in the biosynthesis of surfactin. Similarly, the key gene clusters mlnABCDEFGH and speA, which play roles in the synthesis of macrolactin, were also detected in the FZB42T and LS69 genomes. The 68.2-kb gene clusters associated with the synthesis of bacillibactin were similar in the ZF2, FZB42T and LS69 genomes. The dfnABCDEFGHIJK gene cluster involved in the synthesis of difficidin within the ZF2 genome was collinear with the dfn gene cluster of strains FZB42T and LS69 (Chen et al. 2009; Liu et al. 2017). However, some secondary metabolites were not present in strain DSM 7 T or 168 T, including macrolactin, difficidin, molybdenum cofactor, butirosin, amylolysin and amylocyclicin. Indeed, the genes encoding molybdenum cofactor and butirosin were reported rarely and exhibited very lower similarity (11 and 7%, respectively) in different strains. Butirosin (with the core biosynthetic genes ydhFR and pksF) was reported as a new aminoglycoside antibiotic in Bacillus circulans with activity against gram-positive and gram-negative bacteria (Dion et al. 1972). Molybdenum cofactor (with the core biosynthetic genes metBC and galE) was observed to be associated with nitrogenase (Shah and Brill 1977) and has not been reported in B. velezensis. In summary, the genomic features of ZF2 suggested that it had the potential to be used as a biocontrol agent against plant pathogenic microbes.
Table 3.
Comparative analysis of secondary metabolites clusters of Bacillus velezensis ZF2 identified in genome with plant-associated strains and reference genomes
| Bacillus velezensis ZF2 | Presence ( +) or absence ( − ) of secondary metabolites clusters in Bacillus strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Metabolites | Synthetase | Core gene cluster | Size | Position | Bioactive spectrum | FZB42T | LS69 | DSM7T | 168 T |
| Surfactin | NRPS | srfAABCD, bsdAB | 66,231 bp | 321,977–388,208 | Virus, mycoplasma and fungi | + | + | + | + |
| Butirosin | OtherKS | yhdFR, pksAF, citR, rpoE, cueR | 42,049 bp | 923,781–965.830 | Bacteria, Cyanobacteria | + | + | + | − |
| Macrolactin | TransATPKS | mlnABCDEFGH, kinC, speA | 87,409 bp | 1,385,009–1,472,418 | Bacteria | + | + | − | − |
| Bacillaene | TransATPKS-NRPS | pksCDEJLMNRS, miaB, aprX | 102,336 bp | 1,698,684–1,801,020 | Bacteria | + | + | + | + |
| Fengycin | TransATPKS-NRPS | kdgR, yvrGH, ituABCD, yxiF, bioBFI, fenABCDE,yngI | 138,886 bp | 1,865,753–2,004,639 | Filamentous fungi | + | + | + | + |
| Difficidin | TransATPKS | dfnABCDEFGHIJK, yqkD, lysR, yqiL | 100,585 bp | 2,270,261–2,370,846 | Bacteria | + | + | − | − |
| Bacillibactin | Bacteriocin-NRPS | dhbABEF, besA, ublA, yuxL, lipA | 68,245 bp | 3,000,664–3,068,909 | Microbial competitors | + | + | + | + |
| Teichuronic acid | Saccharide | degSU, tagO, tuaABCEGH, mnaA, ggaA, opuBCD | 56,166 bp | 3,389,585–3,445,751 | Bacteria | + | + | + | + |
| Bacilysin | NRPS | bacABCDE, cysL, ycbL, rfbBCD, spsABCG, ywdF, vpr, nfrA | 73,132 bp | 3,589,054–3,662,186 | Bacteria and Candida albicans | + | + | + | + |
| Citruline | Fatty acid | fabHF | 24,684 bp | 1,080,144–1,104,828 | Bacteria | + | + | + | + |
| Molybdenum cofactor | Putative NRPS | metBC, galE | 17,703 bp | 1,144,555–1,162,258 | Bacteria | + | + | − | − |
| Iturin | TransATPKS-NRPS | xynD, ituABCD, yxjCDEF | 44,809 bp | 1,883,810–1,928,619 | Fungi | + | + | + | + |
| Amylolysin | Lantipeptide | amlFEKRAMT | 9,630 bp | 1,194,266–1,203,896 | Gram-positive bacteria | + | + | + | − |
| Amylocyclicin | Bacteriocin-NRPS | acnBACDEF | 4,171 bp | 3,044,117–3,048,288 | Gram-positive bacteria | + | + | + | − |
Fig. 4.
Comparisons of NRPS, transATPKS and saccharide clusters in ZF2 (above), FZB42T (middle), LS69 (below). Dark red indicates the core biosynthetic genes in different gene clusters among three Bacillus strains. The core biosynthetic genes are labeled in each gene cluster
Mining for functional genes potentially associated with plant–bacteria interactions
Bacillus strains have recently become widely used as biocontrol agents because of their strong ability to tolerate adverse environmental conditions, such as high temperature, pressure and salinity, and to promote beneficial plant–bacteria interactions. These properties were largely attributed to various functional genes which were linked with biocontrol, including those responsible for sporulation, biofilm formation, bacterial colonization, flagella biosynthesis, plant growth promotion, and plant defense induction (Rampelotto 2010). As expected, B. velezensis ZF2 genome contained large numbers of genes coding for these biological functions (Table S7). Specifically, diverse genes for sporulation occurred in the ZF2 genome. The homologous Spo0A (AXY70842.1) served as the master transcription regulator of sporulation in B. subtilis (Virginie et al. 2010). SinR (AXY70880.1) and its antagonist SinI (AXY70879.1) were identified as pleiotropic DNA binding proteins and are essential for sporulation and subtilisin synthesis (Mandic-Mulec et al. 1995). The YqxM-SipW-TasA (AXY70883.1) operon encodes the TasA protein that binds cells together in biofilms (Diego et al. 2010) and is associated with sporulation (Stöver and Driks 1999). In addition, many regulatory genes associated with biofilm formation were detected in the ZF2 genome, such as sigEFGH (encoding RNA polymerase sporulation sigma factor), pgsABC (involved in poly-γ-glutamate synthesis), and escABC (encoding a transporter permease).
To be effective biocontrol agents, B. velezensis strains are required for motility and colonization in plant tissues (Ben et al. 2011; Wu et al. 2015). Interestingly, our results showed that ZF2 possessed swrABC genes, encoding swarming motility proteins, an exopolysaccharide operon (epsA-O) associated with capsular biosynthesis (Hua et al. 2007), and a number of fli and flg genes involved in flagella biosynthesis. All these functions have been suggested to enhance swarming motility and colonization (Emilia et al. 2012; Mordini et al. 2013). Moreover, these genes were highly similar in ZF2 and FZB42T, with an amino acid identity of 97–100% (Table S7). However, many functional genes of ZF2 were not found in B. subtilis 168 T, or with a lower similarity, and the reason may be that the two strains belonged to different species.
Furthermore, a variety of genes encoding proteins associated with plant growth promotion were detected in the ZF2 genome, including putative indole-3-acet-aldehyde (IAA) dehydrogenase (dhaS), IAA acetyl-transferase (ysnE), auxin efflux carrier (ywkB), phytase and nitrilase (yhcX). These genes are responsible for the production of IAA, which has been suggested to contribute to the plant growth-promoting abilities of Bacillus strains (Shao et al. 2015). Strain ZF2 also had various genes encoding proteins involved in the synthesis of 3-hydroxy-2-butanone, including acetolactate decarboxylase (alsD), acetolactate synthase (alsS), a transcriptional regulator (alsR) and 2,3-butanediol dehydrogenase (bdhA) (He et al. 2013), and showed high similarity in ZF2 and FZB42T (95–100% amino acid identity). This compound was reported to improve plant growth and trigger systemic resistance (Renna et al. 1993; Nicholson 2008). In addition, ZF2 also harbored a number of genes associated with biosynthesis of cellulase and protease, including those encoding serine protease (spr, ispA, aprX, yyjD, and yyxA), glucanase (bglC and bglS), galactokinase (galK, galE, and galT), phosphotransferase (lacE and lacF), and numerous intramembrane proteases (ydcA, ydiL, prsW, ypbD, and yyaK). Moreover, these genes displayed a high level of similarity in ZF2 and FZB42T (96 to 100% amino acid identity) (Table S7). These enzymes were suggested to be associated with carbon source and cellulose utilization by Bacillus spp. in plants (Dardanelli et al. 2000). Altogether, these findings demonstrated that B. velezensis ZF2 was adapted to diverse environments and of the potential to promote plant growth.
Conclusions
B. velezensis ZF2, which was isolated from the stem of a healthy cucumber plant, had a broad range of antagonistic activities against 14 plant bacterial and fungal pathogens, and could be used for biological control of cucumber Corynespora leaf spot disease. Whole genome sequencing and comparative genomic analysis confirmed the taxonomic classification for strain ZF2 as a member of B. velezensis. B. velezensis ZF2 harbored 14 gene clusters involved in the production of secondary metabolites that have been shown to possess antimicrobial activities. Furthermore, a number of genes involved in bacterial colonization and plant growth promotion were also present in the ZF2 genome, and all these genes were highly homologous to B. velezensis FZB42T. All these features indicated that strain ZF2 could be a promising biocontrol agent for plant disease control and advance a better understanding of biocontrol mechanisms of B. velezensis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was performed with the support of the National Key Research & Development (R&D) plan (2016YFD0201000); the Science and Technology Innovation Program, Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS); the Key Laboratory of Horticultural Crops Genetic Improvement, Ministry of Agriculture in China (IVF2019ZF01); and the Modern Agro-industry Technology Research System (CARS-25).
Author contributions
SX, XWX, LL, and BJL conceived and designed the experiments. SX, XWX, YRZ, and LL performed the experiments and analyzed the data. SX, LL, and BJL wrote the manuscript. YXS and ALC revised manuscript. All authors have read and approved the final version of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest in the publication.
Nucleotide sequence accession number
The complete genome sequence of Bacillus velezensis ZF2 has been deposited in NCBI under the GenBank accession number CP032154.1. The strain has also been deposited in the China General Microbiological Culture Collection Center (CGMCC) for Type Culture Collection under the accession number 16013.The complete genome sequence of Bacillus velezensis ZF2 has been deposited in NCBI under the GenBank accession number CP032154.1. The strain has also been deposited in the China General Microbiological Culture Collection Center (CGMCC) for Type Culture Collection under the accession number 16013.
Footnotes
Shuai Xu and Xuewen Xie are the co-first author.
Contributor Information
Shuai Xu, Email: xsh1122a@163.com.
Xuewen Xie, Email: xiexuewen@caas.cn.
Yurong Zhao, Email: Z15046656769@163.com.
Yanxia Shi, Email: Shiyanxia813@163.com.
Ali Chai, Email: chaiali@163.com.
Lei Li, Email: caulilei@163.com.
Baoju Li, Email: libaojuivf@163.com.
References
- Abdellaziz L, Chollet M, Abderrahmani A, Béchet M, Yaici L, Chataigné G, Arias AA, Leclère V, Jacques P. Lipopeptide biodiversity in antifungal Bacillus strains isolated from Algeria. Arch Microbiol. 2018;6:1–12. doi: 10.1007/s00203-018-1537-8. [DOI] [PubMed] [Google Scholar]
- Adnan N, Shahid M, Shashidar A, Sarosh B, Johan M, Erik BR. Genome Analysis of Bacillus amyloliquefaciens Subsp plantarum UCMB5113: A Rhizobacterium That Improves Plant Growth and Stress Management. PLoS ONE. 2014;9:e104651. doi: 10.1371/journal.pone.0104651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez F, Castro M, Príncipe A, Borioli G, Fischer S, Mori G, Jofré E. The plant-associated Bacillus amyloliquefaciens strains MEP2 18 and ARP2 3 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J Appl Microbiol. 2015;112:159–174. doi: 10.1111/j.1365-2672.2011.05182.x. [DOI] [PubMed] [Google Scholar]
- Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben F, Hua CX, Anto B, Wilfrid B, Joachim V, Rainer BJ. Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. J Biotechnol. 2011;151:303–311. doi: 10.1016/j.jbiotec.2010.12.022. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak SJ. Improved prediction of signal peptides: SignalP 30. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bochow H, El-Sayed SF, Junge H, Stavropoulou A, Schmiedeknecht G. Use of Bacillus subtilis as biocontrol agent IV Salt-stress tolerance induction by Bacillus subtilis FZB24 seed treatment in tropical vegetable field crops, and its mode of action / Die Verwendung von Bacillus subtilis zur biologischen Bekämpfung IV I. J Plant Dis Protect. 2001;108:21–30. doi: 10.1614/0890-037X(2001)015[0190:EOIAPO]2.0.CO;2. [DOI] [Google Scholar]
- Cai XC, Liu CH, Wang BT, Xue YR. Genomic and metabolic traits endow Bacillus velezensis CC09 with a potential biocontrol agent in control of wheat powdery mildew disease. Microbiol Res. 2017;196:89–94. doi: 10.1016/j.micres.2016.12.007. [DOI] [PubMed] [Google Scholar]
- Caulier S, Gillis A, Colau G, Licciardi F, Liépin M, Desoignies N, Modrie P, Legrève A, Mahillon J, Bragard C. Versatile antagonistic activities of soil-borne Bacillus spp. and Pseudomonas spp. against Phytophthora infestans and other potato pathogens. Front Microbiol. 2018;9:143. doi: 10.3389/fmicb.2018.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Cui Y, Liu Y, Dumenyo CK, Chatterjee AK. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-L-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1002/bit.260460313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R. Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol. 2008;140:27–37. doi: 10.1016/j.jbiotec.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Chen XH, Scholz R, Borriss M, Junge H, Mögel G, Kunz S, Borriss R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J Biotechnol. 2009;140:38–44. doi: 10.1016/j.jbiotec.2008.10.015. [DOI] [PubMed] [Google Scholar]
- Chen L, Shi H, Heng J, Wang D, Bian K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol Res. 2019;218:41–48. doi: 10.1016/j.micres.2018.10.002. [DOI] [PubMed] [Google Scholar]
- Christian R, Jochen B, Xiaohua C, Oleg R, Rainer B. Genome sequence of B amyloliquefaciens type strain DSM7(T) reveals differences to plant-associated B amyloliquefaciens FZB42. J Biotechnol. 2011;155:78–85. doi: 10.1016/j.jbiotec.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Chulze S. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol Res. 2016;192:30–36. doi: 10.1016/j.micres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Cristina RG, Victoria B, Fernando MC, Inmaculada L, Emilia Q. Bacillus velezensis sp nov, a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int J Syst Evol Microbiol. 2005;55:191–195. doi: 10.1099/ijs.0.63310-0. [DOI] [PubMed] [Google Scholar]
- Dardanelli M, Gonzalez P, Ma GN. Synthesis, accumulation and hydrolysis of trehalose during growth of peanut rhizobia in hyperosmotic media. J Basic Microbiol. 2000;40:149. doi: 10.1002/1521-4028(200007)40:3<149::AID-JOBM149>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Das R, Li G, Mai B, An T. Spore cells from BPA degrading bacteria Bacillus sp. GZB displaying high laccase activity and stability for BPA degradation. Sci Total Environ. 2018;640–641:798–806. doi: 10.1016/j.scitotenv.2018.05.379. [DOI] [PubMed] [Google Scholar]
- Diego R, Claudio A, Richard L, Roberto K. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion HW, Woo PWK, Willmer NE, Kern DL, Fusari SA. Butirosin, a New Aminoglycosidic Antibiotic Complex: Isolation and Characterization. Antimicrob Agents Chemother. 1972;2:84–88. doi: 10.1128/aac.2.2.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domka J, Lee J, Bansal T, Wood TK. Temporal gene-expression in Escherichia coli K-12 biofilms. Environ Microbiol. 2007;9:332–346. doi: 10.1111/j.1462-2920.2006.01143.x. [DOI] [PubMed] [Google Scholar]
- Dunlap CA, Kim SJ, Kwon SW, Rooney AP. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp plantarum and 'Bacillus oryzicola' are later heterotypic synonyms of Bacillus velezensis based on phylogenomic. Int J Syst Evol Microbiol. 2015;66:1212–1217. doi: 10.1099/ijsem.0.000858. [DOI] [PubMed] [Google Scholar]
- Déon M, Fumanal B, Gimenez S, Bieysse D, Oliveira RR, Shuib SS, Breton F, Elumalai S, Vida JB, Seguin M. Diversity of the cassiicolin gene in Corynespora cassiicola and relation with the pathogenicity in Hevea brasiliensis. Fungal Biol. 2014;118:32–47. doi: 10.1016/j.funbio.2013.10.011. [DOI] [PubMed] [Google Scholar]
- Emilia G, Sara S, Mara C, Gueye SA, Francesco C, Sonia S. Contribution of surfactin and SwrA to flagellin expression, swimming, and surface motility in Bacillus subtilis. Appl Environ Microbiol. 2012;78:6540–6544. doi: 10.1128/AEM.01341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Blom J, Klenk HP, Borriss R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis Form an "Operational Group B amyloliquefaciens" within the B subtilis Species Complex. Front Microbiol. 2017;8:22. doi: 10.3389/fmicb.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H, Zhanwei Z, Yan L, Xun Z, Yongming D, Microbiology WQ. c Antibacterial Activity and Colonization. Front Microbiol. 2017;8:1973. doi: 10.3389/fmicb.2017.01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikawa T, Sawada H. Genome analysis of the kiwifruit canker pathogen Pseudomonas syringae pv actinidiae biovar 5. Sci Rep. 2016;6:21399. doi: 10.1038/srep21399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman AD, Lewis T, Levin N, Devivo R. Suppressors of a spo0A missense mutation and their effects on sporulation in Bacillus subtilis. Biochimie. 1992;74:679–688. doi: 10.1016/0300-9084(92)90140-A. [DOI] [PubMed] [Google Scholar]
- Harwood Wipat CRA. Sequencing and functional analysis of the genome of Bacillus subtilis strain 168. Febs Lett. 1996;389:84–87. doi: 10.1016/0014-5793(96)00524-8. [DOI] [PubMed] [Google Scholar]
- He P, Hao K, Blom J, Rückert C, Vater J, Mao Z, Wu Y, Hou M, He P, He Y. Genome sequence of the plant growth promoting strain Bacillus amyloliquefaciens subsp. plantarum B9601–Y2 and expression of mersacidin and other secondary metabolites. J Biotechnol. 2013;164:281–291. doi: 10.1016/j.jbiotec.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Hua CX, Alexandra K, Romy S, Andreas E, Kathrin S, Isabelle H, Burkhard M, Hess WR, Oleg R, et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat Biotechnol. 2007;25:1007–1014. doi: 10.1038/nbt1325. [DOI] [PubMed] [Google Scholar]
- Ibtissem G, Gilles V, Christine P. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;36:145–148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris EE, Iglesias DJ, Talon M, Borriss R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe Interact. 2007;20:619–626. doi: 10.1094/MPMI-20-6-0619. [DOI] [PubMed] [Google Scholar]
- Jaruchoktaweechai C, Suwanborirux K, Tanasupawatt S, Kittakoop P, Menasveta P. New macrolactins from a marine Bacillus sp. Sc026. J Nat Prod. 2000;63:984–986. doi: 10.1021/np990605c. [DOI] [PubMed] [Google Scholar]
- Karlovsky P, Thomashow LS, Bonsall RF, Weller DM, Reuben S, Bhinu VS, Swarup S, Ferluga S, Steindler L, Venturi V (2008) Secondary metabolites in soil ecology. Heidelberg, Berlin. 10.1111/j.1439-0434.2009.01592.x
- Kong Q, Shan S, Liu Q, Wang X, Yu F. Biocontrol of Aspergillus flavus on peanut kernels by use of a strain of marine Bacillus megaterium. Int J Food Microbiol. 2010;139:31–35. doi: 10.1016/j.ijfoodmicro.2010.01.036. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland E, Staerfeldt H, Rognes T, Ussery D. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Jaroszewski L, Godzik A. Tolerating some redundancy significantly speeds up clustering of large protein databases. Bioinformatics. 2002;18:77–82. doi: 10.1093/bioinformatics/18.1.77. [DOI] [PubMed] [Google Scholar]
- Lim SM, Yoon MY, Choi GJ, Choi YH, Jang KS, Shin TS, Park HW, Yu NH, Kim YH, Kim JC. Diffusible and Volatile Antifungal Compounds Produced by an Antagonistic Bacillus velezensis G341 against Various Phytopathogenic Fungi. Plant Pathol. 2017;33:488–498. doi: 10.5423/PPJ.OA.04.2017.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Kong Y, Fan Y, Geng C, Peng D, Sun M. Whole-genome sequencing of Bacillus velezensis LS69, a strain with a broad inhibitory spectrum against pathogenic bacteria. J Biotechnol. 2017;249:20–24. doi: 10.1016/j.jbiotec.2017.03.018. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1039/nar/gwk413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandic-Mulec I, Doukhan L, Smith I. The Bacillus subtilis SinR protein is a repressor of the key sporulation gene spo0A. J Bacteriol. 1995;177:4619–4627. doi: 10.1128/jb.177.16.4619-4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Raudales I, De La Cruz-Rodríguez Y, Alvarado-Gutiérrez A, Vega-Arreguín J, Fraire-Mayorga A, Alvarado-Rodríguez M, Balderas-Hernández V, Fraire-Velázquez S. Draft genome sequence of Bacillus velezensis 2A–2B strain: a rhizospheric inhabitant of Sporobolus airoides (Torr) Torr, with antifungal activity against root rot causing phytopathogens. Stand Genomic Sci. 2017;12:73. doi: 10.1186/s40793-017-0289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiyazhagan S, Kavitha K, Nakkeeran S, Chandrasekar G, Manian K, Renukadevi P, Krishnamoorthy AS, Fernando AGD. PGPR mediated management of stem blight of Phyllanthus amarus (Schum and Thonn) caused by Corynespora cassiicola (Berk and Curt) Wei. Arch Phytopathol Plant Protect. 2004;37:183–199. doi: 10.1080/03235400410001730658. [DOI] [Google Scholar]
- Meier-Kolthoff JP, Klenk H-P, Goeker M, Auch AF. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Q, Jiang H, Hao J. Effects of Bacillus velezensis strain BAC03 in promoting plant growth. Biol Control. 2016;98:18–26. doi: 10.1016/j.biocontrol.2016.03.010. [DOI] [Google Scholar]
- Michael R, Ramon RM. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Ishii H, Seko T, Kobori S, Tomita Y. Occurrence of Corynespora cassiicola isolates resistant to boscalid on cucumber in Ibaraki Prefecture, Japan. Plant Pathol. 2010;58:1144–1151. doi: 10.1111/j.1365-3059.2009.02151.x. [DOI] [Google Scholar]
- Mondol M, Shin HJ, Islam MT. Diversity of Secondary Metabolites from Marine Bacillus Species: Chemistry and Biological Activity. Mar Drugs. 2013;11:2846–2872. doi: 10.3390/md11082846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordini S, Osera C, Marini S, Scavone F, Bellazzi R, Galizzi A, Calvio C. The role of SwrA, DegU and P(D3) in fla/che expression in B subtilis. PLoS ONE. 2013;8:e85065. doi: 10.1371/journal.pone.0085065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, McEvoy JL, Chatterjee A, Collmer A, Chatterjee AK. Molecular cloning of an aepA gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp carotovora. Mol Plant Microbe Interact. 1991;4:239–246. doi: 10.1094/MPMI-4-239. [DOI] [Google Scholar]
- Nakamura L, Roberts MS, Cohan FM. Note: Relationship of Bacillus subtilis clades associated with strains 168 and W23: A proposal for Bacillus subtilis subsp subtilis subsp nov and Bacillus subtilis subsp spizizenii subsp nov. Int J Syst Evol Microbiol. 1999;49:1211–1215. doi: 10.1099/00207713-49-3-1211. [DOI] [PubMed] [Google Scholar]
- Nicholson WL. The Bacillus subtilis ydjL (bdhA) gene encodes acetoin reductase/2,3-butanediol dehydrogenase. Appl Environ Microbiol. 2008;74:6832–6838. doi: 10.1128/AEM.00881-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA. 2001;98:12215–12220. doi: 10.1073/pnas.211433198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzini JM, Dunlap CA, Bowman MJ, Chulze SN. Bacillus velezensis RC 218 as a biocontrol agent to reduce Fusarium head blight and deoxynivalenol accumulation: Genome sequencing and secondary metabolite cluster profiles. Microbiol Res. 2016;192:30–36. doi: 10.1016/j.micres.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Pandin C, Coq DL, Deschamps J, Védie R, Rousseau T, Aymerich S, Briandet R. Complete genome sequence of Bacillus velezensis QST713: a biocontrol agent that protects Agaricus bisporus crops against the green mould disease. J Biotechnol. 2018;278:10–19. doi: 10.1016/j.jbiotec.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Peypoux F, Bonmatin JM, Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- Rampelotto PH. Resistance of microorganisms to extreme environmental conditions and its contribution to astrobiology. Sustain Basel. 2010;2:1602–1623. doi: 10.3390/su2061602. [DOI] [Google Scholar]
- Renna MC, Najimudin N, Winik LR, Zahler SA. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rückert C, Blom J, Chen X, Reva O, Borriss R. Genome sequence of B amyloliquefaciens type strain DSM7T reveals differences to plant-associated B amyloliquefaciens FZB42. J Biotechnol. 2011;155:78–85. doi: 10.1016/j.jbiotec.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Sang YK, Sang YL, Weon HY, Sang MK, Song J. Complete genome sequence of Bacillus velezensis M75, a biocontrol agent against fungal plant pathogens, isolated from cotton waste. J Biotechnol. 2017;241:112–115. doi: 10.1016/j.jbiotec.2016.11.023. [DOI] [PubMed] [Google Scholar]
- Santner A, Calderon-Villalobos L, Estelle M. Plant hormones are versatile chemical regulators of plant growth. Nat Chem Biol. 2009;5:301–307. doi: 10.1038/nchembio.165. [DOI] [PubMed] [Google Scholar]
- Shah VK, Brill WJ. Isolation of an iron-molybdenum cofactor from nitrogenase. P Natl Acad Sci USA. 1977;74:3249–3253. doi: 10.2307/66916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Li S, Zhang N, Cui X, Zhou X, Zhang G, Shen Q, Zhang R. Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9. Microb Cell Fact. 2015;14:130. doi: 10.1186/s12934-015-0323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler WV, Turco RF. The impact of chlorothalonil application on soil bacterial and fungal populations as assessed by denaturing gradient gel electrophoresis. Appl Soil Ecol. 2002;21:107–118. doi: 10.1016/S0929-1393(02)00088-4. [DOI] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- Stöver AG, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/JB.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Cui J, Jia X, Wang W. Isolation and Characterization of Bacillus Amyloliquefaciens L-1 for Biocontrol of Pear Ring Rot. Hortic Plant J. 2017;3:10–16. doi: 10.1016/j.hpj.2017.10.004. [DOI] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, Dicuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gwk569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanittanakom N, Loeffler W, Koch U, Jung G. Fengycin–a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J Antibiot. 1986;39:888. doi: 10.7164/antibiotics.39.888. [DOI] [PubMed] [Google Scholar]
- Velusamy P, Gnanamanickam SS. The effect of bacterial secondary metabolites on bacterial and fungal pathogens of rice. Second Metab Soil Ecol. 2008 doi: 10.1007/978-3-540-74543-3_5. [DOI] [Google Scholar]
- Virginie M, Masaya F, Jensen ST, Patrick E, González-Pastor JE, Liu JS, Richard L. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2010;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Vollenbroich D, Pauli G, Ozel M, Vater J. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis. Appl Environ Microbiol. 1997;63:44–49. doi: 10.1007/s11136-005-4325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LT, Lee FL, Tai CJ, Kuo HP. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int J Syst Evol Microbiol. 2008;58:671–675. doi: 10.1099/ijs.0.65191-0. [DOI] [PubMed] [Google Scholar]
- Wu L, Wu HJ, Qiao J, Gao X, Borriss R. Novel Routes for Improving Biocontrol Activity of Bacillus Based Bioinoculants. Front Microbiol. 2015;6:1395. doi: 10.3389/fmicb.2015.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu SBC, Wang L, Hou Y, Mo X, Fu Z, Xia L. Study on Fermentation Medium of Bacillus thuringiensis. Life Sci Res. 2007 doi: 10.16605/j.cnki.1007-7847.2007.04.007. [DOI] [Google Scholar]
- Zhang Y, Fan Q, Loria R. A re-evaluation of the taxonomy of phytopathogenic genera Dickeya and Pectobacterium using whole-genome sequencing data. Sys Appl Microbiol. 2016;39:252–259. doi: 10.1016/j.syapm.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Zimmerman SB, Schwartz CD, Monaghan RL, Pelak BA, Weissberger B, Gilfillan EC, Mochales S, Hernandez S, Currie SA, Tejera E. Difficidin and oxydifficidin: novel broad spectrum antibacterial antibiotics produced by Bacillus subtilis I Production, taxonomy and antibacterial activity. J Antibiol. 1987;40:1692–1697. doi: 10.7164/antibiotics.40.1677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.