Abstract
In this review, we report a new reverse approach (from finding bioactive molecules to separating target compounds in the related plant) by using virtual screening, immobilized enzyme and polyclonal antibody, molecularly imprinted polymers for finding, and separating the active compounds from TCM. This approach is faster and more efficient than the traditional time-consuming approach (from selecting the plant to separating compounds following the bioassay guidance).
Section editors:
Li-He Zhang – School of Pharmaceutical Science, University of Peking, Beijing, China
Kaixian Chen – Drug Discovery and Design Center, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China
Introduction
Traditional Chinese medicine (tcm) (see Glossary) is rich in natural compounds and has been used in China for more than 8000 years [1]. TCM can be considered as a natural combinatorial chemical library, compared with synthetic ones. TCM presents more diversity in structure and bioactivity, and less toxicity [2]. Therefore, it represents an attractive source of new active compounds in drug discovery [3, 4]. The conventional approach to find active compounds in TCM involves selecting a potential plant and isolating compounds following bioassay guidance. This approach has been playing an important part in drug development. However, it is often time-consuming and can contain false positives (see Outstanding issues).
In this review, we report a reverse approach (from finding bioactive molecules to separating target compounds in the related plant) by using virtual screening, immobilized enzymes, polyclonal antibodies and molecularly imprinted polymers (MIP) to find and separate the active compounds from TCM quickly and efficiently.
Computer aided drug design applied to TCM
Computer-aided drug design (CADD) permeates all aspects of modern drug discovery and allows the discovery of new candidates with a desired biological activity more quickly and at a lower cost. The progress and applications of CADD have been presented in references [5, 6, 7].
Up till now, there have been very few articles on applications of CADD to TCM owing to the lack of related databases. Qiao et al. [8] have developed a TCM information system including TCM formulation database, a TCM plant database and a 3D structure database of TCM components (Fig. 1 ). The three databases are linked with each other. The 3D structure database contains 15,000 compounds isolated from TCM. It offers not the only basic molecular properties and optimized 3-D structure of compounds but also detailed information on their herb origin, including basic herbal category (e.g. English name, Latin name and family), effective parts, clinical effects and related formulations of TCM.
Figure 1.
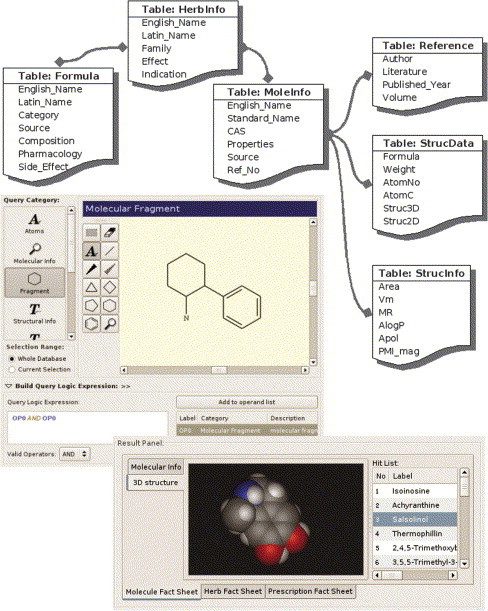
The Chinese Herbal Drug Database (CHDD). CHDD consists of three inter-related modules, compositional molecule (MoleInfo, StrucData and StrucInfo), herb (HerbInfo) and TCM formula (Formula). The upper-right illustrations depict logic relationships among different tables in the database. Whereas the lower-left are screen-shots taken from the latest version of cross-platform CHDD program. Complex query could be generated from the logic query builder and the cross-referenced results are shown in an integrated GUI.
Jiang et al. [9] in collaboration with Neotrident Technology Ltd. (see Table 1 ) developed the Chinese Natural Product Database (CNPD), which contains 45,055 nature products, among which 12,521 entries have been checked. CNPD can provide 2D structures and 3D models, related plant information, TCM property and bioactivity.
Table 1.
Comparison summary table
| Technology 1 | Technology 2 | Technology 3 | |
|---|---|---|---|
|
Name of specific type of technology |
CADDa based on TCMb |
FAC–MSc for screening |
MIP-SPEd for screening and separating |
| Names of specific technologies with associated companies and company Web sites | Accelrys, Inc. http://www.accelrys.com/ | Applied Biosystems http://www.appliedbiosystems.com/ | Applied Biosystems http://www.appliedbiosystems.com/ |
| Neotrident Technique Ltd. http://www.neotrident.com/ |
|||
| Pros | Virtual screening based on TCM database has higher hit rate than that based on chemical database | Only for screening, higher hit rate | Combining the screening, separating and identifying on one single step |
| No false leads |
|||
|
Cons |
High throughput |
Relatively high throughput |
Relatively high throughput |
| References | [9, 10] | [19, 20, 23] | [27, 29] |
Computer-aided drug design.
Traditional Chinese medicine.
Frontal affinity chromatography with mass spectrometry.
Molecularly imprinted polymers in solid phase extraction.
CADD studies derived from TCM typically involve virtual screening, optimization of lead structure, establishment of quantitative structure-activity relationship (QSAR), prediction of drug-likeness, prediction of absorption, distribution, metabolism, elimination/toxicity (ADME/T) and computer aided molecular structure elucidation.
First, information regarding herb mixture related to specific diseases can be retrieved from a TCM formulation database. Second, a 3D structure subset of ingredients related to specific diseases can be constructed through searching all original structures of plants consisting of TCM formulations. Third, this subset is filtered via virtual screening. There are several screening methods in CADD. For example, molecular docking can be used to study the interaction between bioactive compounds and target enzymes when the target 3D structure is known. Pharmacophore searching can be used to search candidate compounds with specific pharmacophoric groups.
When candidate compounds are available, they can be subjected to a bioactivity test. But mostly, the bioactivity test must be applied to the fractional extracts of plants containing the candidate compounds. The compounds with higher activity can then be isolated. The hit rates for screening active compounds from TCM database are correspondingly higher than from the chemical database.
In very rare cases, a natural product like taxol becomes a drug without any modification. Usually, however, the structure of active compounds needs to be modified by CADD (e.g. QSAR or docking) before being synthesized.
Qiao et al. [8] have applied the above method to identify new potential inhibitors of hepatitis C virus (HCV) NS3-NS4A protease. The crystal structure solved by Kim et al. [10] was used as a design target. At first they studied the interaction between compound and target protease via a molecular docking module also developed by their group. Then several high scoring candidate compounds were selected according to an energy and geometry match. Finally, these compounds were extracted from respective plants and subjected to bioactivity test [11]. The structure was modified to improve bioactivity [12] and resulted in nine compounds with IC50 < 1 μm/ml [13]. The binding energy was calculated by the molecular mechanics/Possion–Bolzman surface area (MM/PBSA) method, and the results showed that the binding energies were parallel to IC50 [11].
Jiang et al. [9] performed docking screening (see Table 1) and primary drug-likeness analysis by using CNPD and the 3D model of the eukaryotic K+ channels, and got 14 potential potassium ion (K+) channel blockers. After bioassay tests, four compounds were found that were 20- to 1000-fold more potent that the most extensively used Ik blocker–tetraethlammonion (TEA). Therefore, these compounds might replace TEA as the selective Ik blockers in neurobiology research.
Rapid screening of active compounds directly from TCM
Rapid screening of enzyme inhibitors is a key method for finding drug leads. The most common method used for high-throughput screening (HTS) of enzyme inhibitors involves colorimetric or fluorimetric assays. However, there are numerous difficulties associated with HTS assay development [14].
Enzymes and receptors represent the most common drug targets. Target based drug discovery is an important strategy for developing new agents [15]. TCM preparations are a mixture of a large number of compounds, in which the active targets may be unknown and at low concentrations in a background of other active species, so the screening process might lead to false positives resulting from the sum activity of many weakly active compounds in the case of bioactivity guided isolating and screening.
Biochromatography are widely used for screening the active compounds from TCM [16]. Wang et al. [17] used the immobilized human serum albumin (HBA) and α1-acid glycoprotein (AGP) as the stationary phase for screening active compounds from TCM by using high performance liquid chromatography (HPLC). He et al. [18] applied biomembrane chromatography with immobilized biological cell membranes on silica supports for the study of TCM. Although chromatography is one of the main techniques used in the study of TCM, current chromatography techniques are still unsatisfactory, since it is difficult to distinguish the bioactive component peak from the other peaks [16] and meet the requirements of HTS.
Affinity based screening using frontal affinity chromatography coupled with mass spectrometry (fac–ms) (see Glossary) is a more direct and faster method [19, 20] for screening the active compounds from TCM. It is not only quick and efficient for screening active compounds at low concentration, but also can avoid interference from other active compounds.
FAC–MS was established in 1998 by Schreimer [21]. The application of FAC–MS to a broad range of biological systems, together with its label-free operation, relatively high throughput, ability to rank ligands and determine dissociation constant K d, makes FAC–MS a universal tool enabling convenient and efficient screening for the identification of new potential drug leads [19].
TCM contains many unknown compounds with different unknown concentrations, so the features of a label-free operation, relatively high throughput, ability to rank ligands, and a breakthrough volume that is not related to ligand concentration (when c ≪ K a) makes FAC–MS a very appealing technique for screening active compounds from TCM (see Glossary).
Zhu et al. developed a very efficient and straightforward procedure for analyzing the binding properties of different inhibitors of the epidermal growth factor receptor (EGFR) by combining FAC with a Mariner electrospray ionization mass spectrometry (see Table 1) [22].
Polyclonal antibodies (PcAb) were used in a system, in which the PcAb was raised against piceatannol, which was linked with bovine serum albumin (BSA) to be covalently bound to the chromatographic packing for mimicking the EGFR. Six inhibitors including the half-antigen itself were recognized from the crude extract of Caragana Jubata. The corresponding dissociation constants (K d) for these six analytes were estimated and contrasted to the respective bioactivities (IC50). The binding affinity of the PcAb for the analyte was stronger as the factor K d was smaller, whereas the bioactivity of the molecule was higher as the IC50 was smaller. From the results, it could be concluded that the recognition based on the PcAb was efficient, and the analyte with higher activity was retained on the affinity column longer.
Luo et al.[23] reported the application of FAC–MS for screening active compounds from an extract of Phylantus urinaria L., based on immobilized PcAb that are used to mimic hepatitis C virus (HCV) NS3 protease.
Many articles have been published on the applications of FAC–MS to immobilized enzyme for screening the active compounds from a chemical library [24, 25]. The first report using FAC–MS for TCM was made by Yi et al. They used FAC–MS based on immobilized surface spike protein of SAR-CoV to screen 120 species of TCM and tetra-O-galloy-β-d-glucose (TGG) and luteoline, which bind avidly with the surface spike protein of SARS-CoV, thus can interfere with the entry of the virus to host cell. Using a wild–type SARS-CoV infect system, it was confirmed that TGG exhibits prominent anti-SARS-CoV activity with a 50% effective concentration of 4.5 μm/ml and a selective index of 240.0 [26].
Affinitive screening, separation and online identification of active compounds from TCM by molecularly imprinted polymers
The technique of molecular imprinting was introduced in 1972 by Wulff. Molecularly imprinted polymers are extensively cross-linked polymers containing specific recognition sites with a predetermined selectivity for analytes of interest. The technique involves complexation in a solution of target molecules (template) with functional monomers through either covalent or noncovalent bonds, followed by a polymerization reaction with an excess of cross-linkers. Removal of the templates leaves behind specific recognition sites that are complementary to the template in terms of its shape, size and functionality in the polymer network. These recognition sites enable imprinted polymers to be used as the mimics of enzymes, receptors and antibodies for screening various kinds of compounds from a mixture with abundant interferences.
Up to now, there have been many reviews summarizing the development of MIPs [27, 28, 29], which have covered many aspects from the sorbents for sample preconcentration and stationary phase for separation to bioassays, biosensors and mimics for enzymes, receptors and catalysts.
Theoretically, the more active the inhibitory compound adopted as the template, the more able the resulting imprinted polymer would be for screening other different inhibitors; the microcavities of the corresponding polymer would simulate the binding pocket of the enzyme more successfully owing to the template with higher bioactivity could conjugate to the binding pocket more tightly in terms of its shape, size and functional groups. So MIP is an useful technique for screening and separating the active compounds from TCM.
Steroid receptor mimics, folate receptor mimics and α2-adrenoreceptor mimics have been made by MIPs and used to screen the bioactive compounds from synthetic libraries [29]. Dong et al. [30] used MIP in solid phase extraction (mispe) (see Glossary) of (-)-ephedrine for chromatographic separation of marine and oxymartine from Chinese Ephedra. Li et al. [31] applied fisetin-imprinted polymers for separating the fisetin and analogue quercetin from TCM. All used chromatography to study the behavior of MIPs.
A MIP coupled with MS (see Table 1) was prepared using (E)-piceatannol, a natural potential anti-EGFR inhibitor, as the template and 4-vinylpyridine as the functional monomer by Zhu et al. [32, 33]. The crude extract of this herb was loaded on the MIP column for the binding test, and two different compounds besides the template itself were specifically recognized by the polymer, which were identified as butein and quercetin, possessing potent anti-EGFR tyrosine kinase activities with IC50 values of 10 and 15 μm/ml, respectively. Affinity and selectivity for these inhibitors and another three compounds coexisting with the template in this herb were evaluated in the chromatographic mode. For the first time, the affinity of a MIP was investigated to be correlative to the bioactivities of the analytes.
This work demonstrated that it is feasible to use a MIP for screening and separating active compounds, and online identification of inhibitors directly from the crude extract of the herb, which would be very helpful in discovering lead compounds or drug candidates.
In the same approach, Luo et al. Carried out an efficient separation based on a MIP applied in Phyllanthus urinaria L., from which several novel inhibitors of hepatitis C virus (HCV) NS3 protease were screened out [2].
Xie et al. [34] used a coupled liquid-phase chromatography and mass spectrometry (LC–MS) system, consisting of a combination of a column of MIPs and a MS detector, for affinitive separation and online identification of the antitumor components, harmine and harmaline, from the methanol extract of Peganum nigellastrum seeds. The target binding capacities of the MIPs were evaluated by frontal chromatography.
Isolation of analytes from urine, plasma, bile and animal tissue is important for pharmacokinetic studies There are only a few studies in which the sample was applied directly to the MIP [35]. An anti-quercetin MIP with evident hydrophobic matrix was synthesized using acrylamide (AA) as the functional monomer and 2,2-bis(hydroxymethy)butanol trimethacrylate (TRIM) as the cross linker by Xie et al.[36] The affinity and selectivity were evaluated by liquid chromatography, and the binding sites and the dissociation constants were measured by frontal chromatography. The results showed that MIP-SPE could be used for direct clean up of biological samples for the analysis of functional components in vivo originating from an extract of medicinal herbs. Therefore, MIP is an important tool in study of the pharmacokinetics of TCM.
Conclusion
It is currently estimated that approximately 420,000 plant species exist in nature and over 248,000 species of higher plants have been identified, and from these 12,000 plants are known to have medical properties. However, less than 10% of all plants have been investigated from a phyto-chemical and/or pharmacological point of view [37]. So far, there have been 11,145 medical plants found in China, of which a very small amount of them have been systematically investigated. [38]. So it is necessary for us to carry out drug discovery research by using available information and knowledge and developing new approaches for screening and separating the active compounds directly from TCM.
CADD is a useful approach to utilizing the information and knowledge for finding, optimizing and predicting the properties of active compounds. The main goal for CADD based on TCM is to limit the number of compounds requiring experimental testing while maximizing the hit rate, because it is time-consuming to obtain the pure compounds of TCM. A useful strategy is to use hierarchical multiple filter database screening reported by Wang et al. [39]
The novel application of immobilized enzymes, PcAb and MIP in TCM for effective recognition of active compounds immersed in immersed in afforded us a new approach for the discovery of lead compounds or drug candidates, which will be very beneficial in drug development.
The immobilized enzyme and PcAb methods in combination with FAC–MS are only used for screening. Although both methods have similarly advanced features in recognizing active compounds, the PcAb as a mimic of enzyme has less recognition ability than that of immobilized enzyme. When the immobilized enzyme is not available or not suitable for screening, PcAb can be used as an alternative method.
MIP can be used to mimic the enzyme or receptor for direct screening, separation and online identification of active compounds from TCM, and can speed up drug discovery based on TCM. MIP have proven to be feasible mimics in terms of physical robustness, resistance to elevated temperatures and pressures, and inertness towards acids, bases, metal ions and organic solvents, this material can be well employed in various kinds of samples including some biological and pharmaceutical samples. However, greater amounts (often 1 mmol) of pure template molecule were required for MIP, whereas usually only trace amounts (0.01–0.1 mmol) of half-antigen were required to produce PcAb. Moreover, MIP have difficulty dealing with macromolecules such as proteins and polynucleotides.
CADD, immobilized enzyme, PcAb and MIP are complementary and should be integrated to the multifaceted platform contributing to the early drug discovery process. At first, virtual screening is used to find potential active compounds. Then active fractional parts containing potential compounds are subjected to screening and separation of active compounds by MIP in one single step. We can also screen the active compounds firstly by using immobilized enzymes or PcAb, then separate active compounds using MIP. PcAb and MIP can be used to find substitutes of patent medicine, which are important yet expensive, from TCM.
Outstanding issues
-
•
Although TCM has proven useful for many people throughout history, sometimes it has been neglected in Western research owing to the constituents and the mechanism for TCM being unclear at present, and the different concepts of illness.
-
•
TCM is a complex system. It is time consuming and difficult to separate and identify all constituents. The reverse approach may be useful for finding active compounds immersed in interferences. Synergy is an important fundamental for the mechanism of TCM and involves multiple targets and pathways. Therefore, it is needed to develop approaches for synergy studies.
-
•
The hit rates for virtual screening based on TCM database are corresponding higher than that from chemical database.
-
•
The features of label free: operation; ability to rank ligands; break through volume being unrelated to ligand concentration (when c ≪ K d); availability for multiple targets, make FAC–MS a very efficient approach for screening active compounds in TCM. But sometimes the conformation change should be considered in this process.
Related articles
Drasar, P. and Moravcova, J. (2004) Recent advances in analysis of Chinese medical plants and traditional medicines, J. Chromatogr. B 812, 3–21
Huang X.D. et al. (2004) Strategy for analysis and screening of bioactive compounds in traditional Chinese medicines, J. Chromatogr. B 812, 71–84
Clardy, J. and Walsh, C. (2004) Lessons from natural molecules. Nature 432, 829–837
Jorgensen, W.L. (2004) The many roles of computation in drug discovery. Science 303, 1813–1818
Schriemer, D.C. (2004) Biosensor alternative: frontal affinity chromatography. Anal. Chem. 76, 440A–448A
Links
-
•
Accelrys, Inc.: http://www.accelrys.com/
-
•
Applied Biosystems: http://www.appliedbiosystems.com/
-
•
Neotrident Technique Ltd.: http://www.neotrident.com/
Glossary
FAC–MS: a combination of frontal affinity chromatography with mass spectrometry detection. It is based on the continuous infusion of ligands over a protein target immobilized onto a solid support column, with the eluting ligands detected by MS. When ligands flow through the column, they bind to the target with differing affinities. The breakthrough volume of ligand is dependent on the affinity of the ligand for the target.
Eq. (1) is the basic equation of frontal affinity chromatography:
| (1) |
It also can be shown in other form:
| (2) |
where V is the elution volume of the analyte, c is the concentration of the analyte, V 0 is the elution volume of a controlled substance having no affinity, Lt is the total amount of immobilized ligand, and K d is the dissociation constant. V approaches its maximum value, V m. If c ≪ K d, that is, as c is neglibly small compared with K d, the following equation is obtained:
| (3) |
MIP-SPE (MISPE): the basic concept for the application of MIP in solid phase extraction (MISPE) is that the chromatographic parameters are tuned such that the MISPE column traps only the analytes, or a group of structurally related compounds, whereas the other interfering components in the sample matrix were washed without retention.
TCM: traditional Chinese medicine (TCM) is a medical science governing the theory and practice of traditional medicine. It includes Chinese medication, pharmacology/herbalogy, acupuncture, massage and Qigong. In this article, TCM only is related with traditional Chinese medicinal herbs.
References
- 1.Drasar P. Recent advances in analysis of Chinese medical plant and traditional medicines. J. Chromatogr. B. 2004;812:3–21. doi: 10.1016/j.jchromb.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 2.Xu X.J. Separation and screening of compounds of biological origin using molecularly imprinted polymers. J. Chromatogr. B. 2004;804:61–69. doi: 10.1016/j.jchromb.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Balunas M.J. Drug discovery from medicinal plants. Life Sci. 2005;78:431–441. doi: 10.1016/j.lfs.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 4.Bradbury J. From Chinese medicine to anticancer drug. DDT. 2005;10:1131–1132. doi: 10.1016/S1359-6446(05)03587-7. [DOI] [PubMed] [Google Scholar]
- 5.Stahl M. Integrating molecular design resource within modern drug discovery research: the Roche experience. Drug Disov. Today. 2006;11:326–333. doi: 10.1016/j.drudis.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Jorgensen W.L. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 7.Hou T.J. recent development and application of virtual screening in drug discovery: an Overview. Curr. Pharma. Des. 2004;10:1011–1033. doi: 10.2174/1381612043452721. [DOI] [PubMed] [Google Scholar]
- 8.Qiao X.B. A 3D structure database of compounds from chinese traditional medicine herbs. J. Chem. Inf. Comput. Sci. 2002;42:481–489. doi: 10.1021/ci010113h. [DOI] [PubMed] [Google Scholar]
- 9.Shen J.S. Virtual screening on natural products for discovery active compounds and target information. Curr. Med. Chem. 2003;10:2327–2342. doi: 10.2174/0929867033456729. [DOI] [PubMed] [Google Scholar]
- 10.Kim J.L. Crystal structure of hepatitis C Virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 11.Duan D.L. Antiviral compounds from traditional Chinese medicines Galla Chinese as inhibitors of HCV NS3 protease. Bio. Med. Chem. Lett. 2004;14:6041–6044. doi: 10.1016/j.bmcl.2004.09.067. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y. Search for more effective HCV NS3 protease inhibitors via modification of corilagin. Progress Nature Sci. 2005;15:896–901. [Google Scholar]
- 13.Zuo G.Y. In vitro anti HCV activities of Saxifraga melanocentra and its polyphenolic compounds. Antiviral Chem. Chemother. 2005;16:393–398. doi: 10.1177/095632020501600606. [DOI] [PubMed] [Google Scholar]
- 14.Bogustavasky J. HTS assay development: Is smaller really better? Drug Discov. Dev. 2004;7:37–40. [Google Scholar]
- 15.Deng G. Application of mass spectrometry in early stages of target based drug discovery. J. Pharm. Biomed. Anal. 2006;40:528–538. doi: 10.1016/j.jpba.2005.08.038. [DOI] [PubMed] [Google Scholar]
- 16.Huang X.D. Strategy for analysis and screening of bioactive compounds in traditional Chinese medicine. J. Chromatogr. B. 2004;812:71–84. doi: 10.1016/j.jchromb.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 17.Wang H.L. Fractionation and analysis of Artemisia Capillaris Thumb by affinity chromatography with human serum albumin as stationary phase. J. Chromatogr. A. 2000;870:501–510. doi: 10.1016/s0021-9673(99)01062-6. [DOI] [PubMed] [Google Scholar]
- 18.He J.L. Coating and fusing cell membranes onto a silica surface and their chromatographic characteristics. Chromatography. 2001;54:71–76. [Google Scholar]
- 19.Jacek J. Frontal affinity chromatography with MS detection (FAC–MS) in drug discovery. Drug Discov. Today. 2005;10:409–416. doi: 10.1016/S1359-6446(04)03360-4. [DOI] [PubMed] [Google Scholar]
- 20.Schriemer D.C. Biosensor alternative: frontal affinity chromatography. Anal. Chem. 2004;76:441A–448A. doi: 10.1021/ac041684m. [DOI] [PubMed] [Google Scholar]
- 21.Schriemer D.C. Micro-scale fronted affinity chromatography with mass spectro metric detector: a new method for the screening of compound libraries. Angew. Chem. Int. Ed. Engl. 1998;37:3383–3387. doi: 10.1002/(SICI)1521-3773(19981231)37:24<3383::AID-ANIE3383>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 22.Zhu L.L. Frontal affinity chromatography combined on-line with mass spectrometry: a tool for the binding study of different epidermal growth factor receptor inhibitor. Anal. Chem. 2003;75:6388–6393. doi: 10.1021/ac0341867. [DOI] [PubMed] [Google Scholar]
- 23.Luo H.P. Frontal Immunoaffinity Chromatography with mass spectrometric detection: a method for finding active compounds from traditional Chinese herbs. Anal. Chem. 2003;75:3994–3998. doi: 10.1021/ac034190i. [DOI] [PubMed] [Google Scholar]
- 24.Zhang B. Frontal affinity chromatography coupled to mass spectrometry for screening mixtures of enzyme inhibitors. Anal. BioChem. 2001;299:173–182. doi: 10.1006/abio.2001.5417. [DOI] [PubMed] [Google Scholar]
- 25.Hodgsom R.J. Inhibitor screening using immobilized enzyme reactor chromatography/mass spectrometry. Anal. Chem. 2005;75:7512–7519. doi: 10.1021/ac050761q. [DOI] [PubMed] [Google Scholar]
- 26.Yi L. Small molecules blocking the entry of severe acute respioatory syndrome coronavirus into host cells. J. Virology. 2004;78:11334–11339. doi: 10.1128/JVI.78.20.11334-11339.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kandimalla V.B. Molecular imprinting: a dynamic technique for diverse applications in analytical chemistry. Anal. Bio. Chem. 2004;380:587–605. doi: 10.1007/s00216-004-2793-9. [DOI] [PubMed] [Google Scholar]
- 28.Ye L. Molecularly imprinted polymers as antibody and receptor mimics for assays, sensors and drug discovery. Anal. Bio. Chem. 2004;378:1887–1897. doi: 10.1007/s00216-003-2450-8. [DOI] [PubMed] [Google Scholar]
- 29.Rathbone D.L. Molecularly imprinted polymers in the drug discovery process. Adv. Drug Deliv. Rev. 2005;57:1854–1874. doi: 10.1016/j.addr.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 30.Dong X.C. Molecularly imprinted solid-phase extraction of (-)-ephedrine from Chinese Ephedra. J. Chromatogr. A. 2005;1070:125–130. doi: 10.1016/j.chroma.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Li L. Solid-phase extraction of active component of Chinese traditional medicine fisetin by using molecularly imprinted polymers. Chem. J. Chin. Univ. -Chin. 2006;27:608–611. [Google Scholar]
- 32.Zhu L.L. Application of a molecularly imprinted polymer for the effective recognition of different anti-epidernal growth factor receptor inhibitors. Anal. Chem. 2003;75:6381–6387. doi: 10.1021/ac026371a. [DOI] [PubMed] [Google Scholar]
- 33.Zhu L.L. Selective separation of active inhibitors of epidermal growth factor receptor from Caragana Jubata by molecularly imprinted solid-phase. J. Chromatogr. A. 2003;991:151–158. doi: 10.1016/s0021-9673(03)00207-3. [DOI] [PubMed] [Google Scholar]
- 34.Xie J. Affinitive separation and on-line identification of antitumor compounds from Peganum nigellastrum by coupling a chromatographic column of target analogue imprinted polymer with mass spectrometry. Anal. Chem. 2002;74:2352–2360. doi: 10.1021/ac015755i. [DOI] [PubMed] [Google Scholar]
- 35.Caro E. Application of molecularly imprinted polymers to solid-phase extraction of compounds from environmental and biological samples. Trends Anal. Chem. 2006;25:143–154. [Google Scholar]
- 36.Xie J. Selective extraction of functional compounds derived from herb in plasma by using a molecularly imprinted polymer based of 2,2-bis(hydroxymethyl)butanol trimethacrylate. J. Chromatogr. B. 2003;788:233–242. doi: 10.1016/s1570-0232(02)00796-1. [DOI] [PubMed] [Google Scholar]
- 37.Vuorela P. Nature Products in the Process of Finding New Drug Candidates. Curr. Med. Chem. 2004;11:2375–2389. doi: 10.2174/0929867043365116. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y.Z. Science publishing; 2003. Introduction of Chinese Methods in Study of Modernization of Traditional Chinese Medicines. [Google Scholar]
- 39.Wang J.M. Hierarchical database screening for HIV-1 reverse transcriptase using a pharmacophore model, rigid docking, solvation docking and MM-PB/SA. J. Med. Chem. 2005;48:2432–2444. doi: 10.1021/jm049606e. [DOI] [PubMed] [Google Scholar]


