Abstract
Respiratory bacterial microbiota plays a key role in human health. Lung cancer microbiome is a significant yet an understudied area while bronchiectasis microbiome is often studied. We assessed the bacterial microbiota in the upper and lower respiratory tract of the patients with lung cancer and bronchiectasis against a healthy group and their variations in individuality. 16S rRNA gene based metagenomic sequencing was used to detect entire bacterial community along with conventional aerobic bacterial culturing. In comparison to healthy, increased bacterial diversity was observed in diseased population. Abundance of more than 1% was considered and bacteria were identified in 97% similarity. Only lung cancer patients exhibited bacteria specific to the disease: Corynebacterium tuberculostearicum and Keratinibaculum paraultunense. However, Enterococcus faecalis and Delftia tsuruhatensis were also observed limited to lung cancer and bronchiectasis respectively, in less than 1% but supported with bacterial culturing. In conclusion the disease condition and intra-group variability should be considered in future with larger cohorts to understand individual patient variability highlighting the social habits and gender of the individual.
Electronic supplementary material
The online version of this article (10.1007/s12088-019-00850-w) contains supplementary material, which is available to authorized users.
Keywords: Bacteria, Lung microbiota, 16S metagenomics, Respiratory tract, Lung cancer, Bronchiectasis
Introduction
Microbial population in certain environment is known as its microbiota [1]. The microbiota assumed to play a significant role not only in disease causation, but also in immune mediation and disease progression. The respiratory tract is the main portal of influx of the microorganisms into the human body, thus plays a key role in inhabiting microbiota.
Both developed and developing countries have observed similarity in respiratory diseases where number of cases had been on the rise. Lung cancer and bronchiectasis are two major chronic respiratory diseases where the microorganism management is disturbed. Though, smoking is labelled as the main risk factor for lung cancer, previous history with chronic lung diseases like chronic obstructive pulmonary disease (COPD) or history of an infection with tuberculosis have shown a positive association with lung cancer [2, 3]. Haemophilus sp., Pseudomonas sp. and Staphylococcus aureus had been observed in lower airways of lung cancer patients whereas relative abundance of the phyla Spirochaetae and Synergistetes was reduced in sputum samples [4, 5]. Most recently, Geller et al. [6] discovered that the bacteria inside malignant cells have the potential to inactivate cancer chemotherapeutic drug, gemcitabine specially against class Gammaproteobacteria. This revelation brings a new light into cancer therapy highlighting the necessity of the study of microbiome.
Bronchiectasis, where permanent widening of the airways occurs is another non-communicable, chronic respiratory disease in which the microbial colonisation is increased [7] Here, microbial elimination is disturbed due to impaired mucociliary clearance [8]. Pathogens like Haemophilus influenzae, Pseudomonas aeruginosa, Moraxella catarrhalis and Prevotella sp. had been identified in bronchiectasis lung [9]. Reduced microbial diversity and P. aeruginosa and Veillonella sp. predominance is observed to be associated with increased exacerbation risk [10].
Respiratory microbiome has been evaluated with both culture dependent and culture independent techniques like 16S rRNA gene-based metagenomics sequencing for the bacterial identification. Molecular techniques in identifying respiratory microbiome might be extensively significant, but simultaneously basic laboratory culture is still required to determine the viable pathogens.
In this study, we intended to identify the differences of the bacterial presence in the lungs of the two disease groups of lung cancer and bronchiectasis against a healthy population using 16S rRNA gene-based metagenomics and basic bacterial culturing. The attention was drawn to individuals’ disease condition, sex, recent use of antibiotics, and consumption of alcohol and smoking habits. According to our findings, we have observed certain bacterial species restricted to lung cancer and for bronchiectasis and have witnessed variability where individual statuses are concerned. Currently, there is a lack of studies focusing individual variabilities affecting lung microbiome. The findings suggest that it is compulsory to study patients’ microbiota of these two chronic respiratory diseases, considering every aspect of the individual preferably with larger cohorts.
Methodology
Ethical Approval
The study was approved by the ethical review committee of teaching hospital, Kandy, Sri Lanka. The written informed consent was obtained from the study population prior to sample collection.
Study Group Selection
Sample collection was carried out from October, 2016 to April, 2018, from patients with lung cancer confirmed (LCC) (n = 10), suspected of lung cancer (LCS) (n = 10) and bronchiectasis confirmed (BRC) (n = 07), suspected of bronchiectasis (BRS) (n = 13) attending respiratory unit, Teaching Hospital, Kandy, Sri Lanka. The patients and the healthy population who were confirmed with healthy lung function were selected by the consultant respiratory physician. Healthy population (n = 20) consisted of traffic policemen (HT) (n = 10) and volunteers (HV) (n = 10). A detailed questionnaire which included social habits, medical history and current medical status and use of antibiotics within past 6 months was presented to each participant.
Sample Collection
A total of 120 samples were collected. Oropharyngeal (OP) swab and bronchoalveolar lavage (BAL) respectively representing upper and lower airway was obtained from the patient groups. OP swab and sputum representing the LRT were obtained from the healthy population due to the ethical limitations. The confirmation as to the patients’ disease status was made by the physician considering the symptoms presented, radiological, haematological and cytological data.
16S rRNA Based Metagenomic Sequencing
DNA from the original samples with confirmed disease statuses were isolated from Boom’s method [11] and using QIAamp DNA microbiome kit (QIAGEN, USA). DNA was quantified using Quantifluor Single Tube Fluorometer (Promega Inc.). The sequencing facility was provided by Macrogen Incorporation (Seoul, Korea) (n = 10; LC = 4, BR = 3, HV = 3). Briefly, the quality control and the sequencing procedure were as follows. Samples with a genomic DNA concentration of ≥ 0.1 ng/μL were continued further with library preparation. The sequencing was performed according to the Illumina 16S Metagenomic Sequencing Library preparation guide using recommended primers for V3-V4 (forward primer 5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG 3′ and reverse primer-5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC 3′). The bio samples of this study and the sequenced data are deposited under NCBI BioProject PRJNA477678.
Data Analysis for the Bacterial Community
High quality data were obtained based on Phred scores > 30. The average nucleotide length was 438 bp for the bacterial libraries. Mothur programme was used for identification of chimeric and ambiguous reads and trimming of the sequences. Scythe (v0.994) and Sickle programs are used to remove adapter sequences. Taxonomy assignment and diversity statistics were analysed with QIIME. Operational taxonomy units (OTU) analysis was carried out with a cut-off similarity of 97%. Shannon and Simpson alpha diversity matrices were also analysed with QIIME. The bio samples and the sequenced data were deposited in NCBI database.
Sample Processing and Culture
Part of the collected samples were used for routine bacterial culturing within 24 h of sample collection. One part of the sample was treated according to the modified Petroff’s method [12] for culturing on Lowenstein–Jensen (LJ) medium for isolation of Mycobacterium sp. Briefly, 4% NaOH was added to the sample, vortexed for 10 s and was incubated at room temperature for 15 min. 2 ml of sterile distilled water was added to the tube and centrifuged at 3000 g for 15 min. The supernatants were discarded and the pellets were re-suspended in sterile distilled water. The untreated portion was used to culture on Luria–Bertani (LB) medium for the isolation of bacteria.
Molecular Identification of Isolated Bacteria
Genomic DNA was extracted from isolated organisms using standard CTAB (N-Cetyl-N, N, Ntrimethyl ammonium bromide) method [13]. 16S rRNA gene of 1.5 Kb was amplified with universal bacterial primers [14]. Purified amplified DNA were sequenced from Macrogen, Inc. (Korea). The sequences were analysed (BioEdit v7.6.2.1.) and deposited in NCBI GenBank nucleotide database (accession numbers MF498492.1-MF498510.1, MG733159.1-MG733173.1, MG738354.1).
Rectangular maximum likelihood phylogenetic tree was constructed for identified bacteria using phylogeny.fr platform [15, 16]. Sequences were aligned with MUSCLE (v3.8.31) configured for highest accuracy. Ambiguous regions were removed with Gblocks (v0.91b) [17]. Graphical representation and edition of the phylogenetic tree were performed with TreeDyn (v198.3) [18] The tree was modified with the heatmap and colour coded in iTOL(v3) [19].
Results and Discussion
We intended to identify bacteria in URT and LRT of patients suspected of lung cancer and bronchiectasis, with the aid of culturing and by molecular techniques.
Patient Demographic Data
Demographic details containing status of smoking, alcoholic consumption and antibiotic usage of the study population are as Table 1. The patient demographic data shows that majority of patients are or were smokers and consumers of alcohol and the majority of the bronchiectasis suspected patients had used antibiotics within past 6 months from the time of sample collection.
Table 1.
Population demographic data
| Population | HC | LCS | BRS |
|---|---|---|---|
| N | 20 | 20 | 20 |
| Average age | 46.1 ± 8.8 | 63.5 ± 9.8 | 66.5 ± 11.1 |
| Gender (male:female) | 3:1 | 19:1 | 11:9 |
| Smoking | |||
| Current smoker | 5% | 50% | 0% |
| Ex-smoker | 5% | 35% | 35% |
| Alcoholic consumption | |||
| Current user | 30% | 20% | 0% |
| Ex-user | 5% | 50% | 25% |
| Antibiotic usage (within 6 months) | 10% | 30% | 55% |
N number, HC healthy population, LCS lung cancer suspect, BRS bronchiectasis suspect
16S rRNA Based Metagenomic Sequencing
DNA concentrations of the samples subjected to 16S rRNA based metagenomics sequencing is presented in Table S1. The Illumina MiSeq platform was used for sequencing and the adapter trimmed sequencing data statuses are available in Table S2. Trimmed reads were deposited into the NCBI Sequence Read Archive (SRA) database under the BioProject ID PRJNA477678.
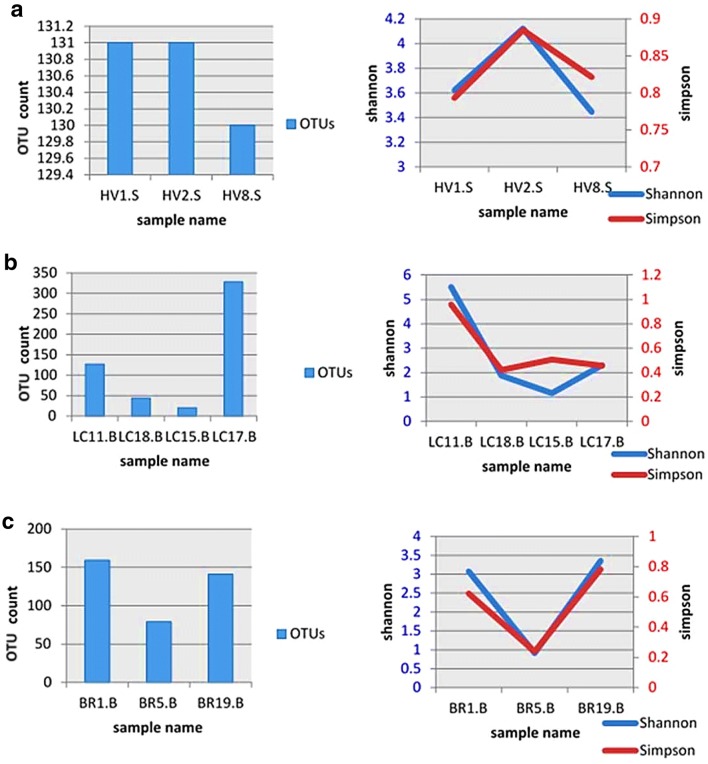
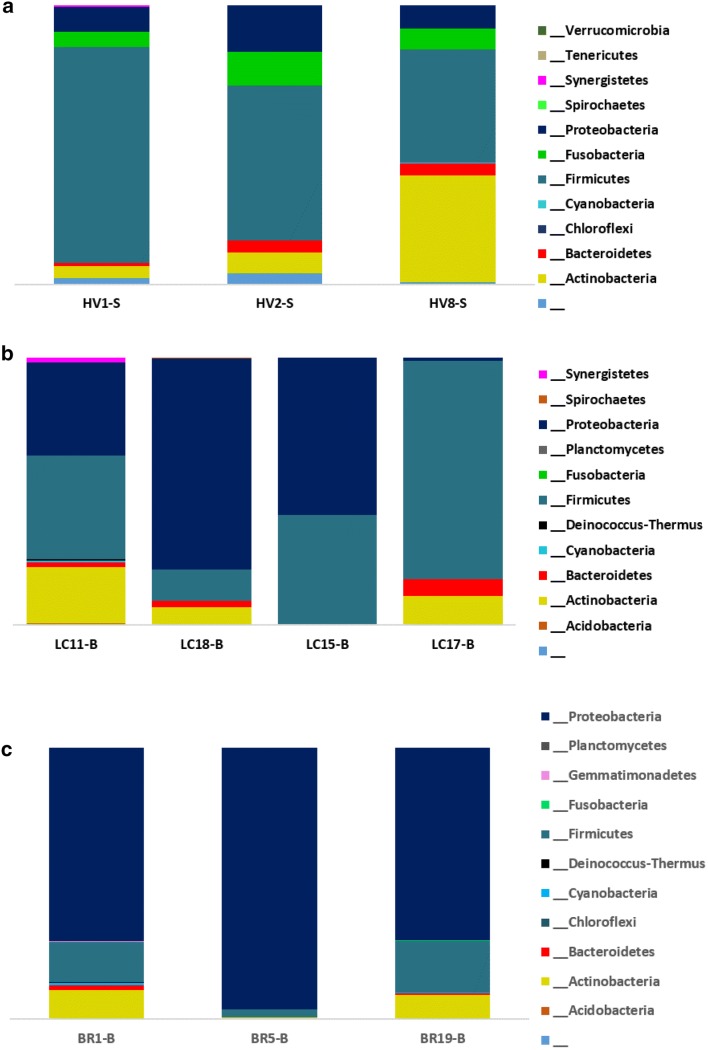
Shannon and Simpson diversity indexes for the three groups are shown in Fig. 1. Shannon diversity acts both as a guide to diversity and evenness of a community. Two samples of healthy group had similar number of OTUs (n = 151), but owned different Shannon indexes as 3.6 and 4.1 (Fig. 1a). The Simpson diversity ranged between 0.79 and 0.88 among the three healthy samples. A wide variation was observed for the diversity among the lung cancer samples as the lowest number of OTUs were recorded as 20 and the highest was 328 (Fig. 1b). Three of the samples exhibited a Shannon diversity ranging 1.15–2.3 while the sample with 127 OTUs had a Shannon diversity of 5.5, which scored 0.95 in Simpson index showing highest diversity (Fig. 1c). BR5-B of bronchiectasis group showed the lowest Shannon diversity and Simpson diversity. According to Simpson’s index BR19-B proved to be most diverse with OTU count of 141. The detected bacterial taxa summary in phylum level are shown in Fig. 2.
Fig. 1.
Shannon and Simpson diversity variation of the bacterial diversity in the three groups. a Healthy population. b Lung cancer population. c Bronchiectasis population
Fig. 2.
Bacterial taxa summary according to the phylum. a Healthy population. b Lung cancer population. c Bronchiectasis population. HV healthy volunteers, LC lung cancer, BR bronchiectasis
Total bacterial species identified in the 16S metagenomics approach is presented in Fig. S1. In the 16S metagenomical approach, the bacterial community among the three healthy samples approximately had similar diversity when considering on phyla. The species with highest abundance ratio was Streptococcus dentisani (31.3%) followed by Rothia mucilaginosa (12%) and S. salivarius (8.1%). Fig. S1.a shows total bacterial species detected in the lungs of the healthy volunteers.
The bacterial OTUs of BAL samples of patients were assessed. Instead of pooling the DNA of patient groups, we sequenced samples individually to get a better insight. Fig. S1.b shows bacteria detected in the lungs of the lung cancer patients. Phylum Proteobacteria and Firmicutes dominated the bacterial composition in BAL samples (Fig. 2b). The three most abundant bacterial species were Achromobacter xylosoxidans, S. sinensis and Staphylococcus sciuri with abundances of 35.30%, 20.50%, and 11.00%, respectively. As Liu et al. [20] observed in their study, genus Streptococcus was seen abundantly in lung cancer patients. Individual group data analysis revealed two bacteria that were limited in lung cancer group: C. tuberculostearicum and K. paraultunense. C. tuberculostearicum has been previously observed in small cell lung cancer [21].
Lung bacterial population in bronchiectasis patients is led by phylum Proteobacteria. Fig. S2.c shows bacteria detected in the lungs of the bronchiectasis patients. P. aeruginosa was observed as the highest abundant species accompanied with Burkholderia lata and S. dentisani as the second and the third. When both disease groups were considered, majority of the OTUs of the two groups belonged to phylum Proteobacteria. The most abundant three species in lung cancer group were not capable of producing colonies in vitro, whereas in bronchiectasis group, P. aeruginosa was proved by the yield of culture isolates too. Interestingly, M. tuberculosis was only evidenced in patient groups but not in healthy population.
Organism Identification by Sanger Sequencing
In culture dependant techniques, no Mycobacterium tuberculosis (MTB) or Non-tuberculous Mycobacteria (NTM) isolates were obtained but a total of sixty bacterial isolates were obtained. Sanger dideoxy sequencing resulted in identifying 19 different organisms, most frequently found bacteria belonging to Enterobacter cloacae (n = 13), P. aeruginosa (n = 9), and Klebsiella pneumoniae (n = 9) among the three populations and the isolated bacteria belonged to two phyla; Proteobacteria and Firmicutes. A complete list of identified culture isolates with NCBI GenBank accession numbers can be found in Table S3.
Bacteria identified by culturing as per the disease condition was studied (Table 2). P. aeruginosa and E. cloacae were common for both disease groups as well as the healthy group, though Enterococcus faecalis was limited to LCC whereas Enterococcus hirae and Delftia tsuruhatensis were limited to BRC. In metagenomic results, while Enterococcus faecalis was limited to lung cancer group and D. tsuruhatensis was limited to bronchiectasis group supporting culturing result, Enterococcus hirae was observed in lung cancer patients.
Table 2.
Organism characterisation of the three groups with confirmed diseases
| Study population | Disease status | Number of cases | Identified organisms | |
|---|---|---|---|---|
| Upper respiratory tract | Lower respiratory tract | |||
| Lung cancer suspects (n = 20) | Lung cancer confirmed | 10 | ||
| Malignancy | 3 | Enterococcus faecalis, Pseudomonas aeruginosa | Enterobacter cloacae | |
| Primary adenocarcinoma | 1 | E. cloacae | ||
| Invasive adenocarcinoma | 1 | E. cloacae | ||
| Moderately differentiated adenocarcinoma | 1 | E. cloacae | ||
| Non-small cell lung cancer | 2 | E. cloacae | P. aeruginosa | |
| Moderately differentiated squamous cell carcinoma | 2 | E. cloacae | Klebsiella pneumoniae | |
| Non-malignancy | n = 10 | |||
| K. pneumoniae | E. cloacae, Bacillus subtilis, B. kochii, Neisseria sp., Morganella morganii | |||
| Bronchiectasis suspects (n = 20) | Bronchiectasis confirmed | n = 7 | ||
| Bronchiectasis | 5 | P. aeruginosa, Enterococcus hirae, Delftia tsuruhatensis | ||
| Cystic Bronchiectasis | 2 | P. aeruginosa | E. cloacae, Staphylococcus saprophyticus | |
| Non-bronchiectasis | n = 13 | |||
| P. aeruginosa, P. stutzeri, B. cereus, Paenibacillus glucanolyticus, K. pneumoniae | P. aeruginosa, K. pneumoniae, E. cloacae, Citobacter koseri | |||
| Healthy group (n = 20) | K. pneumoniae, E. cloacae, S. saprophyticus | K. pneumoniae, Bacillus sp., K. oxytoca, S. aureus, S. pasteuri, E. cloacae complex sp. | ||
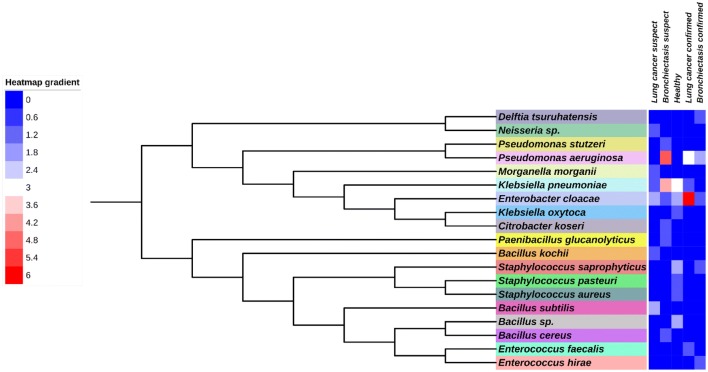
The culturable bacterial respiratory microbiota composed of eleven bacterial genera (Fig. 3). Individually, genera Enterobacter, Klebsiella, Pseudomonas, Enterococcus, Bacillus, Neisseria, and Morganella were isolated from lung cancer suspects, while genera Enterobacter, Klebsiella, Pseudomonas, Enterococcus, Bacillus, Delftia, Paenibacillus, Staphylococcus and Citobacter were present in bronchiectasis suspects. The bacteria in healthy lungs belonged to only four genera, Enterobacter, Klebsiella, Staphylococcus and Bacillus.
Fig. 3.
Phylogenetic tree of the bacterial species identified by culture method using the maximum likelihood method. Reliability for internal branch was assessed using the aLRT test (SH-Like). Leaves are colour coded representing different organism. Heatmap gradient represent the identified number of strains
In our study, it was possible to isolate most of the reported bacterial species in lung microbiome of lung cancer patients. In this study, five bacterial species were isolated from lung cancer suspected patients; Enterococcus faecalis, B. subtilis, B. kochii, Neisseria sp. and M. morganii which had also been previously recorded in lung cancer patients [22]. Enterococcus faecalis, a bacterium we observed in URT of lung cancer confirmed patient, has demonstrated the ability to induce cellular proliferation in oral cancer cells [23]. As in our study, in confirmed lung cancer cases, E. cloacae and P. aeruginosa are commonly observed pathogens. E. cloacae have shown the ability to destroy phagocytes along with epithelial cells leading to metastasis [24] while Bacillus sp. is suspected of favouring toxin penetration to the cells by their toxin production [25]. B. subtilis is commonly observed in gut microbiome. It is a possibility that this could be via the lung-gut axis as described by Marsland et al. [26].
In bronchiectasis confirmed cases, we isolated D. tsuruhatensis and Enterococcus hirae along with previously reported commonly identified bacteria such as P. aeruginosa and E. cloacae. Delftia sp. of Comamonadaceae has not reported to be isolated in bronchiectasis lung microbiome previously. Although Staphylococcus species are commonly observed as commensals, species like Staphylococcus aureus have been observed with bronchiectasis cases [27].
In healthy population, we were able to isolate K. pneumoniae, E. cloacae, Staphylococcus saprophyticus, K. oxytoca, Staphylococcus aureus, S. pastueri and Bacillus sp. It is hypothesised that airway microbiota is subjected to the effect of immunological homeostasis as macrophages and neutrophils phagocytose bacteria in the airways [28, 29].
Bacterial Diversity in Patients
Patient samples with confirmed disease statuses were the only subjects to metagenomics sequencing. The healthy group consisted of a female and two males. Among these, 13 species had more than 1% abundance. Porphyromonas gingivalis, Streptococcus gordonii, N. bacilliformis and N. oralis were observed as limited to male participants with less than 1% abundance. We observed 13 bacterial species that were only seen in the female sample all of which failed to acquire at least 1% abundance. When considering the two males, one was neither a smoker nor an alcoholic and the other also was never a smoker yet presumed consuming alcohol randomly, for 20 years. The profile of the latter observed with higher diversity of bacteria than the former. The effect of alcohol consumption on an individual’s sputum microbiota has not previously studied and from our results an idea is drawn as to whether alcohol consumption might alter the microbial composition of one’s lung. This sample showed some visible deviations of abundance in mutual bacterial species such as R. mucilaginosa (~ 30-fold increase than other two samples), a bacterium known to be associated with pneumonia [30] and S. salivarius, a commonly known commensal.
The lung cancer group was all-males, consisted of an invasive adenocarcinoma patient, a NSCLC patient and two malignant patients. Only three species were found to be common in all samples: Staphylococcus sciuri, A. xylosoxidans and Luteimonas terrae. The invasive adenocarcinoma sample exhibited the most diverse bacterial composition with 67 bacterial species limited to itself. Nocardia pneumoniae, A. israelii, F. periodonticum, L. hackeliae and Treponema socranskii were only observed in non-small cell lung cancer sample. C. jeikeium, Nesterenkonia lacusekhoensis, Propionibacterium acnes, Brevundimonas albigilva and Comamonas denitrificans species were observed in both invasive adenocarcinoma and NSCLC samples but were not present in malignant samples. The most diverse profile was obtained from LC11-B, a sample of an invasive adenocarcinoma patient and the least diversity was seen in LC 15-B, a sample of a patient with primary malignant condition. As per our data, we have observed that as the cancer get more defined, the bacterial diversity increases, which shows the decreasing immunity levels in patients. This can be interpreted since the patient is being immunocompromised, his immune mechanisms in eliminating bacterial influx is impaired.
Bronchiectasis group consisted of one female and two males. The three samples had more than 40 bacterial species in common, of which P. aeruginosa, S. dentisani, Staphylococcus aureus, B. thermoamylovorans, R. xylanophilus and M. aloeverae presented with more than 1% total abundance. The female patient’s sample showed 80 bacterial species confined to the sample whereas among male samples, no specifically limited bacteria were observed.
Immune responses are deemed to alter with sex differences, though mechanisms and reasons for this is not completely understood. Sex difference with microbiota is rather discussed focusing on intestinal microbiome than other body sites. Studies using mice have shown that differences in microbiota may cause in sexually dimorphic immunity [31]. Felagas et al. [32] reviewed immune infection severity and incidents with sex difference in respiratory infections where they concluded that while females are commonly affected with URT infections, males are more susceptible to LRT infections. Though lung microbiome is not discussed with alcohol consumption, Fan et al. [33] observed variations in oral microbiota with the consumption of different alcohols. Thus, gender and social habits of a person may have a significant effect on his/her lung microbiota.
This study is an observational study. The study group involves participants ranging ~ 40 to ~ 60 years and also exhibits high variability in different habits. In order for the results to be more conclusive, future studies will be conducted with subjects with identical socio-demographic criteria.
Conclusion
Specific bacteria that is limited to the lung cancer and bronchiectasis was observed in our results in both routine culturing and in the molecular approach. The results suggest a potentially positive relationship between the bacterial microbiome and these respiratory diseases. This study on two different major non communicable respiratory diseases shows that the necessity of lung microbiome assessment concerning individual profiles is crucial to have a better understanding of the condition. Aside we have observed confirmatory results supporting other research findings. To obtain conclusive results on individual variations are relationship of bacterial microbiota with the diseases, it is important to use molecular methods; 16S rRNA based metagenomics, along with culture dependent methods while recruiting larger cohorts.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to acknowledge the support given by the members of the Respiratory Unit 2 at Teaching Hospital, Kandy and financial assistance provided by National Institute of Fundamental Studies, Kandy, Sri Lanka.
Authors’ Contribution
AE and DNM designed the study. DM did clinically assess the patients and carried out sampling procedures. AE collected the samples, processed and did the experiments with analysis. DNM and NVC supervised the study. AE wrote the manuscript. All authors reviewed the manuscript.
Compliance with Ethical Standards
Conflict of interest
The author(s) declare no competing interests.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Data Availability
Further data of the research can obtain by contacting the corresponding author. The sequences of the identified organisms were deposited in NCBI GenBank nucleotide database (accession numbers MF498492.1-MF498510.1, MG733159.1-MG733173.1, MG738354.1). 16S rRNA gene sequence trimmed reads are deposited in NCBI Sequence Read Archive (SRA) database under the BioProject ID number PRJNA477678. Further details on the organisms and the metagenomics sequencing is provided in electronic supplementary material.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lederberg J, McCray JE (2001) ‘Ome Sweet’ Omics—a genealogical treasury of words | The Scientist Magazine®. In: Sci. https://www.the-scientist.com/commentary/ome-sweet-omics—a-genealogical-treasury-of-words-54889. Accessed 7 March 2019
- 2.Young RP, Hopkins RJ, Christmas T, et al. COPD prevalence is increased in lung cancer, independent of age, sex and smoking history. Eur Respir J. 2009;34:380–386. doi: 10.1183/09031936.00144208. [DOI] [PubMed] [Google Scholar]
- 3.Yu Y-H, Liao C-C, Hsu W-H, et al. Increased lung cancer risk among patients with pulmonary tuberculosis: a population cohort study. J Thorac Oncol. 2011;6:32–37. doi: 10.1097/JTO.0b013e3181fb4fcc. [DOI] [PubMed] [Google Scholar]
- 4.Wu BG, Cahaney CF, Tsay JJ et al (2015) C99 genomics and cancer: has it borne scientific and clinical fruit? Evaluation of the microbiome associated with lung cancer. In: American Thoracic Society 2015 international conference
- 5.Hosgood HD, Sapkota AR, Rothman N, et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ Mol Mutagen. 2014;55:643–651. doi: 10.1002/em.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science. 2017;357:1156–1160. doi: 10.1126/science.aah5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King P. The pathophysiology of bronchiectasis. Int J Chron Obstruct Pulmon Dis. 2009;4:411. doi: 10.2147/COPD.S6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu JC, Modha DE, Gaillard EA. What is the clinical significance of filamentous fungi positive sputum cultures in patients with cystic fibrosis? J Cyst Fibros. 2013;12:187–193. doi: 10.1016/j.jcf.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Purcell P, Jary H, Perry A, et al. Polymicrobial airway bacterial communities in adult bronchiectasis patients. BMC Microbiol. 2014;14:130. doi: 10.1186/1471-2180-14-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rogers GB, Zain NMM, Bruce KD, et al. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc. 2014;11:496–503. doi: 10.1513/AnnalsATS.201310-335OC. [DOI] [PubMed] [Google Scholar]
- 11.Boom R, Sol CJ, Salimans MM, et al. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/JCM.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tripathi K, Tripathi PC, Nema S et al (2014) Modified Petroff’s method: an excellent simplified decontamination technique in comparison with Petroff’s method. Statperson Publications
- 13.Somerville W, Thibert L, Schwartzman K, Behr MA. Extraction of Mycobacterium tuberculosis DNA: a Question of Containment. J Clin Microbiol. 2005;43:2996–2997. doi: 10.1128/JCM.43.6.2996-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lane DJ, Pace B, Olsen GJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dereeper A, Audic S, Claverie J-M, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 18.Chevenet F, Brun C, Bañuls A-L, et al. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H-X, Tao L-L, Zhang J, et al. Difference of lower airway microbiome in bilateral protected specimen brush between lung cancer patients with unilateral lobar masses and control subjects. Int J Cancer. 2018;142:769–778. doi: 10.1002/ijc.31098. [DOI] [PubMed] [Google Scholar]
- 21.Hinic V, Lang C, Weisser M, et al. Corynebacterium tuberculostearicum: a potentially misidentified and multiresistant Corynebacterium species isolated from clinical specimens. J Clin Microbiol. 2012;50:2561–2567. doi: 10.1128/JCM.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berghmans T, Sculier J-P, Klastersky J. A prospective study of infections in lung cancer patients admitted to the hospital. Chest. 2003;124:114–120. doi: 10.1378/chest.124.1.114. [DOI] [PubMed] [Google Scholar]
- 23.Boonanantanasarn K, Gill AL, Yap Y, et al. Enterococcus faecalis enhances cell proliferation through hydrogen peroxide-mediated epidermal growth factor receptor activation. Infect Immun. 2012 doi: 10.1128/IAI.00479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krzymińska S, Mokracka J, Koczura R, Kaznowski A. Cytotoxic activity of Enterobacter cloacae human isolates. FEMS Immunol Med Microbiol. 2009;56:248–252. doi: 10.1111/j.1574-695X.2009.00572.x. [DOI] [PubMed] [Google Scholar]
- 25.Merlos A, Rodríguez P, Bárcena-Uribarri I, et al. Toxins secreted by bacillus isolated from lung adenocarcinomas favor the penetration of toxic substances. Front Microbiol. 2015;6:1301. doi: 10.3389/fmicb.2015.01301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsland BJ, Trompette A, Gollwitzer ES (2015) The gut-lung axis in respiratory disease. In: Annals of the American Thoracic Society. American Thoracic Society, pp S150–S156 [DOI] [PubMed]
- 27.Metersky ML, Aksamit TR, Barker A, et al. The prevalence and significance of staphylococcus aureus in patients with non-cystic fibrosis bronchiectasis. Ann Am Thorac Soc. 2018;15:365–370. doi: 10.1513/AnnalsATS.201706-426OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogg JC, van Eeden S. Pulmonary and systemic response to atmospheric pollution. Respirology. 2009;14:336–346. doi: 10.1111/j.1440-1843.2009.01497.x. [DOI] [PubMed] [Google Scholar]
- 29.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Escalante Yangüela B, Gracia Gutiérrez A, Gracia Tello B, et al. Bilateral bronchopneumonia due to Rothia mucilaginosa. An Sist Sanit Navar. 2017;40:479–483. doi: 10.23938/ASSN.0090. [DOI] [PubMed] [Google Scholar]
- 31.Org E, Mehrabian M, Parks BW, et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7:313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falagas ME, Mourtzoukou EG, Vardakas KZ. Sex differences in the incidence and severity of respiratory tract infections. Respir Med. 2007;101:1845–1863. doi: 10.1016/J.RMED.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Fan X, Peters BA, Jacobs EJ, et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome. 2018;6:59. doi: 10.1186/s40168-018-0448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further data of the research can obtain by contacting the corresponding author. The sequences of the identified organisms were deposited in NCBI GenBank nucleotide database (accession numbers MF498492.1-MF498510.1, MG733159.1-MG733173.1, MG738354.1). 16S rRNA gene sequence trimmed reads are deposited in NCBI Sequence Read Archive (SRA) database under the BioProject ID number PRJNA477678. Further details on the organisms and the metagenomics sequencing is provided in electronic supplementary material.