Abstract
Locked nucleic acid (LNA) is a nucleic acid analogue containing one or more LNA nucleotide monomers with a bicyclic furanose unit locked in an RNA mimicking sugar conformation. This conformational restriction results in unprecedented hybridization affinity towards complementary single stranded RNA and thus, makes LNA uniquely suited for mimicking RNA structures and sequence specific targeting of RNA in vitro or in vivo. The focus of this paper is on LNA-antisense, LNA-modified siRNA (siLNA), and detection and analysis of microRNAs by LNA-modified oligonucleotide probes.
Section editors:
Steve Gullans – RxGen, Inc., New Haven, CT, USA
Robert Zivin – Johnson and Johnson, New Brunswick, NJ, USA
Introduction
The future challenges in diagnostics and treatment of diseases by siRNA and antisense technologies call for novel, improved technologies for in vitro and in vivo targeting of RNA molecules. Locked nucleic acid (LNA) comprises a class of RNA analogues in which the furanose ring of the ribose sugar is chemically locked in an RNA-mimicking conformation by the introduction of a O2′,C4′-methylene linkage [1, 2]. Several studies have demonstrated that LNA-modified oligonucleotides exhibit unprecedented thermal stabilities when hybridized with their RNA target molecules [1, 2, 3, 4]. Thus, an increase in melting temperature (T m value) of +2 to +10°C per LNA monomer against complementary RNA compared to unmodified duplexes has been reported. Here, we describe some basic properties of LNA followed by recent examples of applications based on LNA-mediated RNA targeting in vitro or in vivo.
Hybridization properties of LNA
We have defined LNA as an oligonucleotide containing at least one LNA monomer, that is, one 2′-O,4′-C-methylene-β-d-ribofuranosyl nucleotide (Fig. 1 ) [1, 5]. The LNA monomers adopt an RNA mimicking N-type sugar conformation [5, 6]. The majority of reports on LNA-mediated RNA targeting have utilized mixmer LNA oligonucleotides, that is, LNAs containing a limited number of LNA monomers in combination with other types of monomers, typically DNA but also RNA or 2′-O-Me-RNA monomers. The fact that LNA phosphoramidites and LNA (fully modified or mixmers) oligonucleotides are commercially available, and the compatibility of these LNA constructs with standard modifiers and modifications like phosphorothioate linkages [7], allow use of LNAs in a wide range of applications.
Figure 1.
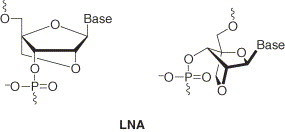
Two representations of the structure of LNA monomers. The key difference between DNA and LNA monomers is the oxymethylene bridge between the C2′ and C4′ atoms of the ribose ring. The resulting bicyclic sugar ring system imposes a locked RNA-like conformation of the ribose ring (as illustrated in the drawing to the right).
A key feature of oligonucleotides containing LNA nucleotides is the very high thermal stability of duplexes towards complementary RNA [1, 2, 3, 5, 7, 8]. Notably, this increase in affinity goes hand in hand with preserved or even improved, Watson-Crick base pairing selectivity. Examples of melting temperatures for LNAs complexed with RNA are shown in Fig. 2 . LNA:LNA base pairing is also very strong [9] (see http://www.exiqon.com/ for LNA design tools to avoid extensive self-complementarity). The largest affinity increase per LNA monomer and the optimum mismatch discrimination is achieved for short DNA oligonucleotides with a single or several dispersed LNA monomers.
Figure 2.

Some examples of melting temperatures (Tm values) for hybridization of LNA and DNA oligonucleotides to complementary RNA sequences [5, 33]. LNA monomers are red, DNA monomers are black and RNA monomers are green.
Susceptibility of LNA to nucleases
The RNA-targeting capacity of LNA makes it attractive for diagnostic and therapeutic applications, which implies contact with various nuclease-containing media. Complete stability against the 3′-exonuclease snake venom phosphodiesterase (SVPD) was reported for a fully modified LNA [10] while a significant increase in 3′-exonucleolytic stability was observed by blocking the 3′-end with two LNA monomers. In a study of oligonucleotide stability in serum, LNA mixmers were found to be very stable and LNA/DNA/LNA gapmers (see section below for definition of gapmers) to be significantly more stable than DNA alone [11]. Furthermore, Kurreck et al. [12] reported that DNA oligonucleotides with LNA nucleotides at the ends are more stable in human serum than the corresponding oligonucleotides composed of phosphothioate DNA gaps with 2′-O-methyl-RNA flanks. DNAse I endonuclease degradation of end-modified 30mer dsDNA showed that incorporation of one or two terminal LNA nucleotides ensures markedly increased stability [13]. From these studies it is clear that LNA nucleotides impose significant protection against nucleolytic degradation, especially if more than one LNA nucleotide is incorporated into an oligonucleotide.
RNase H-mediated cleavage of the RNA target strand of a heteroduplex formed with an antisense oligonucleotide is considered beneficial towards efficient gene silencing. Because only very few fully modified oligonucleotide analogues support RNase H-mediated cleavage, so-called gapmers, that is, oligonucleotides composed of modified segments flanking a central DNA (or phosphorothioate DNA) segment that is compatible with RNase H activity, is often used. Wahlestedt et al. [11] found that an LNA/DNA/LNA gapmer, with a six-nucleotide DNA gap, elicited substantial RNase H activity. However, no RNase H-mediated cleavage was observed with a fully modified 11mer LNA or with an 11mer LNA/DNA mixmer [14]. Kurreck et al. [12] and Elmén et al. [15] have investigated various LNA/DNA mixmers and gapmers and found that a gap of six DNA nucleotides is necessary for noteworthy RNase H activity, and that a gap of seven DNA nucleotides allows complete RNase H activity. In accordance herewith, Frieden et al. [16] concluded that a DNA gap size between 7 and 10 nucleotides is optimal for LNA/DNA/LNA antisense gapmers. The overall conclusion is that antisense LNA oligonucleotides can be designed to elicit RNase H activity while still containing LNA monomers for improved binding and target accessibility [17].
LNA-antisense for disease intervention
The term ‘antisense’ is generally used for nucleic acid based approaches that interferes, in a sequence selective way, with the processing of RNA from its transcription via mRNA to protein or with the effects of other forms of functional RNA. By virtue of their unprecedented RNA binding characteristics, oligonucleotides containing LNA nucleotides are obvious candidates for antisense-based gene silencing, and many previous successful LNA antisense studies have already been the subject of detailed reviews [4, 17].
The first in vivo antisense experiment with LNA-antisense involved two different LNA sequences targeting delta opioid receptor (DOR) mRNA in the central nervous system of living rats [11]. Upon direct injection of the antisense LNA oligonucleotides into the brain a dose-dependent and highly efficacious knockdown of DOR was induced both for an LNA/DNA mixmer and an LNA/DNA/LNA gapmer. In another in vivo study, a fully modified LNA, targeting the RNA polymerase II gene product, inhibited tumor growth in mice and appeared nontoxic at doses less than 5 mg/kg/day [18]. These studies indicate LNA to be a much more potent class of antisense agents than the corresponding phosphorothioate DNA oligomers.
Several studies in vitro or in cells support the usefulness of LNA-antisense for gene silencing (e.g. [19, 20]). LNA-antisense constructs targeting the RNA moiety of telomerase are potent and selective inhibitors [20] with IC50 values of 10 nM for a 13mer fully modified LNA and a 13mer LNA/DNA mixmer. By the introduction of two terminal phosphorothioate linkages, the potency was increased further by tenfold but this was accompanied by a decreased match versus mismatch discrimination. Notably, even very short 8mer fully modified LNAs are potent inhibitors (IC50 values of 2 and 25 nM with and without terminal phosphorothioate linkages, respectively). Also in cells upon employing transfection, the 13mer LNA oligomers induced inhibition of more than 80% of the telomerase activity. No alteration of cell morphology was observed seven days after transfection of 8mer LNAs [20]. These results suggest that short LNAs may provide adequate affinity and selectivity for biologically relevant RNA targeting in cells.
Viral RNA is another important target for LNA-antisense. The effects of different LNAs on the interactions of the HIV-1 trans-activation responsive element (TAR) have recently been published [21]. Binding of oligonucleotides to TAR can inhibit Tat-dependent transcription thereby blocking full-length HIV transcription and hence viral replication, and various LNA oligonucleotides were transfected into HeLa cells and derivatives with a minimum length of 12 residues showed 50% inhibition using nanomolar concentration of LNAs [21]. This study underlines the potential of LNA-antisense oligonucleotides for in vivo targeting of RNA using non-RNaseH dependent approaches. Elmén et al. [15] have demonstrated that LNA/DNA mixmers enhance the inhibition of HIV-1 genome dimerization relative to DNA oligonucleotides and have shown efficient uptake of the LNA/DNA mixmers in a T-cell line and that they can inhibit replication of a clinical HIV-1 isolate.
It is clear that LNA-antisense, either LNA/DNA/LNA gapmers for RNase H activation or LNA mixmers for RNase H independent activity, represents a favorable approach for gene silencing in vitro or in vivo, and reports indicate that LNA-antisense rivals siRNA as the method of choice [22]. Recently, a LNA-antisense drug directed against Bcl-2 expression to treat chronic lymphocytic leukemia has entered Phase I/II clinical trials (http://www.santaris.com/).
LNA-modified siRNA (siLNA)
The discovery of RNA interference (RNAi), in which double-stranded RNA leads to the degradation of homologous RNA [23], has drawn much attention because it mediates potent gene silencing. RNAi relies on a complex cellular mechanism that has probably evolved for protection against viral attack and mobile genetic elements. A crucial step in the RNAi mechanism is the generation of short interfering RNAs (siRNAs; double-stranded RNAs that are about 22 nt long). One study has directly compared the inhibitory effect of siRNA, LNA/DNA/LNA gapmers, phosphorothioate-DNA and 2′-O-methyl-RNA on the expression of vanilloid receptor subtype 1 in cells [22]. Both siRNA and LNA/DNA/LNA gapmers were found to be very efficient with the siRNA slightly more potent than the LNA/DNA/LNA gapmers.
siRNAs are themselves candidate molecules for incorporation of modified nucleotides for improved biostability and RNA targeting. LNA monomers have been shown to be tolerated by the RNAi machinery and to provide thermal stability [24]. The exact positioning and the overall number of LNA monomers will be very crucial for optimizing LNA-containing siRNA. To address this issue, Elmén et al. [25] systematically modified siRNA duplexes with LNA monomers and showed that siLNAs have substantially enhanced serum half-life compared to the corresponding siRNAs. Moreover, they provided evidence that the use of siLNAs reduce sequence-related off-target effects. In their study, Elmén et al. [25] reported on improved efficacy of siLNAs on certain RNA motifs relevant to targeting the SARS-CoV virus. These results emphasize the promise of siLNA in converting siRNA from a functional genomics technology into a therapeutic platform.
MicroRNA detection using LNA probes
MicroRNAs (miRNAs) are a novel class of short endogenous non-coding RNAs that act as post-transcriptional modulators of gene expression [26, 27]. To date more than 1650 miRNAs have been identified in invertebrates, vertebrates and plants. Growing evidence shows that miRNAs exhibit a wide variety of regulatory functions and exert significant effects on cell growth, development and differentiation.
Recent studies have shown that human miRNA genes are frequently located in cancer-associated genomic regions, and perturbed miRNA expression patterns have been observed in many human cancers [28, 29, 30]. These findings have emphasized the potential of miRNA detection and expression profiling in cancer diagnostics. However, the detection of mature miRNAs has been technically challenging because of their small size and therefore calls for robust and improved technologies. In a recent paper, Valoczi et al. [31] describe highly efficient detection of miRNAs by northern blot analysis using LNA-modified oligonucleotide probes and demonstrate their significantly improved sensitivity by detecting different miRNAs in animals and plants. Besides being efficient as northern probes, LNA-modified oligonucleotide probes have proven highly useful for in situ localization of miRNAs in cells and tissues. In a recent study, the temporal and spatial expression patterns of 115 conserved vertebrate miRNAs were determined in zebrafish embryos by in situ hybridizations using LNA-modified oligonucleotide probes [32]. Most miRNAs were expressed in a highly tissue-specific manner during segmentation and later stages, but not early in development. Thus these miRNAs may not be essential for tissue establishment but rather play crucial roles in the maintenance of tissue identity or in differentiation [32].
Conclusion
The remarkable hybridization properties of LNA, both with respect to affinity and specificity, position LNA as an enabling molecule for molecular biology research, biotechnology and RNA targeting in vivo. The facts that LNA phosphoramidites and LNA oligomers are commercially available and that LNA nucleotides can be freely mixed with, for example, DNA, RNA and 2′-O-Me-RNA monomers and standard probes, make LNA a highly flexible tool. LNA-antisense rivals siRNA for gene silencing, and targeting of non-coding RNAs, for example, microRNAs can be achieved very efficiently using LNA probes.
Acknowledgment
The Wilhelm Johannsen Centre for Functional Genome Research and the Nucleic Acid Center are funded by The Danish National Research Foundation.
References
- 1.Koshkin A.A. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 2.Obika S. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 3.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 4.Petersen M., Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 5.Singh S.K. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998;4:455–456. [Google Scholar]
- 6.Obika S. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine novel bicyclic nucleosides having a fixed C3′-endo sugar puckering. Tetrahedron Lett. 1997;38:8735–8738. [Google Scholar]
- 7.Kumar R. The first analogues of LNA (locked nucleic acids): Phosphorothioate-LNA and 2′-thio-LNA. Biorg. Med. Chem. Lett. 1998;8:2219–2222. doi: 10.1016/s0960-894x(98)00366-7. [DOI] [PubMed] [Google Scholar]
- 8.Wengel J. Synthesis of 3′-C- and 4′-C-branched oligonucleotides and the development of locked nucleic acid (LNA) Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 9.Koshkin A.A. LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc. 1998;120:13252–13253. [Google Scholar]
- 10.Frieden M. Nuclease stability of LNA oligonucleotides and LNA–DNA chimeras. Nucleosides Nucleotides Nucleic Acids. 2003;22:1041–1043. doi: 10.1081/NCN-120022731. [DOI] [PubMed] [Google Scholar]
- 11.Wahlestedt C. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc. Natl. Acad. Sci. U S A. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurreck J. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crinelli R. Design and characterization of decoy oligonucleotides containing locked nucleic acids. Nucleic Acids Res. 2002;30:2435–2443. doi: 10.1093/nar/30.11.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sørensen M.D. α-l-ribo-Configured locked nucleic acid (α-l-LNA): synthesis and properties. J. Am. Chem. Soc. 2002;124:2164–2176. doi: 10.1021/ja0168763. [DOI] [PubMed] [Google Scholar]
- 15.Elmén J. Locked nucleic acid containing antisense oligonucleotides enhance inhibition of HIV-1 genome dimerization and inhibit virus replication. FEBS Lett. 2004;578:285–290. doi: 10.1016/j.febslet.2004.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieden M. Expanding the design horizon of antisense oligonucleotides with α-l-LNA. Nucleic Acids Res. 2003;31:6365–6372. doi: 10.1093/nar/gkg820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jepsen J.S., Wengel J. LNA-antisense rivals siRNA for gene silencing. Curr. Opin. Drug Discov. Dev. 2004;7:188–194. [PubMed] [Google Scholar]
- 18.Fluiter K. In vivo tumor growth inhibition and biodistribution studies of locked nucleic acid (LNA) antisense oligonucleotides. Nucleic Acids Res. 2003;31:953–962. doi: 10.1093/nar/gkg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen J.B. Antisense knockdown of PKC-alpha using LNA-oligos. Nucleosides Nucleotides Nucleic Acids. 2003;22:1607–1609. doi: 10.1081/ncn-120023045. [DOI] [PubMed] [Google Scholar]
- 20.Elayadi A.N. Implications of high-affinity hybridization by locked nucleic acid oligomers for inhibition of human telomerase. Biochemistry. 2002;41:9973–9981. doi: 10.1021/bi025907j. [DOI] [PubMed] [Google Scholar]
- 21.Arzumanov A. A structure–activity study of the inhibition of HIV-1 tat-dependent trans-activation by mixmer 2′-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), α-l-LNA, or 2′-thio-LNA residues. Oligonucleotides. 2003;13:435–453. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]
- 22.Grünweller A. Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2′-O-methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res. 2003;31:3185–3193. doi: 10.1093/nar/gkg409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fire A. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 24.Braasch D.A. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;8:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 25.Elmén J. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambros V. MicroRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 27.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.Calin G.A. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Valoczi A. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wienholds E. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 33.Jacobsen N. Direct isolation of poly(A)# RNA from 4 M guanidine thiocyanate lused cell extracts using locked nucleic acid oligo(T) capture. Nucleic Acids Res. 2004;32:e64. doi: 10.1093/nar/gnh056. [DOI] [PMC free article] [PubMed] [Google Scholar]


