Abstract
A novel cellobiohydrolase (CBH)-generating fungi have been isolated and categorized as Schizophyllum commune KMJ820 based on morphology and rDNA gene sequence. Cellulose powder was used as carbon source, the total enzyme activity was 11.51 U/ml is noted; which is among the highest amounts of CBH-generating microbes studied. CBH have been purified to homogenize, with pursual of serial chromatography using S. commune supernatants and two different CBHs were found; CBH 1 and 2. The filtered CBHs showed greater activity (Vmax = 51.4 and 20.8 U/mg) in contrast to CBHs from earlier studies. The MW (molecular weights) of S. commune CBH 1 and 2 were verified to be approximately 50 kDa and 150 kDa, respectively, by size exclusion chromatography. Even though CBHs have been evaluated from other sources, but S. commune CBH is prominent in comparison to other CBHs by its high enzyme activity.
Electronic supplementary material
The online version of this article (10.1007/s12088-019-00843-9) contains supplementary material, which is available to authorized users.
Keywords: Schizophyllum commune, Cellobiohydrolase, Catalytic efficiency, Enzyme purification, Enzyme production
Introduction
Cellulose are known to be one of the highly abundant polymer around the world and they are extremely hard to degrade [1]. There are number of enzymes assisting in cellulose degradation, these are referred as cellulases. The cellulase enzyme are widely used in numerous industrial applications such as food treating, paper and pulp and also, they were integrated with other processes for conversion of reducing sugars to alcohols [2]. Cellulolytic enzyme is also known to be one of the primary components in microbial film formations noted in aquatic systems, fouling of pipe and in dental plaques. In spite of remarkable usage in cellulase based products; but because of their poor structural stability, these are often led to discarding as waste [3, 4]. The conversion of cellulose into reducing sugars such as glucose, and cellobiose are one of the major limitation in cellulose biofuel generating processes [5, 6]. In general, the industrial cellulose conversion practices consist of thermal, mechanical and acidic treatment. But however, the use of enzyme for conversion of cellulose has gained attention due to their eco-friendly nature, high net energy, low thermal necessity, and low processing conditions. But however, the generation of enzyme can be cost effective and time-consuming [7, 8]. Therefore, a better understanding of cellulase performance can guide towards the decrement in enzyme production cost and thereby boosting the industrial application.
The conversion of cellulose to reducing sugars and related components, can occur naturally. Filamentous fungi are one of the major source of cellulases and hemicellulases. Cellobiohydrolases (CBHs) are important components in the multienzyme cellulase complexes [9] and they disrupt the crystalline nature of cellulose and release cellobiose by pursing exo-type attack [1, 10–16]. On the basis of resemblance in amino acid sequence, the CBHs are divided into three major glycoside hydrolase groups (GH6, GH7, and GH48). Among these groups, the GH7 is known to be completely generated by fungal strains and it contains cellobiohydrolases and endoglucanases [17]. In general fungi produces two different types of CBHs (CBHI and CBHII), which are differentiated on the basis of their sequence similarity and active sites. The CBHs can solely accomplish the entire solubilization of cellulose crystals, without requirement of additional enzymes [9, 18, 19].
Therefore, in this study an effective CBH-generating fungi has been isolated and categorized as S. commune KMJ820 (KACC 93084P). Also, we have tested the influence of substrate to boost CBH generation by S. commune. In further we have characterized the CBH on the basis of its physiological and kinetic factors.
Materials and Methods
Isolation of CBH Generating Fungi
For isolation of CBH generating microbes, the soil samples are gathered from Sorak Mountain (Republic of Korea) as pointed in our earlier studies. The collected samples are cleansed/washed in sterile liquids (0.9% saline). In further the aliquots were dispersed on agar plate containing potato dextrose and was grown for 3 days. Preliminary examination for CBH yielding fungi were conducted out in agar petri dishes consisting of 10 mM of 4-methylumbelliferyl-β-d-glucuronide (MUG) as described previously [20]. On basis of fluorescence noted under UV, a total of 20 strains were transferred into a growth medium (3 ml) encompassing peptone (8 g/l), yeast extract (2 g/l) and cellulose (20 g/l) and grown at 28 °C with mixing at 200 rpm for 5 days. Later, the CBH performance of the fungi was examined on basis of enzyme activity as described previously [21, 22]. After preliminary investigations, the fungi with the best CBH performance was carefully chosen.
Growth of S. commune
For growing, the fungi S. commune KMJ820 was inoculated into 100 ml of potato dextrose solution in a conical flask. Preliminary culture of 5 ml was injected into 200 ml of carbon containing solution during fermenter studies. Influence of carbon on CBH generation were examined following 6 days of development in flasks having a medium comprised of 20 g/l of carbon source. For fermenter microbial growth, the S. commune were inoculated into 100 ml of potato dextrose browth. Pre cultures of 50 ml were injected into 3-l solution in a 7-l fermenter. This culture mixture included peptone 8 g/l, yeast extract 2 g/l, and cellulose powder 20 g/l. Additional operating conditions were similar as described in fermenter culture.
Enzyme Assay
CBH activities were analyzed using pNPC (p-nitrophenyl-d-cellobiopyranoside; substrate). Enzyme reaction mixture (1 ml) consisted 100 µl of enzyme and 0.1 mM substrate (pNPC) in 100 mM of sodium acetate buffer buffer (pH 5.0) with an incubation time of 15 min at 50 °C with later addition of 2 M Na2CO3 to the reaction blend. The quantity of p-nitrophenol generated was evaluated in UV spectrophotometer at a wavelength of A415. 1 U of pNPC-hydrolyzing activity is described as amount of enzyme corresponding to release 1 µmol of p-nitrophenol for every minute. The CBH is also analyzed with cellobiose or cello-oligosaccharides as substrate by quantifying the quantity of glucose produced using GOD-POD method [23]. The protein amount was calculated by Bradford assay [24], by using BSA (bovine serum albumin) as a standard. The purification of CBH was followed as described previously [23].
Analyzing the Molecular Mass of CBH
To determine the molecular weight of CBH, sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was operated as specified previously [25]. Protein bands were observed with the help of Coomassie brilliant blue R-250 dye. The molecular weight of purified enzyme was verified by size exclusion chromatography utilizing a BioLogic FPLC system (Bio-Rad) equipped with SuperoseTM column. The CBH was eluted with 200 µM buffer (pH 5.0) at a flow rate of 1 ml/min.
Establishing pH and Temperature Optima
The optimum pH of CBH activity was considered by incubating the purified enzyme at 50 °C for 15 min in various buffers: sodium acetate buffer (100 mM, pH 3–6), phosphate buffer (100 mM, pH 6–8). To point the optimal temperature, the enzyme was incubated in sodium acetate buffer (100 mM, pH 5) for 15 min at various temperatures starting from 40 to 70 °C.
Determination of Kinetic Parameters
The values for Michaelis constant (Km) and maximum velocity (Vmax) were established for CBHs by incubating in 100 mM buffer (pH 5.0) at 50 °C using pNPC at various concentrations ranging from 0.2 to 12 mM. Km and Vmax values are noted using nonlinear regression. The specificity of purified enzyme for various substrates was calculated under standard assay conditions.
Impact of Metals and Reagents on CBH
The impacts of different metals and chemicals at 0.1 mM on CBH performance was characterized by pre-incubating the CBH with certain reagents in 20 mM buffer at optimized pH and temperature conditions for 30 min. CBH activity was further calculated at 50 °C for 15 min in the existence of metals or other chemicals. The performance of CBH assayed in deficiency of metals and chemicals was established as 100%.
Results and Discussion
Detection of the Unique Microbe for CBH Generation
Amongst 340 strains examined for CBH generation, twenty strains were chosen on the basis of fluorescence noticed with agar plates covering 10 mM of MUG. Among twenty selected strains, an effective CBH-generating microbes was isolated for further study. The identification of isolated fungus was carried out in the ITS rDNA region and output sequence were presented to GenBank. The strain revealed the greatest identity (47%) with S. commune. Phylogenetic interactions were recommended by employing taxonomic tree analysis on the basis of comparison between the unknown sequence of microbes with preestablished sequence of microbe (Fig. S1).
Optimization of Carbon Sources for CBH Generation
Numerous carbon resources (cellulose powder, carboxymethyl cellulose, Avicel, rice straw, sugarcane bagasse, tangerine peel) were analyzed for CBH generation by S. commune KMJ820. As exhibited in Fig. 1a, cellulose powder was observed to be optimal carbon resource for three cellulase production. In order to analyze, the impact of preliminary influence of carbon source on S. commune KMJ820 development and CBH generation in culture; a numerous dilutions of cellulose powder were examined. The strain generated an elevated level of CBH (11.5 U/ml) when 2% of cellulose powder was employed as the carbon source (Fig. 1b). The elevated enzymatic activity noted in crude extract may be ascribed to industrial use of cellulase to produce bioethanol.
Fig. 1.
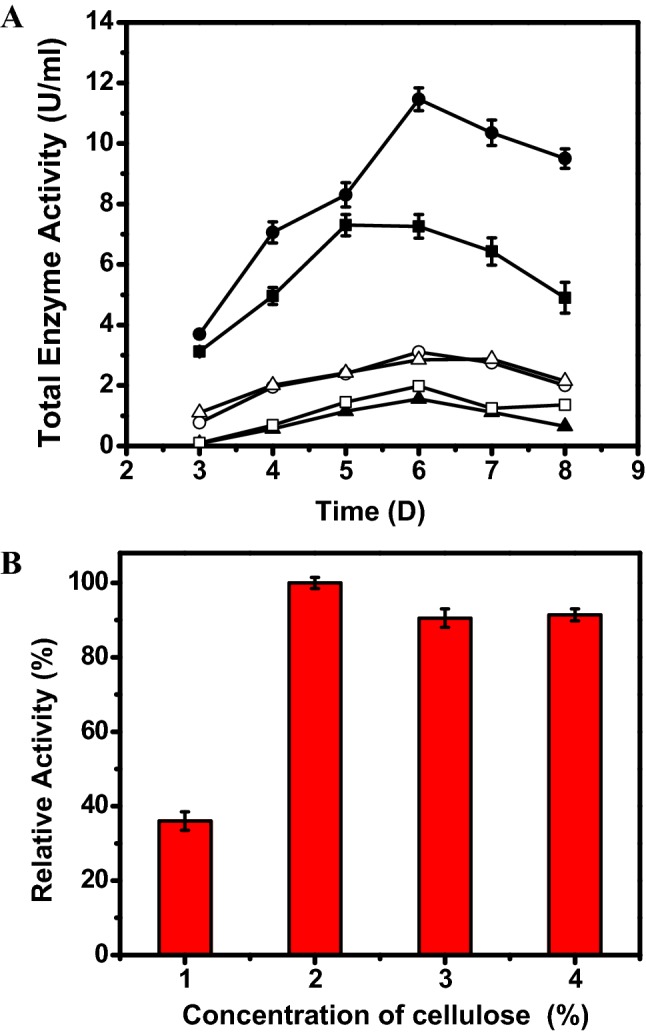
Optimization of carbon source on total enzyme activity of cellobiohydrolases in Schizophyllum commune KMJ820 culture. Cellulose powder (filled circle), carboxymethyl cellulose (open circle), Avicel (filled square), rice straw (open square), sugarcane bagasse (filled triangle), tangerine peel (open triangle) (a). Influence of cellulose concentration on total enzyme activity of cellobiohydrolases in S. commune KMJ820 culture (b)
Purification of CBH from S. commune Culture
CBHs were purified as defined in the previous sections, and results are exhibited in Table 1. As stated above, there were two CBHs. It was revealed when the crude extract enzyme was exposed to SDS-PAGE and they were divided by first DEAE chromatography step (DEAE chromatography-1). Fractionation with ammonium sulfate precipitation boosted the enzyme specific activity of about 1.1-fold, with nearly 40% enhancement in CBH activity. The CBH effective fractions are injected to a DEAE Sepharose column, and CBH was separated using 100 mM NaCl. At this stage, CBH 2 from S. commune was split. To purify CBH 1, a subsequent hydroxyapatite step was executed, and it yielded two peaks comprising protein; one was large and the other was minor. The large peak showed CBH activity. FPLC elution of second trial of DEAE ion exchange chromatography (DEAE chromatography-2) column delivered an effective CBH protein peak. These separation techniques for CBH 1 and 2 helped in achieving a specific activities of 37.6 U/mg and 18.1 U/mg, respectively. As noted in Fig. 2, the purified enzyme seemed as a one band on SDS-PAGE.
Table 1.
Purification of cellobiohydrolases (CBH) from Schizophyllum commune KMJ820
| Step | Total protein (mg) | Specific activity (U/mg) | Total activity (U/ml) | Yield (%) | Purification fold | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CBH 1 | CBH 2 | CBH 1 | CBH 2 | CBH 1 | CBH 2 | CBH 1 | CBH 2 | CBH 1 | CBH 2 | |
| Crude extract | 463.4 | 463.4 | 11.51 | 11.51 | 5335 | 5335 | 100 | 100 | – | – |
| Ammonium sulfate precipitation | 162.9 | 162.9 | 12.84 | 12.84 | 2092 | 2092 | 39.22 | 39.22 | 1.12 | 1.12 |
| DEAE chromatography-1 | 33.30 | 3.00 | 19.57 | 18.12 | 651.8 | 54.32 | 12.22 | 1.02 | 1.70 | 1.57 |
| Hydroxyapatite | 2.66 | – | 21.37 | – | 56.82 | – | 1.07 | – | 1.86 | – |
| DEAE chromatography-2 | 0.22 | – | 37.63 | – | 8.34 | – | 0.16 | – | 3.27 | – |
Fig. 2.
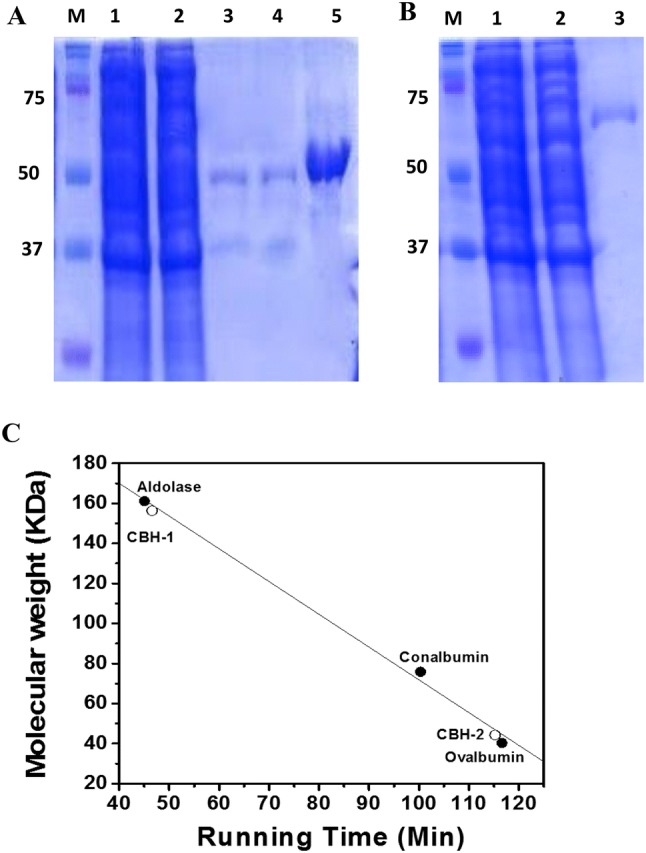
Analysing the molecular weight of cellobiohydrolase 1 (a), and cellobiohydrolase 2 (b), isolated from Schizophyllum commune KMJ820. Lane M marker, lane 1 crude extract, lane 2 ammonium sulfate precipitative fraction, lane 3 DEAE ion exchange portion, lane 4 hydroxyapatite portion, lane 5 DEAE ion exchange portion. Analysing the indigenous molecular weight of S. commune KMJ820, cellobiohydrolases by gel filtration chromatography (c)
Characterization of CBH Refined from S. commune Culture
Size exclusion chromatography of enzyme exhibited a proportioned peak suggesting native Mr of two CBHs. So, it was shown that CBH 1 is monomer, whereas CBH 2 is trimer (Fig. 2). The optimal pH for both CBHs was 5 (Fig. S2). The ideal temperature for hydrolysis with CBH 1 and 2 were 55 °C and 50 °C, respectively (Fig. S3). Early velocities with enzyme were created in standard assay mixture at optimum pH. The enzyme kinetics examined have demonstrated hyperbolic saturation curved, and consequent double-reciprocal plots are linearly increased. The pNPC concentration was differed from 0 to 12 mM for both CBH 1 and 2. Figure 3 indicates the kinetics of CBH by rising pNPC concentrations. Lineweaver–Burk plot (Fig. 3, inset) were achieved by transformation of pNPC over standard assay conditions. CBH1 has exhibited a Km and Vmax of 2.0 mM and 51.4 U/mg protein, respectively. Whereas, CBH 2, had exhibited a Km of 1.4 mM and Vmax of 20.8 U/mg protein accordingly. The impacts of different metals and reagents at 100 µm on CBH activity are determined with preincubation of CBH using particular chemicals in buffer at optimized pH for 30 min. As shown in Table S1, for both CBHs, ZnCl2 and CuCl2 (each at 0.1 mM) had hindered CBH activity.
Fig. 3.
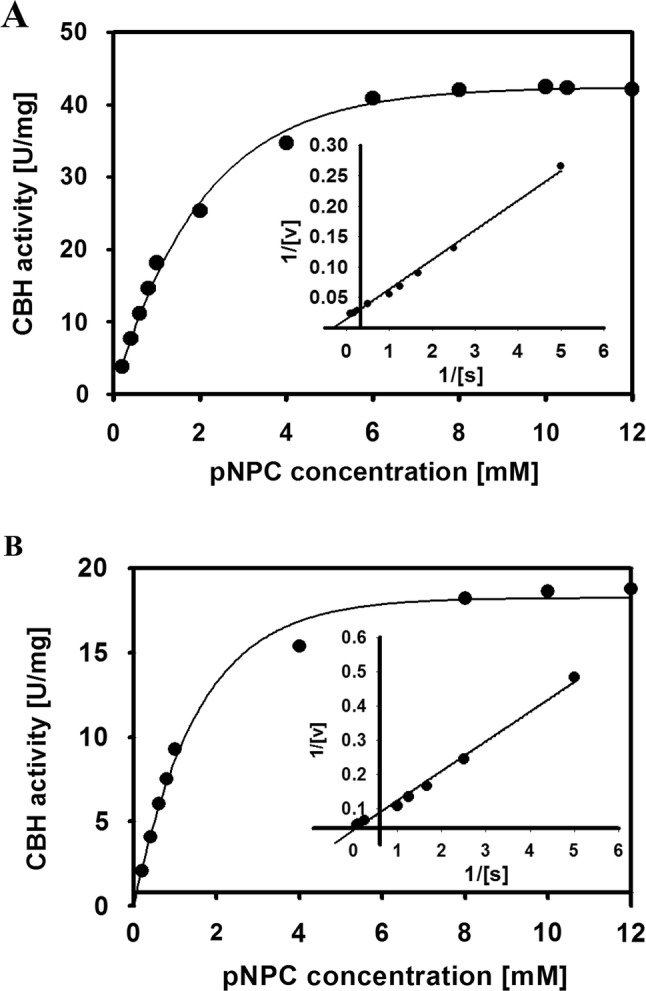
Influence of substrate concentration on cellobiohydrolase 1 (a), and cellobiohydrolase 2 (b), from Schizophyllum commune KMJ820. The inset figure point towards Lineweaver–Burk plot of cellobiohydrolase
Table 2 points a difference in the characteristics of different CBHs from various sources. S. commune CBH 1 had an enzymatic activity and Vmax of 37.6 U/mg and 51.4 U/mg for pNPC. These enzymatic characteristics from CBH can be essential for establishing the elevated reducing sugar generation in saccharification noted in S. commune (data not shown). The extracellular CBH 1 and 2 purified from S. commune were monomer and trimer, respectively, with molecular weight of about 50 and 150 kDa. The CBH activities were hindered when 0.1 mM of Zn and Cu ion were added. It indicates the presence of alkaline and acidic based amino groups may represent essential catalytic domains in the active sites of these enzymes [11]. These activities of CBH in can be stabilized/enhanced by immobilization of enzyme on nano particles [31–37] and thereby can assist in integrative biotransformation of waste to achieving high calorific value-added products [38–40].
Table 2.
Properties of cellobiohydrolases (CBH) isolated from several sources
| MW (kDa) | Quaternary structure | Opt. pH | Opt. temp (°C) | Specific activity (U/mg) | Kma (mM) | Vmaxa (U/mg) | References | |
|---|---|---|---|---|---|---|---|---|
| Dichomitus squalens Ex 1 | 39 | Monomer | 5 | 60 | 13.64 | NR | NR | [26] |
| D. squalens Ex 2 | 36 | Monomer | 5 | 60 | 12.06 | NR | NR | [26] |
| Irpex lacteus Ex 1 | 53 | NR | 5 | 50 | 33.2 | NR | NR | [27] |
| I. lacteus Ex 2 | 56 | NR | 5 | 50 | 34.0 | NR | NR | [27] |
| Talaromyces emersonii CBH IA | 66 | Monomer | 3.6 | 78 | 7.7 | 2.1 | 9.2 | [28] |
| T. emersonii CBH IB | 56 | Monomer | 4.1 | 66 | 2.4 | 0.8 | 3.8 | [28] |
| T. emersonii CBH II | 56 | Monomer | 3.8 | 68 | NR | NR | NR | [28] |
| Trametes versicolor | 55 | Monomer | 5 | 40 | 0.8 | 0.58 | 1 | [29] |
| Penicillium occitanis CBH I | 60 | NR | 4–5 | 60 | 1.09 | 1 | NR | [30] |
| P. occitanis CBH II | 55 | NR | 4–5 | 65 | 0.03 | 5 | NR | [30] |
| Schizophyllum commune KMJ820 CBH 1 | 50 | Monomer | 5 | 55 | 37.6 | 2.0 | 51.4 | (This study) |
| S. commune KMJ820 CBH 2 | 150 | Trimer | 5 | 50 | 18.1 | 1.4 | 20.8 | (This study) |
NR not reported
aKinetic constraints of CBHs were exhibited using pNPC
Conclusion
In conclusion, an effective CBH-generating strain was isolated and categorized as S. commune KMJ820. The effective purification and categorization of CBHs generated by S. commune characterized a CBH displaying the highest specific activity. Further studies on the CBH from S. commune are needed to evaluate its capacity in saccharification and bioethanol generation. Our findings can assist in large scale production of glucose or ethanol by biological methods.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was supported by Brain Pool Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2019R1C1C1009766, NRF-2019H1D3A2A01060226, 2019R1F1A1063131). This research was supported by 2018 KU Brain Pool fellowship of Konkuk University.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
In-Won Kim, Email: Inwon@konkuk.ac.kr.
Vipin Chandra Kalia, Email: vckaliaku@gmail.com.
Jung-Kul Lee, Email: jkrhee@konkuk.ac.kr.
References
- 1.Lee K-M, Kalyani D, Tiwari MK, Kim T-S, Dhiman SS, Lee J-K, Kim I-W. Enhanced enzymatic hydrolysis of rice straw by removal of phenolic compounds using a novel laccase from yeast Yarrowia lipolytica. Bioresour Technol. 2012;123:636–645. doi: 10.1016/j.biortech.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 2.Kalyani D, Lee K-M, Kim T-S, Li J, Dhiman SS, Kang YC, Lee J-K. Microbial consortia for saccharification of woody biomass and ethanol fermentation. Fuel. 2013;107:815–822. doi: 10.1016/j.fuel.2013.01.037. [DOI] [Google Scholar]
- 3.Jeya M, Kalyani D, Dhiman SS, Kim H, Woo S, Kim D, Lee J-K. Saccharification of woody biomass using glycoside hydrolases from Stereum hirsutum. Bioresour Technol. 2012;117:310–316. doi: 10.1016/j.biortech.2012.03.047. [DOI] [PubMed] [Google Scholar]
- 4.Singh DN, Sood U, Singh AK, Gupta V, Shakarad M, Rawat CD, Lal R. Genome sequencing revealed the biotechnological potential of an obligate thermophile Geobacillus thermoleovorans strain RL isolated from hot water spring. Indian J Microbiol. 2019;59:351–355. doi: 10.1007/s12088-019-00809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeya M, Nguyen N-P-T, Moon H-J, Kim S-H, Lee J-K. Conversion of woody biomass into fermentable sugars by cellulase from Agaricus arvensis. Bioresour Technol. 2010;101:8742–8749. doi: 10.1016/j.biortech.2010.06.0556. [DOI] [PubMed] [Google Scholar]
- 6.Jeya M, Zhang Y-W, Kim I-W, Lee J-K. Enhanced saccharification of alkali-treated rice straw by cellulase from Trametes hirsuta and statistical optimization of hydrolysis conditions by RSM. Bioresour Technol. 2009;100:5155–5161. doi: 10.1016/j.biortech.2009.05.040. [DOI] [PubMed] [Google Scholar]
- 7.Kumar V, Patel SKS, Gupta RK, Otari SV, Gao H, Lee J-K, Zhang L. Enhanced saccharification and fermentation of rice straw by reducing the concentration of phenolic compounds using an immobilized enzyme cocktail. Biotechnol J. 2019;14:1800468. doi: 10.1002/biot.201800468. [DOI] [PubMed] [Google Scholar]
- 8.Baek S-H, Kim S, Lee K, Lee J-K, Hahn J-S. Cellulosic ethanol production by combination of cellulase-displaying yeast cells. Enzyme Microb Technol. 2012;51:366–372. doi: 10.1016/j.enzmictec.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Teeri TT. Crystalline cellulose degradation: new insight into the function of cello-biohydrolases. Trends Biotechnol. 1997;15:160–167. doi: 10.1016/S0167-7799(97)01032-9. [DOI] [Google Scholar]
- 10.Kumar A, Patel SK, Mardan B, Pagolu R, Lestari R, Jeong S-H, Kim T, Haw JR, Kim S-Y, Kim I-W. Immobilization of xylanase using a protein–inorganic hybrid system. J Microbiol Biotechnol. 2018;28:638–644. doi: 10.4014/jmb.1710.10037. [DOI] [PubMed] [Google Scholar]
- 11.Shin K, Kim YH, Jeya M, Lee J-K, Kim Y-S. Purification and characterization of a thermostable cellobiohydrolase from Fomitopsis pinicola. J Microbiol Biotechnol. 2010;20:1681–1688. doi: 10.4014/jmb.1008.08009. [DOI] [PubMed] [Google Scholar]
- 12.Xu Q, Sun M, Ning J, Fang S, Ye Z, Chen J, Fu R. The core role of Bacillus subtilis and Aspergillus fumigatus in pile-fermentation processing of Qingzhuan Brick Tea. Indian J Microbiol. 2019;59:288–294. doi: 10.1007/s12088-019-00802-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurya S, Rashk-E-Eram, Naik SK, Choudhary JS, Kumar S. Heavy metals scavenging potential of Trichoderma asperellum and Hypocrea nigricans isolated from acid soil of Jharkhand. Indian J Microbiol. 2019;59:27–38. doi: 10.1007/s12088-018-0756-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MR, Gao H, Prabhu P, Bugada LF, Roth C, Mutukuri D, Yee CM, Lee L, Ziff RM, Lee J-K, Wen F. Elucidating structure–performance relationships in whole-cell cooperative enzyme catalysis. Nat Catal. 2019;2:809–819. doi: 10.1038/s41929-019-0321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JK, Kang YC, Dhiman S, Li J, Ranjitha R, Chandra SS, Inventors (2017) Simultaneous pretreatment and saccharification of biomass using fungal consortium and method of preparing biofuel using the same. U.S. Patent 9,631,207
- 16.Sateesh L, Rodhe AV, Naseeruddin S, Yadav KS, Prasad Y, Rao LV. Simultaneous cellulase production, saccharification and detoxification using dilute acid hydrolysate of S. spontaneum with Trichoderma reesei NCIM 992 and Aspergillus niger. Indian J Microbiol. 2012;52:258–262. doi: 10.1007/s12088-011-0184-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards IP, Upchurch RA, Zak DR. Isolation of fungal cellobiohydrolase I genes from sporocarps and forest soils by PCR. Appl Environ Microbiol. 2008;74:3481–3489. doi: 10.1128/AEM.02893-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JK, Kim TS, Jagtap SS, Cha MH, Lee JI, Roh HD (2017) Cellulase-producing novel strain and saccharification method using the same. U.S. Patent Application 15/483,337
- 19.Jagtap SS, Woo SM, Kim T-S, Dhiman SS, Kim D, Lee J-K. Phytoremediation of diesel-contaminated soil and saccharification of the resulting biomass. Fuel. 2014;116:292–298. doi: 10.1016/j.fuel.2013.08.017. [DOI] [Google Scholar]
- 20.Daenen L, Saison D, Sterckx F, Delvaux FR, Verachtert H, Derdelinckx G. Screening and evaluation of the glucoside hydrolase activity in Saccharomyces and Brettanomyces brewing yeasts. J Appl Microbiol. 2008;104:478–488. doi: 10.1111/j.1365-2672.2007.03566.x. [DOI] [PubMed] [Google Scholar]
- 21.Singh RK, Zhang Y-W, Nguyen N-P-T, Jeya M, Lee J-K. Covalent immobilization of β-1,4-glucosidase from Agaricus arvensis onto functionalized silicon oxide nanoparticles. Appl Microbiol Biotechnol. 2011;89:337–344. doi: 10.1007/s00253-010-2768-z. [DOI] [PubMed] [Google Scholar]
- 22.Jagtap SS, Dhiman SS, Kim T-S, Li J, Lee J-K, Kang YC. Enzymatic hydrolysis of aspen biomass into fermentable sugars by using lignocellulases from Armillaria gemina. Bioresour Technol. 2013;133:307–314. doi: 10.1016/j.biortech.2013.01.118. [DOI] [PubMed] [Google Scholar]
- 23.Jeya M, Joo A-R, Lee K-M, Tiwari MK, Lee K-M, Kim S-H, Lee J-K. Characterization of β-glucosidase from a strain of Penicillium purpurogenum KJS506. Appl Microbiol Biot. 2010;86:1473–1484. doi: 10.1007/s00253-009-2395-8. [DOI] [PubMed] [Google Scholar]
- 24.Singh RK, Singh R, Sivakumar D, Kondaveeti S, Kim T, Li J, Sung BH, Cho B-K, Kim DR, Kim SC, Kalia VC, Zhang Y-HPJ, Zhao H, Kang YC, Lee J-K. Insights into cell-free conversion of CO2 to chemicals by a multienzyme cascade reaction. ACS Catal. 2018;8:11085–11093. doi: 10.1021/acscatal.8b02646. [DOI] [Google Scholar]
- 25.Zhang L, Guo Z, Chen J, Li J, Chen F, He Y, Gao U, Li Y, Guan X, Kang YC, Lee J-K. An artificial synthetic pathway for acetoin, 2,3-butanediol, and 2-butanol production from ethanol using cell free multi-enzyme catalysis. Green Chem. 2018;20:230–242. doi: 10.1039/C7GC02898A. [DOI] [Google Scholar]
- 26.Rouau XOE. Purification and properties of two enzymes from Dichomitus squalens which exhibit both cellobiohydrolase and xylanase activity. Carbohydr Res. 1986;145:279–292. doi: 10.1016/S0008-6215(00)90435-X. [DOI] [Google Scholar]
- 27.Hamada N, Ishikawa K, Fuse N, Kodaira R, Shimosaka M, Amano Y, Kanda T, Okazaki M. Purification, characterization and gene analysis of exo-cellulase II (Ex-2) from the white rot basidiomycete Irpex lacteus. J Biosci Bioeng. 1999;87:442–451. doi: 10.1016/S1389-1723(99)80092-9. [DOI] [PubMed] [Google Scholar]
- 28.Limam FCS, Ghrir R, Marzouki N. Two cellobiohydrolases of Penicillium occitanis mutant Pol 6: purification and properties. Enzym Microbial Technol. 1995;17:340–346. doi: 10.1016/0141-0229(94)00033-6. [DOI] [Google Scholar]
- 29.Tuohy MG, Walsh DJ, Murray PG, Claeyssens M, Cuffe MM, Savage AV, Coughlan MP. Kinetic parameters and mode of action of the cellobiohydrolases produced by Talaromyces emersonii. Biochim Biophys Acta. 2002;1596:366–380. doi: 10.1016/S0167-4838(01)00308-9. [DOI] [PubMed] [Google Scholar]
- 30.Lahjouji K, Storms R, Xiao Z, Joung KB, Zheng Y, Powlowski J, Tsang A, Varin L. Biochemical and molecular characterization of a cellobiohydrolase from Trametes versicolor. Appl Microbiol Biotechnol. 2007;75:337–346. doi: 10.1007/s00253-006-0824-5. [DOI] [PubMed] [Google Scholar]
- 31.Kumar A, Park GD, Patel SKS, Kondaveeti S, Otari S, Anwar MZ, Kalia VC, Singh Y, Kim SC, Cho B-K, Sohn J-H, Kim D-R, Kang YC, Lee J-K. SiO2 microparticles with carbon nanotube-derived mesopores as an efficient support for enzyme immobilization. Chem Eng J. 2019;359:1252–1264. doi: 10.1016/j.cej.2018.11.052. [DOI] [Google Scholar]
- 32.Patel SKS, Lee JK, Kalia VC. Nanoparticles in biological hydrogen production: an overview. Indian J Microbiol. 2018;58:8–18. doi: 10.1007/s12088-017-0678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel SKS, Ray S, Prakash J, Wee JH, Kim S-Y, Lee JK, Kalia VC. Co-digestion of biowastes to enhance biological hydrogen process by defined mixed bacterial cultures. Indian J Microbiol. 2019;59:154–160. doi: 10.1007/s12088-017-0643-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A, Kim I-W, Patel SKS, Lee J-K. Synthesis of protein-inorganic nanohybrids with improved catalytic properties using Co3(PO4)2. Indian J Microbiol. 2018;58:100–104. doi: 10.1007/s12088-017-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel SKS, Gupta RK, Kumar V, Mardina P, Lestari R, Kalia VC, Choi M-S, Lee J-K. Influence of metal ions on the immobilization of β-glucosidase through protein–inorganic hybrids. Indian J Microbiol. 2019;59:370–374. doi: 10.1007/s12088-019-0796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel SKS, Jeon MS, Gupta RK, Jeon Y, Kalia VC, Kim SC, Cho B-K, Kim DR, Lee J-K. Hierarchical macro-porous particles for efficient whole-cell immobilization: application in bioconversion of greenhouse gases to methanol. ACS Appl Mater Interfaces. 2019;11:18968–18977. doi: 10.1021/acsami.9b03420. [DOI] [PubMed] [Google Scholar]
- 37.Patel SKS, Kondaveeti S, Otari SV, Pagolu RT, Jeong SH, Kim SC, Cho BK, Kang YC, Lee JK. Repeated batch methanol production from a simulated biogas mixture using immobilized Methylocystis bryophila. Energy. 2018;145:477–485. doi: 10.1016/j.energy.2017.12.142. [DOI] [Google Scholar]
- 38.Kondaveeti S, Kim I-W, Otari S, Patel SKS, Pagolu R, Losetty V, Kalia VC, Lee J-K. Co-generation of hydrogen and electricity from biodiesel process effluents. Int J Hydrog Energy. 2019;44:27285–27296. doi: 10.1016/j.ijhydene.2019.08.258. [DOI] [Google Scholar]
- 39.Kondaveeti S, Mohanakrishna G, Pagolu R, Kim I-W, Kalia VC, Lee J-K. Bioelectrogenesis from raw algal biomass through microbial fuel cells: effect of acetate as Co-substrate. Indian J Microbiol. 2019;59:22–26. doi: 10.1007/s12088-018-0769-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondaveeti S, Patel SKS, Pagolu R, Li J, Kalia VC, Choi M-S, Lee J-K. Conversion of simulated biogas to electricity: sequential operation of methanotrophic reactor effluents in microbial fuel cell. Energy. 2019 doi: 10.1016/j.energy.2019.116309. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


