Abstract
Protein microarrays are versatile tools for parallel, miniaturized screening of binding events involving large numbers of immobilized proteins in a time- and cost-effective manner. They are increasingly applied for high-throughput protein analyses in many research areas, such as protein interactions, expression profiling and target discovery. While conventionally made by the spotting of purified proteins, recent advances in technology have made it possible to produce protein microarrays through in situ cell-free synthesis directly from corresponding DNA arrays. This article reviews recent developments in the generation of protein microarrays and their applications in proteomics and diagnostics.
Keywords: high throughput, in situ synthesis, protein microarray, proteomics
Protein microarray technology has rapidly developed into one of the most active areas in biotechnology today, and is being increasingly applied for the high-throughput discovery of molecular interactions, profiling of protein expression and monitoring of protein modifications. While genome sequencing and annotations provide comprehensive information on protein-encoding genes, and DNA arrays analyze gene expression at the mRNA level, these technologies need to be complemented by functional proteomic studies in order to better understand protein activities, expression levels and interactions at the system level. Owing to alternative mRNA splicing and a multitude of post-translational modifications (PTMs), the number of cellular protein species far exceeds that of the genes. In addition, expression levels of individual proteins can range over as much as six orders of magnitude when cells are exposed to different environments [1]. It is clear that, for large-scale analyses, proteomics studies require sensitive, high-throughput tools with a wide dynamic range of detection. One such tool is the protein microarray, consisting of several thousands of protein species immobilized on a solid surface in miniaturized features, enabling sensitive, high-throughput screening with economical use of samples and reagents. In contrast to screening systems such as yeast two-hybrid, the in vitro nature of protein arrays allows for the control of the interaction conditions, protein concentrations, PTMs and specific cofactor requirements [2].
The generation of a large diversity of proteins, such as the array elements, presents a major challenge for protein microarray systems, despite the development of high-throughput protein-production methods. For many applications, the proteins also need to remain stable and functional on the array surfaces during long-term storage, which can pose an additional difficulty. A recent advance to overcome these problems is to produce protein microarrays through in situ cell-free synthesis directly from arrayed DNA templates [3–5]. This article reviews the conventional methods, as well as more recent developments, in protein microarray technologies, together with novel applications of these technologies in medical and proteomics studies.
Types of protein microarrays
Currently, three types of protein microarrays are employed: functional protein arrays, analytical or capture protein arrays and reverse-phase protein arrays. Functional protein arrays display folded and active proteins and are designed to assay functional properties [6–8]. They are used for screening molecular interactions, studying protein pathways, identifying targets for PTMs and analyzing enzymic activities. On analytical or capture arrays, affinity reagents (e.g., antibodies) or antigens (that may be nonfolded) are arrayed for profiling the expression of proteins [9,10] or for the quantification of antibodies [11] in complex samples such as serum. Applications of antibody arrays include biomarker discovery and monitoring of protein quantities and activity states in signaling pathways. Antigen arrays are applied for profiling antibody repertoires in autoimmunity, cancer, infection or following vaccination. Moreover, antigen arrays are tools for controlling the specificity of antibodies and related affinity reagents. Reverse-phase arrays comprise cell lysates [12,13] or serum samples [14]. Analysis is multiplexed by probing replicates of the arrays with different antibodies. Reverse-phase arrays are particularly useful for studying the changes in the expression of specific proteins and protein modifications during disease progression and, thus, are primarily applied for biomarker discovery.
Generation of protein microarrays
Figure 1 shows the two general strategies for making protein microarays. In conventional approaches, individual proteins are expressed by recombinant technologies and are then purified and spotted onto a solid support, often glass slides. By contrast, in the newer in situ arrays, proteins are directly produced onto the array surface through cell-free protein synthesis.
Figure 1. The two strategies for making protein microarrays.
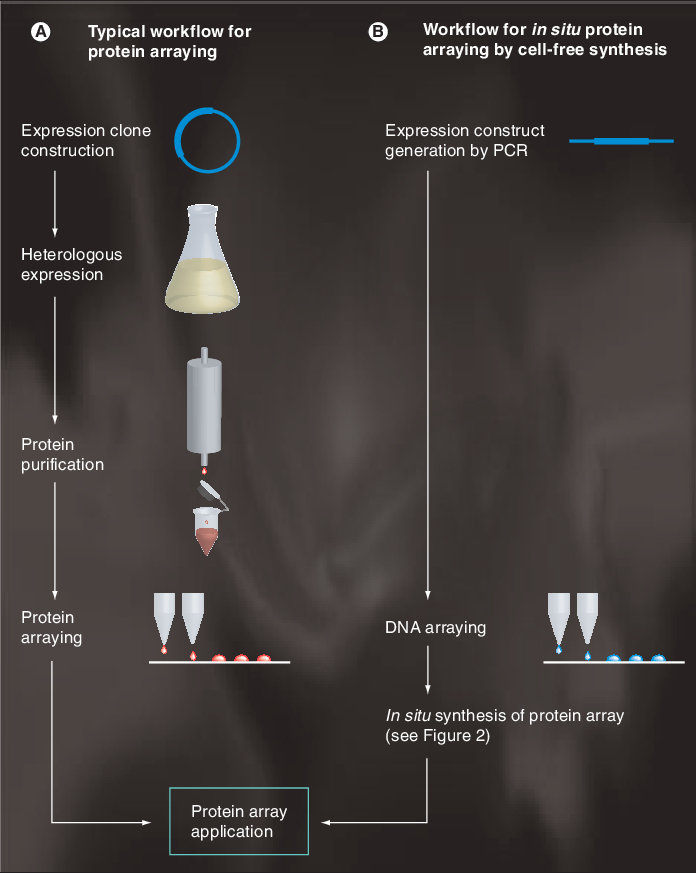
(A) Conventional method. (B)In situ synthesis of protein microarrays.
Conventional methods
Production of proteins
The most commonly used strategies for the production of proteins for arrays are heterologous expression in Escherichia coli[15] or by cell-free systems [16]. The choice of protein production method depends on the downstream applications, in which protein yields, folding and modifications must be taken into account. Clearly, high-throughput formats are desirable for generating proteins.
The advantages of using E. coli are rapidity, low cost and ease of adaptation to high-throughput production. However, some eukaryotic proteins are poorly expressed in E. coli or are produced, but at low solubility, and eukaryotic PTMs are lacking in prokaryotic cells. Screening assays have been developed for the rapid assessment of protein solubility, including using reporter proteins to directly monitor protein expression and correct folding [17–19]. Cell-free protein synthesis provides an effective and rapid alternative by using PCR DNA fragments as templates, circumventing the need for E. coli cloning [16,20]. Cell-free methods are capable of making proteins that are difficult to express in in vivo systems. Cell-free lysates can be prepared from different species, allowing protein synthesis to be carried out in prokaryotic or eukaryotic systems and at defined temperatures. These open systems can also be adjusted through the addition of further components, so as to provide environments suitable for protein folding or modifications [20]. In conventional methods, proteins expressed by E. coli or cell-free systems are purified prior to spotting onto the array surface. Purification is usually performed using affinity tags attached at either the N- or C-terminus, of which the most popular tags are hexahistidine (His6) and glutathione-S-transferase (GST) [3,21]. By contrast, by spotting unpurified tagged proteins onto capture surfaces, the immobilization and purification steps can be combined into a single step [22].
Surface chemistry
Proteins are often arrayed onto functionalized glass slides, using four main physicochemical principles, which may be combined: adsorption, covalent binding, affinity interaction by specific tags and physical entrapment into gels [23].
Surface adsorption is generally mediated by electrostatic charges (e.g., on polylysine-coated slides) or hydrophobic interactions. Despite its simplicity, the main drawback of this method is the possibility of denaturating proteins on the surface. Nevertheless, studies have suggested that surface-adsorbed proteins may still retain their functionality [24]. Surface adsorption is convenient where native protein structure and function are not essential; for example, antigen arrays.
Covalent binding of proteins to derivatized surfaces is a more efficient and robust approach [25]. The surfaces usually carry reactive groups, such as epoxides, aldehydes, succinimidyl esters or isothiocyanates, which react with nucleophilic groups (e.g., amino, thiol or hydroxyl groups) of amino acid residues. Covalent immobilization via random attachment also tends to denature arrayed proteins.
Affinity interaction by specific tags provides a means of immobilizing proteins in a defined orientation on a tag-capture surface, often retaining full protein activity. Examples include the use of biotin, His6- and GST-tagged proteins binding to streptavidin, Ni-NTA and glutathione-coated slides, respectively [3,26]. Proteins fused with a carbohydrate-binding module have been successfully immobilized on a cellulose-coated surface [27]. Affinity-capture surfaces are also capable of immobilizing tagged proteins directly from complex mixtures without prior purification [22]. Recently, an enzymatic method has been described for covalent immobilization of tagged proteins directly from cell lysates [28].
The use of 3D matrices (hydrogels) to entrap proteins into a structured environment maintains the function of arrayed proteins. Usually, a layer of a polymer, such as polyacrylamide, agarose or gelatine, covers the slide and provides a porous structure filled with an aqueous buffer. The matrix can be additionally modified with functional groups for covalent or affinity immobilization of proteins. Gel matrices permit the arraying of enzymes, while conserving their activities [29].
A systematic study comparing eight different commercially available array surfaces, combined with detection by different fluorescent dyes, has revealed critical parameters affecting the reproducibility and reliability of protein microarrays [30].
Arraying of proteins onto solid surfaces
Robotic spotting of proteins is generally carried out by either contact printing with solid or split metal pins, or via noncontact delivery in an ink-jet printer-like fashion. Unlike arraying of DNA samples, procedures for spotting functional proteins must ensure that they are hydrated, stable and folded during the process. However, the distributed volumes of solutions are typically in the subnanoliter range and the spots are therefore prone to drying rapidly by evaporation. Buffers for protein arraying therefore need to combine physiological conditions and compatibility with both the surface-immobilzation mechanism and the robotic-arraying process in terms of viscosity and surface tension. Typical buffer additives for protein stabilization against drying are hydroxylated cryoprotectants (e.g., glycerol, polyethylene glycol, trehalose and sucrose), but their effectiveness must be evaluated with the surface of choice [31,32]. Several new non-nucleophilic substances have recently been identified as buffer additives that improve spot morphology without interfering with immobilization [33]. An innovative method for preventing droplet evaporation was demonstrated by spotting under a layer of water-immiscible liquid fluorocarbon, coupled with a liquid-bridge, noncontact spotting technology [34]. Depending on the types of slides and the viscosity of the buffer used, printing density can generally be up to 20,000 individual features on a standard microscope slide, corresponding to a center-to-center spacing of spots in the range of 250 µm [35].
In situ synthesis
To avoid separate expression and purification of individual proteins by cell-based methods, cell-free expression has been exploited to produce protein microarrays by synthesising proteins directly onto the surface from arrayed DNAs (Figure 2). A number of different methods have been developed [36].
Figure 2. Methods for in situ synthesis of protein microarrays.
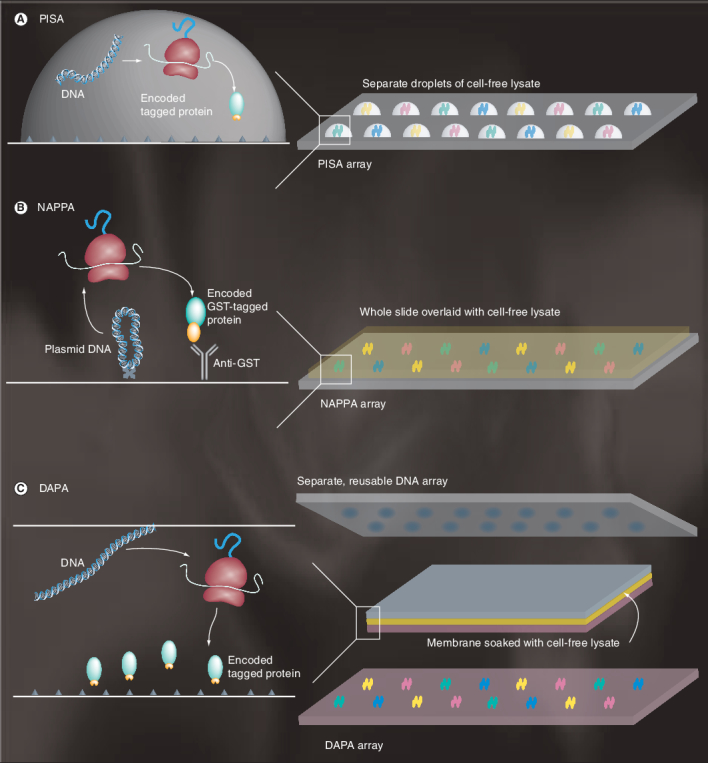
(A) PISA. (B) NAPPA. (C) DAPA.
DAPA: DNA array to protein array; GST: Glutathione-S-transferase; NAPPA: Nucleic acid programmable protein array; PISA: Protein in situ array.
Protein in situ array
In the protein in situ array (PISA) method, PCR DNA constructs and the cell-free expression system are codistributed onto protein-capture surfaces, so that newly synthesized proteins are captured in situ and unbound lysate material can be washed away (Figure 2A)[5]. PISA first demonstrated the advantages of ‘on-chip’ synthesis of proteins, notably the ability to convert DNA directly into functional proteins in an array format, without separate E. coli cloning, expression and purification. PISA also avoids the problem of maintaining protein stability on the array surface in long-term storage, since the proteins are only expressed as and when the protein array is needed.
The PISA concept was first exemplified using an engineered double-His6 tag, which permits single-step immobilization of the tagged proteins onto Ni-NTA-coated surfaces from the cell-free lysate [5,37]. A highly miniaturized version, based on a multiple spotting technique (MIST), has been developed, allowing in situ synthesis of proteins on the surface in subnanoliter volumes (350 pl). This makes it possible to produce protein microarrays with a potential density of up to 13,000 spots per slide [38].
Nucleic acid programmable protein array
Instead of codistributing soluble DNA and cell-free lysate as in PISA, the nucleic acid programmable protein array (NAPPA) method creates a protein array from immobilized DNA templates [4,39]. Plasmid DNA is immobilized together with a protein-capturing antibody onto a glass slide, which is then covered with a rabbit reticulocyte lysate cell-free system to express the proteins. The newly synthesized proteins become trapped by the antibody colocalized at each spot (Figure 2B). NAPPA has been applied to produce protein arrays (up to 2000 spots per slide) identifying immune-response signatures of breast cancer autoantibodies in patient sera [39,40] and to isolate target proteins with agonist activities from a cDNA library [41]. As the proteins immobilized by NAPPA remain on the same surface as the DNA array, the technology does not generate a ‘pure’ protein array, but rather one in which individual proteins are colocalized with both their encoding DNA and the general capture reagent.
DNA array to protein array
Recently, a novel system has been introduced that converts a reusable DNA array into multiple protein array copies (Figure 2C)[3]. Unlike NAPPA, the DNA array to protein array method generates pure protein arrays on a separate surface. Cell-free protein synthesis is carried out within a membrane filter sandwiched between two glass slides. One of the slides carries an array of immobilized PCR DNA fragments encoding tagged proteins, while the other slide is coated with a specific tag-capture reagent. Individual proteins are synthesized in parallel from the DNA template array, diffuse through the membrane filter and become immobilized on the capture surface, creating a protein array corresponding to the DNA array (but with a mirror-image layout). Fully consistent with the concept that diffusion is the mechanism of protein transport to the capture slide, the protein-density profiles of DAPA protein spots are well-defined Gaussian distributions. DAPA is capable of producing at least 20 copies of the same protein array by the repeated use of a single DNA array template [3]. This technique may find particular use for the repeated printing of protein arrays in laboratories that do not have access to microarray spotters. DAPA has been validated by arraying a number of different proteins, including antibody fragments, green fluorescent protein and transcription factors [3].
In situ puromycin capture from mRNA arrays
A puromycin-capture strategy has been described to fabricate protein arrays by capturing nascent polypeptides in situ[42]. It requires extra steps to make mRNA–ssDNA hybrid molecules, and protein yields are limited by the amount of mRNA spotted.
Detection
Readout technologies to detect interactions of probes on protein microarrays can be grouped into label-based and label-free methods. To date, approaches based on fluorescent labeling are the most widely used, with fluorophores either directly attached to the interactors or introduced on secondary reagents, such as in sandwich-assay formats. Label-free approaches, by contrast, measure an inherent property of the interactors themselves, such as mass or dielectric properties. Fluorescence-based methods use widely available reagents and instrumentation, but the covalent introduction of fluorophores may affect the protein function or epitope being monitored. Label-free technologies circumvent this problem and may allow real-time monitoring, but they require more sophisticated equipment. To date, the sensitivity of the routinely used label-based methods is higher than that of label-free approaches [43].
Label-based methods
Current microarray scanners are typically equipped for fluorophore excitation at 532 and 633 nm, allowing dual color fluorescence detection in the Cy3 and Cy5 ranges. Scanners with a wider range of excitation wavelengths, including 488 nm suitable for GFP-fusion proteins, are also becoming increasingly available.
Direct labeling of proteins for interaction screens is usually by covalent conjugation of N-hydroxysuccinimide ester-linked fluorescent dyes to primary amines of the protein N-terminus or ε-NH2 of lysine residues. Alternatively, thiol-reactive dyes are used to couple to cysteine residues. These methods can be applied to purified proteins as well as to complex mixtures (e.g., sera). For sample comparison, ratiometric two-color labeling strategies are used, analogous to those used for expression profiling on DNA microarrays [9,44]. Random chemical conjugation of dyes may interfere with protein function.
Cell-free expression systems provide alternatives for site-directed introduction of fluorophores at the protein N- or C-terminus. Using a modified initiator tRNA, the N-terminal methionine can be replaced by a fluorophore–amino acid conjugate, with labeling efficiencies of 67% in an E. coli cell-free system [45]. C-terminal labeling proceeds through the incorporation of fluorophore-coupled puromycin derivatives, with labeling efficiencies of up to 90% [46,47].
Besides direct coupling of fluorophores, primary and secondary immunofluorescent staining protocols are often applied [48,49]. In antibody arrays, such sandwich-detection strategies provide a second level of target recognition, significantly increasing the specificity and sensitivity (down to picogram per milliliter) of target detection. Enhanced sensitivity can also be achieved with a number of amplification systems. Tyramide signal amplification uses peroxidase-coupled reagents to catalyze the coupling of reactive fluorophore derivatives to tyrosine residues in the vicinity [3,50] and can detect femtogram amounts of protein. DNA-based technologies, such as rolling circle amplification (RCA) [10,51–53], as well as proximity ligation of oligonucleotides coupled to antibodies recognizing different epitopes [54,55], provide high levels of amplification, essentially converting from protein to DNA detection. Quantum dots are emerging as the new-generation fluorophores for ultrasensitive readout of binding events [13,56]. Recently, single-walled carbon nanotubes coupled to antibodies were used as multicolor Raman labels for the highly sensitive detection of proteins arrayed on Raman-scattering gold surfaces [57]. It is likely that novel combinations of existing tools will continue to enhance the detection sensitivity of protein microarrays.
Label-free strategies
Current label-free methods applicable to array formats include mass spectrometry (MS), optical biosensors on metal surfaces (e.g., surface plasmon resonance [SPR] and nanohole arrays) and conductance biosensors on carbon nanotube and nanowire surfaces [43].
Mass spectrometry offers intriguing possibilities through its ability to directly identify proteins, and is ‘hypothesis-free’ compared with identification via the use of specific antibodies. The direct interfacing of 3D polymer-based protein arrays to MALDI-MS for the detection of antigen–antibody interactions and enzymatic activities has been demonstrated [58]. Porous silicon surfaces structured with arrays of nanovials have been applied for rapid tryptic digestion of biomarker proteins, subsequently identified by MALDI [59].
Biosensor-based approaches detect the binding of a protein to a sensor surface by virtue of the change of the local refractive index or conductivity on the surface. They typically enable the detection of real-time interactions to determine association and dissociation kinetics [60]. Based on the principle of SPR, grating-coupled SPR [61] allows higher degrees of multiplexing, with up to 400 arrayed elements [62]. A recently developed interferometric biosensor, based on optical-phase differences due to molecular associations, allows for real-time kinetics with picogram per square millimeter sensitivity [63]. Nanohole-array sensors utilize the optical properties of light transmission through 150–200-nm diameter holes in metal surfaces, which are susceptible to changes in the local refractive index brought about by molecular interactions [64]. Biosensors of carbon nanowires and nanotubes, which detect changes in conductivity on the surface upon binding of molecules, can also be used to detect molecular interactions with proteins, small molecules and nucleic acids [65,66].
Applications
Screening molecular interactions
Interactions of proteins, both with other proteins and with nonprotein partners (e.g., nucleic acids, small molecules and lipids), form the basis of many cellular pathways. Global identification of these interactions greatly contributes to our understanding of signal transduction and its aberration in disease, which, in turn, facilitates target discovery and drug design. With regard to identifying protein interactions and networks, protein microarrays will not generally be the sole definitive technique, but are likely to be complementary and confirmatory to a range of other methods, from isolation and characterization of protein complexes by pulldown and MS, to yeast two-hybrid screening and similar in vivo systems. Nevertheless, protein arrays provide the benefit of studying defined individual protein components under controllable conditions.
Protein–protein interactions
Functional proteome arrays provide a high-throughput system for exploring protein interaction networks, as first exemplified by the landmark application of a yeast proteome array [26]. One-on-one screening of interactions by functional protein microarrays often identifies novel interactions. For example, screening of 49 arrayed coiled-coil domains from human leucine zipper transcription factors against each of the domains in their soluble form led to the discovery of a number of previously unknown pairwise interactions. Their relative binding strengths, as determined on the array surface, were also in good agreement with those from solution-based studies [67]. In another study based on in situ arraying by NAPPA, a protein functional array containing 29 proteins involved in DNA replication initiation was probed with each of these 29 proteins in turn. A total of 110 pairwise interactions were identified, of which 63 were previously unknown [4]. Novel interactions with calmodulin and calmodulin-related proteins in the presence of calcium have been identified through the use of high-density functional protein microarrays prepared from 1133 open reading frames of Arabidopsis spp.[68]. The regulatory functions of the proteins for a wide range of targets and cellular activities were revealed. Unknown interactions were also identified directly from cell lysates using arrays of functional protein-interaction domains, such as SH2, SH3, PDZ, WW, FF, PH, FHA and 14.3.3 [24]. In an approach based on parallel capture of native protein complexes from lysates of differentially stimulated T cells, peptide microarrays were used for the analysis of signaling-dependent protein-interaction networks [48]. Functional arrays of nuclear hormone receptors and coactivators were used for the systematic detection of receptor dimerization, functional classification of recruited coactivators and pharmacological characterization of receptor ligands [69]. Recently, an interesting example has demonstrated the use of human protein microarrays for identifying polyanion-binding proteins [70].
Protein–DNA/RNA interactions
Arrayed proteins, such as transcription factors, can be probed with DNA or RNA, enabling simultaneous identification of both the nucleic acid-binding proteins and their target sequences [71]. The first proteomic array employed for the analysis of DNA-binding events contained 5800 yeast proteins. After screening with fluorescently labeled genomic ssDNA or dsDNA, 273 proteins that bound at least one of the DNA forms were identified [26]. In another assay, genomic DNA from Saccharomyces cerevisiae was used to probe proteome chips, identifying 200 DNA-binding proteins, including known transcription factors. This report also identified a novel DNA-binding protein with a metabolic enzyme activity directly regulating eukaryotic gene expression [72]. Recently, commercial protein microarrays have been used for the large-scale search of protein–DNA interactions [73]. A microarray composed of 282 known and predicted yeast transcription factors detected numerous pairings of transcription factors and their binding DNA sequences. Target genes for one previously uncharacterized DNA-binding protein were identified and shown to be involved in the stress response and oxidative phosphorylation [74]. The sequence-specific binding to DNA of two bacterial transcription factors (ArgR repressor protein and cAMP-binding protein) and the DNA-replication protein (DnaA) have also been demonstrated on protein microarrays [75]. Using 33P-labeled DNA as the probe, p53 DNA-binding variants were identified on functional arrays, and their relative affinities were simultaneously measured [76].
Protein–lipid interactions
A screen for protein–lipid interactions was performed on functional yeast proteome arrays [26]. Probing with several phosphoinositides pointed to six new proteins associating with signaling lipids.
Protein–small molecule interactions
The first example of using a protein array to analyze protein–small molecule interactions was between FKBP12 and three fluorophore-coupled small molecules, showing differences in the affinity of their interactions [77]. Binding of GTP to GTP-binding protein on functional arrays has also been demonstrated [78]. A significant finding was that three different G-protein-coupled receptors arrayed together with their associated membrane lipids retained high-affinity, selective interactions with small-molecule ligands, demonstrating the functionality of arrayed membrane proteins [79]. Screening of yeast protein microarrays with small molecules led to the discovery of a target protein required for the suppression of rapamycin growth arrest [80]. It is anticipated that, in the future, functional protein arrays will be further explored for screening a variety of biological and synthetic small molecules for their potential as drug candidates [81].
Profiling & quantification of protein expression
Antibody and antigen arrays are ideal ‘capture’ tools for profiling protein expression or immune responses, with applications in diagnostics and biomarker discovery.
Antibody arrays
Arrays comprising large numbers of immobilized antibodies of different specificities are a sensitive means for qualitative and quantitative determination of protein expression [9,82–84]. Antibody arrays can be used to capture specific proteins from a variety of biological samples (e.g., lysates of cultured cells, tissue lysates and body fluids), enabling the identification of possible diagnostic signatures of disease-associated proteins [10,85,86], and have been applied with success to cancer studies. A recombinant antibody (scFv) microarray allowed profiling of proteins from the sera of pancreatic adenocarcinoma patients, producing a protein signature of 19 nonredundant proteins that could discriminate cancer patients from healthy individuals [87]. In a further example of antibody array application to biomarker discovery, 29 out of 378 proteins were identified as having significant changes in their expression profiles in biopsies of human squamous cell lung cancer carcinoma compared with normal tissues [88]. The recent development of antibody microarrays in suspension using color-coded beads offers a fast, flexible, sensitive and multiplexed means for screening large numbers (>200) of clinical serum samples [89].
The content of antibody arrays, in terms of their range and quality, is a critical issue with regard to their applications. Antibody arrays used in protein profiling must recognize the native conformations. Antibodies have different affinities for their specific targets, so the analysis of ligand binding in parallel and on the same surface may require the optimization of antibody concentrations prior to spotting. The concentration range of the target proteins and the conditions under which different ligands bind may also require optimization of buffers. In addition, cross-reactivity of antibodies can cause a variable background. Protein mutations, modifications and epitope changes may result in the failure of antibody arrays to detect these proteins. To help reduce these problems, the ‘Antibodypedia’ database has been established, providing details of individual antibody validation from publicly available resources [90].
Antigen & reverse-phase arrays
Antigen arrays are used to profile antibody responses in autoimmunity, cancer, infection or following vaccination. Proteome-wide antigen arrays generated through cell-free synthesis of 185 proteins from vaccinia virus [91,92] or 1700 proteins from Francisella tularensis have been used to study humoral immune responses to infection with these agents [93]. They produced specific antibody profiles in sera from vaccinated or infected humans and animals that, in turn, facilitated the rational design of vaccines and diagnostic antigens [94]. Other examples are the identification of immunodominant antigens from Yersinia pestis[95] and potential vaccine candidates from Plasmodium falciparum (a prominent malaria pathogen) proteins [96].
Profiling autoantibodies has an important potential for biomarker discovery, with further uses in diagnostics and therapy selection. For example, the patterns of antibody reactivity in multiple sclerosis sera identified by antigen microarrays, comprising heat-shock proteins, lipid autoantigens and CNS proteins, distinguished as different forms of the disease [97]. A remarkable ‘myelin proteome’ antigen microarray has been used to identify ‘epitope spreading’ through profiling the evolution of autoantibody responses that, in turn, guided the design of tolerizing DNA vaccines for encephalomyelitis [98]. To address autoimmune events associated with neurological disorders, cerebrospinal fluid, rather than serum, has also been studied on antigen microarrays [99]. As well as in the established autoimmune diseases themselves, profiling autoantibodies may be a future means of cancer diagnosis, and the antigens identified may also be potential immunotherapeutics [100]. A conformational protein array containing 329 folded and functional autoantigens was capable of detecting serum antibody profiles from non-small-cell lung cancer patients [101]. Reverse-phase antigen arrays, prepared by directly spotting various dilutions of lysates from 60 human cancer cell lines, have led to the identification of two promising pathological markers distinguishing colon from ovarian adenocarcinomas [50]. Reverse-phase arrays allowed profiling of protein levels from as few as three cell protein equivalents in samples of primary leukemia and hematopoetic stem cells [102].
Antigen arrays are also an ideal method for profiling specificity and cross-reactivity of affinity reagents such as antibodies. A proteome microarray comprising 5000 different yeast proteins was used to screen 11 polyclonal and monoclonal antibodies for a comparative profiling of antibody crossreactivity [103]. A commercial human-protein array comprising approximately 10,000 recombinant proteins has been found to be particularly useful for the rapid characterization of antibody specificity [104].
Studying functional protein pathways
The activity of signal transduction pathways can be measured using arrays by determining the amounts and activation state of the signaling proteins downstream of receptors. This was demonstrated for the ErbB receptor tyrosine kinase pathway using lysates of human tumor cell lines and antibody microarrays. This approach also enabled the characterization of the inhibitory profile of a small-molecule inhibitor on the pathway level [105]. To facilitate our understanding of pathways and their changes in disease, the effects of drugs and growth factors on the PTMs of specific proteins in normal and tumor cells have been studied by antibody microarrays [106,107]. A reverse-phase microarray approach has been successfully applied to discover distinct pathway-activation differences among breast cancer cell lines, demonstrating the use of array-based analysis for multiple pharmacodynamic biomarkers responding to targeted kinase inhibitors [108]. Additional signaling pathways or defects can be ascertained by exposing tumor cells or other diseased tissues to cytokines, chemokines or other bioactive molecules, using antibody arrays [85] or reverse-phase arrays [102]. In the future, it should be possible to reconstitute the complex pathways connecting the genome and the proteome in normal and diseased states through combining protein microarrays with genomic-profiling technologies. However, undertaking multiplexed quantitative analysis of protein–ligand interactions on microarrays, even with purified individual domains of the same functional families, requires careful standardization to avoid pitfalls due to dissimilar behavior of the proteins and variable densities of active proteins. These considerations and the danger of relying on single concentration determinations have been discussed with respect to binding of phosphopeptides by an arrayed set of SH2 domains [2].
Monitoring post-translational modifications
Protein microarrays provide a high-throughput method for analysis of PTMs such as glycosylation and phosphorylation.
Protein glycosylation
Protein microarrays allow large numbers of different glycoproteins to be studied in a single experiment [109]. Currently, lectin microarrays are used for high-throughput identification of protein glycosylation [110], while glycoprotein microarrays can be probed with a variety of lectins in turn to reveal glycosylation patterns in sera from healthy controls and patients with chronic pancreatitis and pancreatic cancer [111].
Protein phosphorylation
While phosphorylation controls much of the protein activity in living cells, linking particular kinases with specific substrates has been problematic. With functional protein microarrays, novel kinase substrates have been identified. For example, an array of 1690 proteins from Arabidopsis thaliana was screened with MPK3 and MPK6, respectively, followed by antiphosphotyrosine antibodies, leading to the identification of 48 and 39 candidate substrates for MPK3 and MPK6, respectively [112]. In another study illustrating the power of functional yeast proteome arrays, substrate screening identified candidate targets for each of the 87 yeast kinases [113]. Such discovery approaches are complemented by reverse-phase and antibody microarrays for the parallel analysis of known protein phosphorylations. Using reverse-phase microarrays, site-specific phosphorylation of 62 signaling proteins from IL-2 -stimulated T cells was detected, revealing differential protein phosphorylation [114]. Antibody microarrays have been employed for the time-resolved monitoring of cell-signaling pathways by quantifying both the amount and the phosphorylation status of signaling proteins [115].
Enzyme arrays
Activities of phosphatases, cysteine proteases and serine hydrolases have been studied on array surfaces [116] and by using suicide inhibitors, which attach covalently to the active site [117]. Sulfation reactions can also be studied on a chip surface [118]. Significantly, enzymes constituting an entire functional pathway can be identified by enzyme arrays. This has been achieved by immobilizing mixtures of enzymes in different ratios on an array surface. Activities and efficiencies of different enzymes were analyzed by product detection. Two-step sequential reactions, catalyzed by either luciferase or nucleoside diphosphate kinase, and a five-step pathway for trehalose synthesis, have been demonstrated [119]. This approach provides a means of optimizing enzyme proportions for maximal pathway-output efficiency. An interesting application using metabolizing enzyme arrays for toxicologic screening of drugs has been described with arrayed P450 isozymes [120]. After exposing the array to a candidate drug, a cell monolayer was grown on the P450 drug array and analyzed for the proportion of living and dead cells.
Epitope mapping & vaccine development
Capture microarrays provide suitable systems for mapping protein epitopes recognized by antibodies or other affinity reagents. By arraying all overlapping peptides of defined length taken from a protein sequence, linear epitopes with the minimal sequence requirement for molecular recognition can be identified. Individual amino acids within an epitope can be assessed for their contribution through the arraying of protein mutants. A library-scanning strategy was described in which each individual position in a 25-mer peptide was randomized by the other 19 amino acids to generate 25 peptide libraries [121]. All libraries were then spotted and probed by the antibody. The detected signals indicated the role of particular residues in epitope recognition.
Identification of protective epitopes is essential for the design of effective vaccines. As there is no in silico method that can accurately predict the target proteins or epitopes that stimulate protective antibody immunity, screening with protein microarrays is useful for identifying immunodominant antigens from complex pathogens. In one such example, the immunogenic epitopes of the SARS coronavirus were identified after screening 52 human-sera samples from infected individuals on capture arrays of viral proteins [122,123].
Expert commentary
Although a variety of array formats are increasingly used in basic research, biotechnology and medical applications, protein microarrays have not yet reached their full potential, particularly in academic research, for a number of possible reasons. Current commercial array production methods are generally based on separate expression and purification of proteins; thus, they are prohibitively expensive, with costs of approximately US$1000 per slide. Typically, academic applications require customized arrays displaying proteins of interest rather than complete proteome arrays. Moreover, protein arrays for species other than humans and the most widespread model organisms are not commercially available. Another problem is their limited shelf life, especially for functional protein arrays. We expect that the in situ methods for protein-array generation will contribute to changing this situation, especially approaches such as PISA, NAPPA and DAPA, which offer cheaper and more flexible methods for academic laboratories. While small numbers of customized array slides can be economically produced through in situ synthesis when required, conventional methods that rely on spotting purified proteins are only cost effective if large batches of arrays are produced. The DAPA technique is particularly useful for laboratories without microarraying equipment as it can generate multiple copies of the same protein microarray using a single DNA array template. In current clinical applications, antigen microarrays and reverse-phase arrays are widely applied for profiling antibody repertoires from individual patient samples.
Five-year view
Protein microarray technology will continue to be optimized in the coming years. The growing collections of cloned genes or gene sequences will facilitate the generation of ‘proteome arrays’ for many species. Ongoing improvements in high-throughput protein expression and purification methods will enhance protein quality and quantity, increasing the functionality of arrayed proteins, with cell-free systems becoming a widely used alternative. Technologies for making protein arrays by in situ synthesis will improve and accelerate array generation. Further optimization of surface chemistries and structures, immobilization methods and readout technologies will allow for the generation of arrays with densities in the submicrometer scale.
In proteomic applications, important areas of use will include high-throughput functional in vitro screens of molecular interaction networks, while for medical applications, arrays will be developed for diagnostics and the identification of relevant biomarkers. The increasing availability of affinity reagents through international initiatives such as ProteomeBinders will provide exciting possibilities in profiling cancer proteomes, to monitor characteristic protein signatures in patient sera or tumor extracts, leading to earlier diagnosis and prognosis before and during therapy [201]. It is expected that the multidisciplinary collaboration of scientists from biotechnology, biochemistry, material sciences, physics and bioinformatics will ensure the continued development of robust and reliable protein microarrays, their widespread uptake and novel applications.
Key issues
• High-throughput protein production is desirable for making protein arrays.
• Arraying proteins in a defined orientation is a preferred approach, as it retains the protein activity on the array surfaces.
• While current array readouts are mostly based on fluorescence, label-free detection methods have particular areas of use.
• An emerging alternative to conventional protein arraying is the in situ synthesis and arraying of proteins by cell-free systems, as in the protein in situ array, nucleic acid programmable protein array and DNA array to protein array technologies.
• Functional protein microarrays are increasingly used for high-throughput screening of molecular interactions, monitoring post-translational modifications and analyzing protein functions.
• Antibody arrays are valuable in profiling protein expression, thereby enabling the identification of biomarkers. Antigen arrays are efficient tools for mapping protein epitopes and assist in the identification of vaccine candidates.
• Reverse-phase protein arrays are particularly useful for the screening of changes in the expression level of biomarkers and pathway targets involved in disease.
• Enzyme arrays provide a means for the large-scale analysis of enzymatic activities. Pathway output efficiency can be enhanced by controlling the enzyme proportions spotted onto the array surface.
Financial & competing interests disclosure
The Babraham Institute is an institute of the Biotechnology and Biological Sciences Research Council (BBSRC), UK. This review is an activity of the ProteomeBinders consortium, a Research Infrastructure Coordination Action supported by the EU 6th Framework Programme. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest •• of considerable interest
- 1.Pandey A, Mann M. Proteomics to study genes and genomes. Nature 405(6788), 837–846 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Gordus A, MacBeath G. Circumventing the problems caused by protein diversity in microarrays: implications for protein interaction networks. J. Am. Chem. Soc. 128(42), 13668–13669 (2006). [DOI] [PubMed] [Google Scholar]
- 3.He M, Stoevesandt O, Palmer EA, Khan F, Ericsson O, Taussig MJ. Printing protein arrays from DNA arrays. Nat. Methods 5(2), 175–177 (2008). [DOI] [PubMed] [Google Scholar]; •• Describes the DNA array to protein array technology for making protein arrays directly from arrayed DNA templates on demand, which is capable of generating multiple copies of the same protein array by repeated use of a single DNA array.
- 4.Ramachandran N, Hainsworth E, Bhullar B et al. Self-assembling protein microarrays. Science 305(5680), 86–90 (2004). [DOI] [PubMed] [Google Scholar]; •• Describes the nucleic acid programmable protein array procedure to convert a DNA array into a protein array.
- 5.He M, Taussig MJ. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilization (PISA method). Nucleic Acids Res. 29(15), E73–E73 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]; • First introduces the idea of in situ protein-array synthesis.
- 6.Merkel JS, Michaud GA, Salcius M, Schweitzer B, Predki PF. Functional protein microarrays: just how functional are they? Curr. Opin. Biotechnol. 16(4), 447–452 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Zhu H, Snyder M. Protein chip technology. Curr. Opin. Chem. Biol. 7(1), 55–63 (2003) [DOI] [PubMed] [Google Scholar]
- 8.Dietrich HR, Knoll J, van den Doel LR et al. Nanoarrays: a method for performing enzymatic assays. Anal. Chem 76(14), 4112–4117 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Hamelinck D, Zhou H, Li L et al. Optimized normalization for antibody microarrays and application to serum-protein profiling. Mol. Cell. Proteomics 4(6), 773–784 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon-Cardo C. Profiling bladder cancer using targeted antibody arrays. Am. J. Pathol. 168(1), 93–103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann M, Schrenk M, Döttinger A et al. Expanding assay dynamics: a combined competitive and direct assay system for the quantification of proteins in multiplexed immunoassays. Clin. Chem. 54(6), 956–963 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Paweletz CP, Charboneau L, Bichsel VE et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene 20(16), 1981–1989 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Geho D, Lahar N, Gurnani P et al. Pegylated, steptavidin-conjugated quantum dots are effective detection elements for reverse-phase protein microarrays. Bioconjug. Chem. 16(3), 559–566 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Janzi M, Odling J, Pan-Hammarstrom Q et al. Serum microarrays for large scale screening of protein levels. Mol. Cell. Proteomics 4(12), 1942–1947 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Graslund S, Nordlund P, Weigelt J et al. Protein production and purification. Nat. Methods 5(2), 135–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goshima N, Kawamura Y, Fukumoto A et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5(12), 1011–1017 (2008). [DOI] [PubMed] [Google Scholar]; •• Reports the use of a cell-free system to express the human proteome. In total, 33,275 clones have been constructed, of which 13,364 were tested by a cell-free, wheatgerm expression system.
- 17.Waldo GS. Genetic screens and directed evolution for protein solubility. Curr. Opin. Chem. Biol. 7(1), 33–38 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Lesley SA, Graziano J, Cho CY, Knuth MW, Klock HE. Gene expression response to misfolded protein as a screen for soluble recombinant protein. Protein Eng. 15(2), 153–160 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat. Biotechnol. 19(2), 131–136 (2001). [DOI] [PubMed] [Google Scholar]
- 20.He M. Cell-free protein synthesis: applications in proteomics and biotechnology. N. Biotechnol. 25(2–3), 126–232 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Braun P, Hu Y, Shen B et al. Proteome-scale purification of human proteins from bacteria. Proc. Natl Acad. Sci. USA 99(5), 2654–2659 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinhauer C, Wingren C, Khan F, He M, Taussig MJ, Borrebaeck CA. Improved affinity coupling for antibody microarrays: engineering of double-(His)-6-tagged single framework recombinant antibody fragments. Proteomics 6(15), 4227–4234 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Cretich M, Damin F, Pirri G, Chiari M. Protein and peptide arrays: recent trends and new directions. Biomol. Eng. 23(2–3), 77–88 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. A protein-domain microarray identifies novel protein-protein interactions. Biochem. J. 367(Pt 3), 697–702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateo C, Fernandez-Lorente G, Abian O, Fernandez-Lafuente R, Guisan JM. Multifunctional epoxy supports: a new tool to improve the covalent immobilization of proteins. Biomacromolecules 1(4), 739–745 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Zhu H, Bilgin M, Bangham R et al. Global analysis of protein activities using proteome chips. Science 293(5537), 2101–2105 (2001). [DOI] [PubMed] [Google Scholar]; •• Ground-breaking paper that reports the first use of a yeast proteomic array for large-scale screening and identification of calmodulin and phospholipid-binding proteins.
- 27.Ofir K, Berdichevsky Y, Benhar I et al. Versatile protein microarray based on carbohydrate-binding modules. Proteomics 5(7), 1806–1814 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Wong LS, Thirlway J, Micklefield J. Direct site-selective covalent protein immobilization catalyzed by a phosphopantetheinyl transferase. J. Am. Chem. Soc. 130(37), 12456–12464 (2008). [DOI] [PubMed] [Google Scholar]; • Recent report describing the covalent immobilization of tagged proteins on solid surfaces from cell lysates.
- 29.Kiyonaka S, Sada K, Yoshimura I, Shinkai S, Kato N, Hamachi I. Semi-wet peptide/protein array using supramolecular hydrogel. Nat. Mater. 3(1), 58–64 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Guilleaume B, Buness A, Schmidt C et al. Systematic comparison of surface coatings for protein microarrays. Proteomics 5(18), 4705–4712 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Wu P, Grainger DW. Comparison of hydroxylated print additives on antibody microarray performance. J. Proteome Res. 5(11), 2956–2965 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusnezow W, Jacob A, Walijew A, Diehl F, Hoheisel JD. Antibody microarrays: an evaluation of production parameters. Proteomics 3(3), 254–264 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Mace CR, Yadav AR, Miller BL. Investigation of non-nucleophilic additives for the reduction of morphological anomalies in protein arrays. Langmuir 24(22), 12754–12757 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann M, Sjodahl J, Stjernstrom M, Redeby J, Joos T, Roeraade J. Non-contact protein microarray fabrication using a procedure based on liquid bridge formation. Anal. Bioanal. Chem 393(2), 591–598 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Bertone P, Snyder M. Advances in functional protein microarray technology. FEBS J. 272(21), 5400–5411 (2005). [DOI] [PubMed] [Google Scholar]
- 36.He M, Stoevesandt O, Taussig MJ. In situ synthesis of protein arrays. Curr. Opin. Biotechnol. 19(1), 4–9 (2008). [DOI] [PubMed] [Google Scholar]; • Review article for in situ synthesis of protein microarrays.
- 37.Khan F, He M, Taussig MJ. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on Ni-nitrilotriacetic acid surfaces. Anal. Chem. 78(9), 3072–3079 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Angenendt P, Kreutzberger J, Glokler J, Hoheisel JD. Generation of high density protein microarrays by cell-free in situ expression of unpurified PCR products. Mol. Cell. Proteomics 5(9), 1658–1666 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Ramachandran N, Raphael JV, Hainsworth E et al. Next-generation high-density self-assembling functional protein arrays. Nat. Methods 5(6), 535–538 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson KS, Ramachandran N, Wong J et al. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J. Proteome Res. 7(4), 1490–1499 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rolfs A, Montor WR, Yoon SS et al. Production and sequence validation of a complete full length ORF collection for the pathogenic bacterium Vibrio cholerae Proc. Natl Acad. Sci. USA 105(11), 4364–4369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Uses an in situ procedure to produce high-density protein microarrays from the genome collection of open reading frame clones for Vibrio cholerae. After screening with cell lysate, the protein microarray identified Toll-like receptor agonists.
- 42.Tao SC, Zhu H. Protein chip fabrication by capture of nascent polypeptides. Nat. Biotechnol. 24(10), 1253–1254 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Ramachandran N, Larson DN, Stark PR, Hainsworth E, LaBaer J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 272(21), 5412–5425 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Miller JC, Zhou H, Kwekel J et al. Antibody microarray profiling of human prostate cancer sera: antibody screening and identification of potential biomarkers. Proteomics 3(1), 56–63 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Mamaev S, Olejnik J, Olejnik EK, Rothschild KJ. Cell-free N-terminal protein labeling using initiator suppressor tRNA. Anal. Biochem. 326(1), 25–32 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Nemoto N, Miyamoto-Sato E, Yanagawa H. Fluorescence labeling of the C-terminus of proteins with a puromycin analogue in cell-free translation systems. FEBS Lett. 462(1–2), 43–46 (1999). [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi J, Nemoto N, Sasaki T et al. Rapid functional analysis of protein–protein interactions by fluorescent C-terminal labeling and single-molecule imaging. FEBS Lett. 502(3), 79–83 (2001). [DOI] [PubMed] [Google Scholar]
- 48.Stoevesandt O, Köhler K, Wolf S, Andre T, Hummel W, Brock R. A network analysis of changes in molecular interactions in cellular signaling. Mol. Cell. Proteomics 6(3), 503–513 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Kattah MG, Alemi GR, Thibault DL, Balboni I, Utz PJ. A new two-color Fab labeling method for autoantigen protein microarrays. Nat. Methods 3(9), 745–751 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Nishizuka S, Charboneau L, Young L et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc. Natl Acad. Sci. USA 100(24), 14229–14234 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nallur G, Luo C, Fang L et al. Signal amplification by rolling circle amplification on DNA microarrays. Nucleic Acids Res. 29(23), E118 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schweitzer B, Roberts S, Grimwade B et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 20(4), 359–365 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mullenix MC, Wiltshire S, Shao W, Kitos G, Schweitzer B. Allergen-specific IgE detection on microarrays using rolling circle amplification: correlation with in vitro assays for serum IgE. Clin. Chem. 47(10), 1926–1929 (2001). [Google Scholar]
- 54.Melin J, Jarvius J, Larsson C, Soderberg O, Landegren U, Nilsson M. Ligation-based molecular tools for lab-on-a-chip devices. N. Biotechnol. 25(1), 42–48 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Gustafsdottir SM, Schallmeiner E, Fredrikkson S et al. Proximity ligation assays for sensitive and specific protein analyses. Anal. Chem. 345(1), 2–9 (2005) [DOI] [PubMed] [Google Scholar]
- 56.Ghazani AA, Lee JA, Klostranec J et al. High throughput quantification of protein expression of cancer antigens in tissue microarray using quantum dot nanocrystals. Nano. Lett. 6(12), 2881–2886 (2006) [DOI] [PubMed] [Google Scholar]
- 57.Chen Z, Tabakman SM, Goodwin AP et al. Protein microarrays with carbon nanotubes as multicolor Raman labels. Nat. Biotechnol. 26(11), 1285–1292 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Gavin IM, Kukhtin A, Glesne D, Schabacker D, Chandler DP. Analysis of protein interaction and function with a 3-dimensional MALDI-MS protein array. Biotechniques 39, 99–107 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Finnskog D, Jaras K, Ressine A et al. High-speed biomarker identification utilizing porous silicon nanovial arrays and MALDI-TOF mass spectrometry. Electrophoresis 27(5–6), 1093–1103 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Kanda V, Kariuki JK, Harrison DJ, McDermott MT. Label-free reading of microarray-based immunoassays with surface plasmon resonance imaging. Anal. Chem. 76(24), 7257–7262 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Homola J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 377(3), 528–539 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Unfricht DW, Colpitts SL, Fernandez SM, Lynes MA. Grating-coupled surface plasmon resonance: a cell and protein microarray platform. Proteomics 5(17), 4432–4442 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Ozkumur E, Needham JW, Bergstein DA et al. Label-free and dynamic detection of biomolecular interactions for high-throughput microarray applications. Proc. Natl Acad. Sci. USA 105(23), 7988–7992 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ji J, O’Connell JG, Carter DJ, Larson DN. High-throughput nanohole array based system to monitor multiple binding events in real time. Anal. Chem. 80(7), 2491–2498 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Cui Y, Wei Q, Park H, Lieber CM. Nanowire nanosensors for highly sensitive and selective detection of biological and chemical species. Science 293(5533), 1289–1292 (2001). [DOI] [PubMed] [Google Scholar]
- 66.Li Y, Yang HH, You QH, Zhuang ZX, Wang XR. Protein recognition via surface molecularly imprinted polymer nanowires. Anal. Chem. 78(1), 317–320 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300(5628), 2097–2101 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Popescu SC, Popescu GV, Bachan S et al. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl Acad. Sci. USA 104(11), 4730–4735 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim SH, Tamrazi A, Carlson KE, Katzenellenbogen JA. A proteomic microarray approach for exploring ligand-initiated nuclear hormone receptor pharmacology, receptor selectivity, and heterodimer functionality. Mol. Cell. Proteomics 4(3), 267–277 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Salamat-Miller N, Fang J, Seidel CW, Assenov Y, Albrecht M, Middaugh CR. A network-based analysis of polyanion-binding proteins utilizing human protein arrays. J. Biol. Chem. 282(14), 10153–10163 (2007). [DOI] [PubMed] [Google Scholar]
- 71.Sakanyan V. High-throughput and multiplexed protein array technology: protein–DNA and protein–protein interactions. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 815(1–2), 77–95 (2005). [DOI] [PubMed] [Google Scholar]
- 72.Hall DA, Zhu H, Zhu X, Royce T, Gerstein M, Snyder M. Regulation of gene expression by a metabolic enzyme. Science 306 (5695), 482–484 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Stansfield HE, Kulczewski BP, Lybrand KE, Jamieson ER. Identifying protein interactions with metal-modified DNA using microarray technology. J. Biol. Inorg. Chem. 14(2), 193–199 (2009). [DOI] [PubMed] [Google Scholar]
- 74.Ho SW, Jona G, Chen CT, Johnston M, Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc. Natl Acad. Sci. USA 103(26), 9940–9945 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Snapyan M, Lecocq M, Guevel L, Arnaud MC, Ghochikyan A, Sakanyan V. Dissecting DNA–protein and protein–protein interactions involved in bacterial transcriptional regulation by a sensitive protein array method combining a near-infrared fluorescence detection. Proteomics 3(5), 647–657 (2003). [DOI] [PubMed] [Google Scholar]
- 76.Boutell JM, Hart DJ, Godber BL, Kozlowski RZ, Blackburn JM. Functional protein microarrays for parallel characterisation of p53 mutants. Proteomics 4(7), 1950–1958 (2004). [DOI] [PubMed] [Google Scholar]
- 77.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science 289(5485), 1760–1763 (2000). [DOI] [PubMed] [Google Scholar]
- 78.Schweitzer B, Predki P, Snyder M. Microarrays to characterize protein interactions on a whole-proteome scale. Proteomics 3(11), 2190–2199 (2003). [DOI] [PubMed] [Google Scholar]
- 79.Fang Y, Frutos AG, Lahiri J. Membrane protein microarrays. J. Am. Chem. Soc. 124(11), 2394–2395 (2002). [DOI] [PubMed] [Google Scholar]
- 80.Huang J, Zhu H, Haggarty SJ et al. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl Acad. Sci. USA 101(47), 16594–16599 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salcius M, Michaud GA, Schweitzer B, Predki PF. Identification of small molecule targets on functional protein microarrays. Methods Mol. Biol. 382, 239–248 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Korf U, Henjes F, Schmidt C et al. Antibody microarrays as an experimental platform for the analysis of signal transduction networks. Adv. Biochem. Eng. Biotechnol. 110, 153–175 (2008). [DOI] [PubMed] [Google Scholar]
- 83.Pavlickova P, Schneider EM, Hug H. Advances in recombinant antibody microarrays. Clin. Chim. Acta 343(1–2), 17–35 (2004). [DOI] [PubMed] [Google Scholar]
- 84.Chaga GS. Antibody arrays for determination of relative protein abundances. Methods Mol. Biol. 441, 129–151 (2008). [DOI] [PubMed] [Google Scholar]
- 85.Sreekumar A, Nyati MK, Varambally S et al. Profiling of cancer cells using protein microarrays: discovery of novel radiation-regulated proteins. Cancer Res. 61(20), 7585–7593 (2001). [PubMed] [Google Scholar]
- 86.Li S, Sack R, Vijmasi T et al. Antibody protein array analysis of the tear film cytokines. Optom. Vis. Sci. 85(8), 653–660 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ingvarsson J, Wingren C, Carlsson A et al. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics 8(11), 2211–2219 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Bartling B, Hofmann HS, Boettger T et al. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer 49(2), 145–154 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Schwenk JM, Gry M, Rimini R, Uhlen M, Nilsson P. Antibody suspension bead arrays within serum proteomics. J. Proteome Res. 7(8), 3168–3179 (2008). [DOI] [PubMed] [Google Scholar]
- 90.Björling E, Uhlen M. Antibodypedia, a portal for sharing antibody and antigen validation data. Mol. Cell. Proteomics 7(10), 2028–2037 (2008). [DOI] [PubMed] [Google Scholar]
- 91.Davies DH, Liang X, Hernandez JE et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl Acad. Sci. USA 102(3), 547–552 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davies DH, Molina DM, Wrammert J et al. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 7(10), 1678–1686 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Eyles JE, Unal B, Hartley MG et al. Immunodominant Francisella tularensis antigens identified using proteome microarray. Proteomics 7(13), 2172–2183 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Davies DH, McCausland MM, Valdez C et al. Vaccinia virus H3L envelope protein is a major target of neutralizing antibodies in humans and elicits protection against lethal challenge in mice. J. Virol. 79(18), 11724–11733 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li B, Jiang L, Song Q et al. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect. Immun. 73(6), 3734–3739 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Doolan DL, Mu Y, Unal B et al. Profiling humoral immune responses to P. falciparum infection with protein microarrays. Proteomics 8(22), 4680–4694 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quintana FJ, Farez MF, Viglietta V et al. Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc. Natl Acad. Sci. USA 105(48), 18889–18894 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robinson WH, Fontoura P, Lee BJ et al. Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis. Nat. Biotechnol. 21(9), 1033–1039 (2003). [DOI] [PubMed] [Google Scholar]
- 99.Roche S, Dauvilliers Y, Tiers L et al. Autoantibody profiling on high-density protein microarrays for biomarker discovery in the cerebrospinal fluid. J. Immunol. Methods 338(1–2), 75–78 (2008). [DOI] [PubMed] [Google Scholar]
- 100.Chatterjee M, Draghici S, Tainsky MA. Immunotheranostics: breaking tolerance in immunotherapy using tumor autoantigens identified on protein microarrays. Curr. Opin. Drug Discov. Devel. 9(3), 380–385 (2006). [PubMed] [Google Scholar]
- 101.Gnjatic S, Wheeler C, Ebner M et al. Seromic analysis of antibody responses in non-small cell lung cancer patients and healthy donors using conformational protein arrays. J. Immunol. Methods 341(1–2), 50–8 (2009). [DOI] [PubMed] [Google Scholar]
- 102.Tibes R, Qiu Y, Lu Y et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 5(10), 2512–2521 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Michaud GA, Salcius M, Zhou F et al. Analyzing antibody specificity with whole proteome microarrays. Nat. Biotechnol. 21(12), 1509–1512 (2003). [DOI] [PubMed] [Google Scholar]; • First report using a proteome array comprising 5000 different yeast proteins to screen the specificity of antibodies.
- 104.Kijanka G, Ipcho S, Baars S et al. Rapid characterization of binding specificity and cross-reactivity of antibodies using recombinant human protein arrays. J. Immunol. Methods 340(2), 132–137 (2009). [DOI] [PubMed] [Google Scholar]
- 105.Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc. Natl Acad. Sci. USA 100(16), 9330–9335 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ivanov SS, Chung AS, Yuan ZL et al. Antibodies immobilized as arrays to profile protein post-translational modifications in mammalian cells. Mol. Cell. Proteomics 3(8), 788–795 (2004). [DOI] [PubMed] [Google Scholar]
- 107.Kopf E, Shnitzer D, Zharhary D. Panorama Ab Microarray Cell Signaling kit: a unique tool for protein expression analysis. Proteomics 5(9), 2412–2416 (2005). [DOI] [PubMed] [Google Scholar]
- 108.Boyd ZS, Wu QJ, O’Brien C et al. Proteomic analysis of breast cancer molecular subtypes and biomarkers of response to targeted kinase inhibitors using reverse-phase protein microarrays. Mol. Cancer Ther. 7(12), 3695–3706 (2008). [DOI] [PubMed] [Google Scholar]
- 109.Kung LA, Snyder M. Proteome chips for whole-organism assays. Nat. Rev. Mol. Cell. Biol. 7(8), 617–622 (2006). [DOI] [PubMed] [Google Scholar]
- 110.Pilobello KT, Slawek DE, Mahal LK. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Natl Acad. Sci. USA 104(28), 11534–11539 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhao J, Patwa TH, Qiu W et al. Glycoprotein microarrays with multi-lectin detection: unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J. Proteome Res. 6(5), 1864–1874 (2007). [DOI] [PubMed] [Google Scholar]
- 112.Feilner T, Hultschig C, Lee J et al. High throughput identification of potential Arabidopsis mitogen-activated protein kinases substrates. Mol. Cell. Proteomics 4(10), 1558–1568 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Ptacek J, Devgan G, Michaud G et al. Global analysis of protein phosphorylation in yeast. Nature 438(7068), 679–684 (2005). [DOI] [PubMed] [Google Scholar]; •• Reports a large-scale analysis of protein phosphorylation by the use of proteome chips. Over 4000 phosphorylation events, involving 1325 different proteins, were identified in a single screen.
- 114.Chan SM, Ermann J, Su L, Fathman CG, Utz PJ. Protein microarrays for multiplex analysis of signal transduction pathways. Nat. Med. 10(12), 1390–1396 (2004). [DOI] [PubMed] [Google Scholar]; •• Reports the first description of the use of reverse-phase protein microarrays to monitor site-specific phosphorylation of many signaling proteins from stimulated living cells.
- 115.Korf U, Derdak S, Tresch A et al. Quantitative protein microarrays for time-resolved measurements of protein phosphorylation. Proteomics 8(21), 4603–4612 (2008). [DOI] [PubMed] [Google Scholar]
- 116.Chen GY, Uttamchandani M, Zhu Q, Wang G, Yao SQ. Developing a strategy for activity-based detection of enzymes in a protein microarray. ChemBioChem 4(4), 336–339 (2003). [DOI] [PubMed] [Google Scholar]
- 117.Funeriu DP, Eppinger J, Denizot L, Miyake M, Miyake J. Enzyme family-specific and activity-based screening of chemical libraries using enzyme microarrays. Nat. Biotechnol. 23(5), 622–627 (2005). [DOI] [PubMed] [Google Scholar]; • Describes an interesting thematic enzyme-microarray format to characterize family-specific enzymes on array surfaces.
- 118.Cha T, Guo A, Zhu XY. Enzymatic activity on a chip: the critical role of protein orientation. Proteomics 5(2), 416–419 (2005). [DOI] [PubMed] [Google Scholar]
- 119.Jung GY, Stephanopoulos G. A functional protein chip for pathway optimization and in vitro metabolic engineering. Science 304(5669), 428–431 (2004). [DOI] [PubMed] [Google Scholar]; •• Reports the use of protein arrays to reconstitute an enzyme pathway, demonstrating that entire pathways can be studied by protein microarrays.
- 120.Lee MY, Park CB, Dordick JS, Clark DS. Metabolizing enzyme toxicology assay chip (MetaChip) for high-throughput microscale toxicity analyses. Proc. Natl Acad. Sci. USA 102(4), 983–987 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Poetz O, Ostendorp R, Brocks B et al. Protein microarrays for antibody profiling: specificity and affinity determination on a chip. Proteomics 5(9), 2402–2411 (2005). [DOI] [PubMed] [Google Scholar]
- 122.Lu DD, Chen SH, Zhang SM et al. Screening of specific antigens for SARS clinical diagnosis using a protein microarray. Analyst 130(4), 474–482 (2005). [DOI] [PubMed] [Google Scholar]
- 123.Chen Z, Pei D, Jiang L et al. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 50(6), 988–995 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 201.ProteomeBinders www.proteomebinders.org


