Abstract
Pseudomonas aeruginosa is considered as foremost cause of hospital acquired infections due to its innate and plasmid mediated resistance to multiple antibiotics making it a multi drug resistant (MDR) pathogen. Biofilm formation is a pathogenic mechanism harbored by this pathogen which further elevates its resistance to antibiotics and host defense system. The aim of the present study was to evaluate the biofilm forming potential and distribution of pslA gene in multi drug resistant Pseudomonas aeruginosa isolates obtained from different clinical samples. A total of 200 different clinical samples were collected after obtaining written consent from the patients. The samples were subjected to isolation and identification of P. aeruginosa by standard microbiological procedures. Confirmation of isolates was done by polymerase chain reaction targeting oprL gene. Kirby Bauer method was performed for detection of MDR isolates. Congo red agar (CRA) test and Microtiter plate assay (MPA) for observing the biofilm forming ability and amplification of pslA gene was also performed on MDR isolates. The results showed that from 200 samples 52 (26 %) were P. aeruginosa and among them 20 (38.46 %) were MDR isolates. The CRA showed 23 (44.23 %) while MPA detected 49 (94.23 %) isolates as biofilm producers while all the MDR isolates showed biofilm formation by MPA method. The pslA gene was detected in all biofilm forming isolates while 90 % in MDR P. aeruginosa. It was concluded that biofilm forming P. aeruginosa are more resistant to tested antibiotics and biofilm formation is strongly associated with presence of pslA gene.
Keywords: multi drug resistant, biofilm formation, Pseudomonas aeruginosa, pslA gene
Introduction
Pseudomonas aeruginosa is a threatening and emerging public health problem throughout the world particularly in developing countries. It is still a main cause of mortality and morbidity in humans (Hirsch and Tam, 2010[10]). It is ranked 4th among the nosocomial pathogens and difficult to treat due to its resistant behavior against different antibiotic drugs (Khan et al., 2015[17]). Pseudomonas aeruginosa is a foremost cause of nosocomial infections, including urinary tract infections, pneumonia and bacteremia. It is also found in patients having burns, surgical, pus and accidental wounds (Khalid et al., 2017[15]). These infections become severe when the patients have impaired immune system (Pagani et al., 2004[23]).
Biofilms are structurally complex surface connected populations in which bacterial cells are enclosed by extra cellular polymeric substances (EPS) produced by their own self. These EPS are mostly exopolysaccharides, extracellular deoxyribonucleic acid and proteins (Ryder et al., 2007[28]). Biofilm formation contributes to pathogenesis of P. aeruginosa both in acute as well as chronic infection in clinical settings (Schaber et al., 2007[31]). The bacteria residing in biofilms are much more resistant to antimicrobial agents and host immune response compared to their planktonic counterparts, leading to prolonged or chronic infections which are difficult to eradicate (Mah et al., 2003[20]). Biofilm forming bacteria produce couple of extracellular polymeric matrices which attached the bacterial community together within the biofilm. The key components of biofilm matrix are polysaccharides because they contribute towards overall architecture of biofilm and make the bacterial population resistant to antibacterials (Wozniak et al., 2003[34]). There are about three exopolysaccharides reported to be involved in P. aeruginosa biofilm formation which includes alginate, Psl, and Pel (Ghafoor et al., 2011[7]). Among these exopolysaccharides, Psl is a mannose-rich polymer having an essential role in preliminary steps of biofilm formation (Ma et al., 2006[19]).
The biofilm production retards the antimicrobial therapy against bacteria because the biofilm develops a barrier which reduces the drug penetration leading to treatment failure as well as hindering the recognition of the microorganisms by immune system (Gil-Perotin et al., 2012[8]). In view of the above mentioned facts, the objective of the current study was to phenotypically and genotypically evaluate the biofilm production ability of multi drug resistant Pseudomonas aeruginosa from clinical samples.
Materials and Methods
Sample collection
A total (n=200) clinical samples were collected aseptically from different healthcare centers of Punjab, Pakistan consisting of burn wounds, surgical wounds, sputum, urine and blood samples which were shifted to laboratory as soon as possible. The samples were collected during January 2018 to December 2018 by obtaining the written consent from the patients after explaining them the purpose and objectives of the study.
Ethical statement
This study was approved by Institutional Ethics Review Committee of Government College University Faisalabad, Pakistan under code GCUF/ERC/18/03B, and the samples were collected in accordance with international safety rules and ethical standards.
Isolation and identification of Pseudomonas aeruginosa isolates
The samples were streaked on the plates containing Pseudomonas cetrimide agar (Oxoid, Uk) and incubated at 37 °C overnight aerobically. Isolates were identified on the basis of their colony morphology, cultural and biochemical characteristics (Ijaz et al., 2019[13]).
Molecular confirmation of Pseudomonas aeruginosa isolates
All the identified isolates of Pseudomonas aeruginosa were confirmed by Polymerase chain reaction (PCR) using specific primer set targeting oprL gene (oprL-F 5'-ATGGAAATGCTG AAATTCGGC-3') and (oprL-R 5'-CTTCTTCAGCTCGACGCGACG-3') with the product size of 504 bp. First the DNA of all the bacterial isolates was extracted with the help of GeneJET Genomic DNA Purification Kit (Thermo Scientific, UK). The DNA was subjected to PCR using the conditions as described by Douraghi et al. (2014[4]). The PCR products were visualized in 1 % ethidium bromide stained agarose gel under (Slite 200W, Taiwan) Gel documentation system.
Antimicrobial susceptibility testing
The antibiotic susceptibility pattern of P. aeruginosa clinical isolates was evaluated by Kirby-Bauer disc diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI) (Ilyas et al. 2016[14]) based on the zone of inhibition. The cultures were streaked on Mueller Hinton agar plates and incubated at 37 °C. The zone of inhibition was measured after cultures had been incubated overnight. The test was performed using standard antibiotic discs Ceftriaxone (30 μg), Gentamicin (10 μg), Ciprofloxacin (5 μg), Amikacin (30 μg), Amoxicillin clavulanic acid (30 μg) Ceftazidime (30 μg), Doripenem (10 μg), Meropenem (10 μg), Imipenem (10 μg) and Aztreonam (30 μg) (Oxoid).
Phenotypic characterization of biofilm production
Biofilm formation was determined in vitro using Congo red agar and microtiter plate assay. The CRA preparation, inoculation and observation of results were done as described by Freeman et al. (1989[5]). Briefly, the inoculated plates having Congo red agar were incubated overnight aerobically at 37 °C to obtain isolated bacterial colonies. CRA-positive isolates appeared as black colonies, while CRA-negative strains remained red.
In microtiter plate assay, overnight cultures were adjusted and diluted using tryptic soy broth (100 folds). Aliquots (250 μL) of each isolate suspension were then inoculated into a flat-bottom 96-well plate and incubated for 22-24 hours at 37 °C. Each well was washed twice with 250 μL of sterile saline followed by shaking to remove poorly adherent or non-adherent bacteria. The plates were then dried using heat which favors the fixation of biofilm. Each well was then stained with 250 μL of crystal violet (0.1 %) for 15-20 mins. After washing and drying of plates, the dye bound to the adherent cells in each well was dissolved with 250 μL of 50 % acetone. The optical density (OD) of the biofilms was measured at 594 nm with ELISA reader (BioRad, USA) as described by Stepanovic et al. (2000[32]).
Detection of the pslA gene
The PCR amplification of pslA gene was done with primers: (pslA-F, 5'-TGGGTCTTCAAGTTCCGCTC-3') and (pslA-R, 5'-ATGCTGGTCTTGCGGATGAA-3') generating an amplicon of 119 bp (Maita and Boonbumrung, 2014[21]). P. aeruginosa (PAO1) was used as positive control for the pslA gene. The PCR reaction mixture was prepared (25 μL) and following thermal conditions were applied. The preliminary denaturation was performed at 94 °C for 5 minutes. Then 35 cycles were performed (denaturation at 94 °C for 30 seconds, annealing at 52 °C for 40 seconds and extension at 72 °C for 50 seconds). The final extension was done for 10 minutes at 72 °C in a thermocycler device (BioRad, USA).
Results
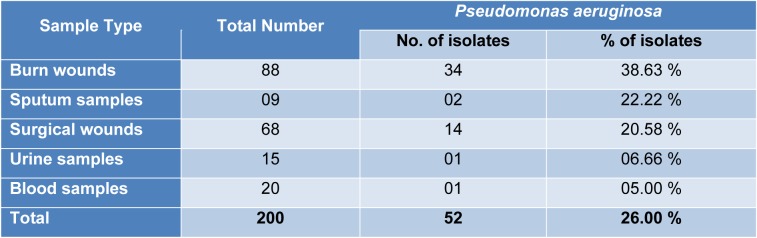
In the present study, a total of 200 clinical samples were collected from different hospitals of Punjab, Pakistan. The results showed that Pseudomonas aeruginosa was identified from (n=52) samples on the basis of biochemical tests and PCR method making the cumulative prevalence of 26 %. The distribution of P. aeruginosa isolates on the basis of type of samples was elaborated in Table 1(Tab. 1).
Table 1. Distribution of Pseudomonas aeruginosa clinical isolates according to the sample type.
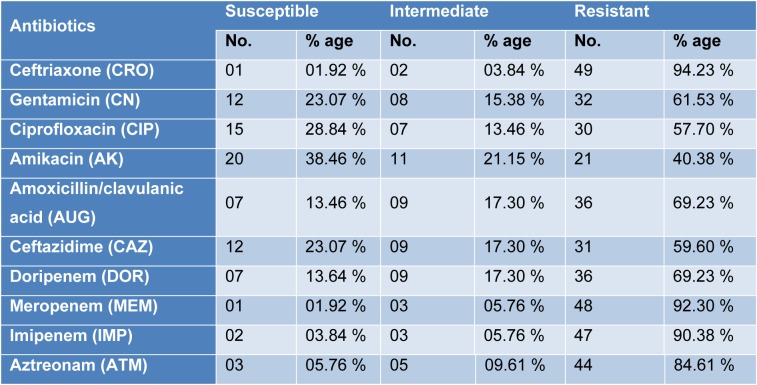
All the isolates of P. aeruginosa were subjected to antimicrobial susceptibility testing and highest resistance was found against Ceftriaxone (94.23 %), Meropenem (92.30 %), Imipenem (90.38 %) and Aztreonam (84.61 %) while the isolates were found most susceptible to Amikacin (38.46 %) and Ciprofloxacin (28.84 %). Among all the isolates, 20 (38.46 %) were found multi drug resistant (MDR) Pseudomonas aeruginosa as shown in Table 2(Tab. 2).
Table 2. Antimicrobial susceptibility patterns of Pseudomonas aeruginosa isolates.
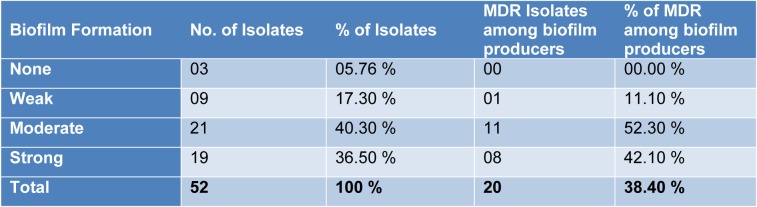
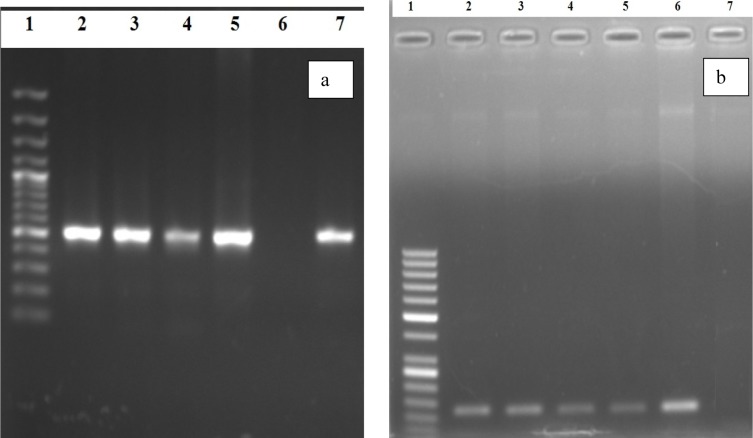
On the basis of Congo red agar test 23 (44.23 %) samples showed biofilm forming potential while when the Microtiter plate assay (MPA) was applied 49 (94.23 %) samples showed variable degree of biofilm production. Highest percentage was found (40.30 %) of moderate biofilm producing Pseudomonas aeruginosa followed by strong biofilm producers (36.50 %). All the (n=20) MDR P. aeruginosa isolates showed biofilm formation by MPA as shown in Table 3(Tab. 3). The pslA gene was detected in all biofilm forming isolates while (18/20) 90 % in MDR Pseudomonas aeruginosa isolates with polymerase chain reaction showing a 119 bp band on agarose gel as shown in Figure 1(Fig. 1).
Table 3. Association between biofilm forming and multi drug resistant isolates of Pseudomonas aeruginosa.
Figure 1. Polymerase chain reaction based detection of Pseudomonas aeruginosa oprL (a) and psl A (b) genes.
PCR based detection of (a) oprL (504 bp); Lane 1: 100 bp ladder, Lane 2-5: Positive Samples, Lane 6: Negative Control, Lane 7: Positive control for oprL genes confirming Pseudomonas aeruginosa; (b) psl A gene (119 bp); Lane 1: 50 bp ladder, Lane 2-5: Positive Samples, Lane 6: Positive Control, Lane 7: Negative Control for psl A gene
Discussion
Pseudomonas aeruginosa is an opportunistic pathogen having greatest concern for infections in various types of wounds, immunocompromised patients, and intensive-care units' patients (ICUs) (Bukholm et al., 2002[2]). Biofilm-producing P. aeruginosa have a key role in pathogenesis because biofilm decreased the penetration of antibiotics in bacteria and are difficult to remove from the surfaces. In the present study total 200 clinical samples were collected and the results showed 52 (26 %) found positive for Pseudomonas aeruginosa with most prominent 38.63 % in burn wound samples. The finding of our research was very close to the previous observations (20.05 %) by Samad et al. (2017[29]) in Peshawar and (22.2 %) by Hussain et al. (2017[12]) in Islamabad while our results were lower than the finding of Ijaz et al. (2019[13]) having 38 % prevalence of P. aeruginosa. Similarly the results of Khan et al. (2014[16]), Qureshi et al. (2018[26]) and Ijaz et al. (2019[13]) are consistent with our findings that P. aeruginosa was prominently found in burn wound/pus. The high prevalence of Pseudomonas aeruginosa is mainly due to elevated level of contamination, poor management of wounds and lack of sanitary facilities in hospital environment while the formation of pus in burn and surgical wounds serve as favorable condition for the growth of pathogens and transmission of infections (Procop et al., 2017[25]).
Multi drug resistant (MDR) refers to bacteria which show resistance to minimum two specific representatives of minimum two classes of antibiotics (Park et al., 2011[24]). The results of antimicrobial susceptibility pattern showed highest resistance of P. aeruginosa isolates towards Ceftriaxone (94.23 %), Meropenem (92.30 %), Imipenem (90.38 %) and Aztreonam (84.61 %) which are in accordance with the findings of Qureshi et al. (2018[26]) and Ijaz et al. (2019[13]) in Pakistan. Similarly Wang et al. (2010[33]) from China and Rodriguez-Martinez et al. (2009[27]) from France observed resistance towards beta lactams and carbapenems respectively. Among the positive Pseudomonas aeruginosa isolates 20 (38.46 %) were detected multi drug resistant (MDR). The results are lower than the observations of Maita and Boonbumrung (2014[21]) with 51 % and Ijaz et al. (2019[13]) with 58.6 % MDR strains of P. aeruginosa. The reasons behind such a resistance are the inappropriate or irrational use of antibiotics, mutation in the genome of P. aeruginosa and environmental conditions of the specific area.
In the present study, two different methods for detection of biofilm formation in P. aeruginosa were used and compared. It was observed that Congo red agar (CRA) test detected 23 (44.23 %) while the Microtiter plate assay (MPA) detected 49 (94.23 %) isolates as biofilm producers. The efficacy of MPA is better than CRA was also reported by Lima et al. (2017[18]) in Brazil and Samad et al. (2019[30]) in Pakistan. These results indicate that the biofilm formation carried out in liquid media gives more reliable results.
Among the total (n=52) Pseudomonas aeruginosa isolates, 49 (94.23 %) were detected to be positive for pslA gene and from the (n=49) biofilm forming Pseudomonas aeruginosa, all the isolates were found positive for pslA gene. Similarly among the 20 MDR P. aeruginosa, all were found positive for biofilm production by MPA but 18 (90 %) were found positive for pslA gene. Very similar results were previously obtained by Heydari and Eftekhar (2015[9]) and Abootaleb et al. (2020[1]) in Iran showing 100 % presence of pslA gene in biofilm forming P. aeruginosa while Ghadaksaz et al. (2014[6]) showed 83.7 % and Maita and Boonbumrung (2014[21]) showed 94 % prevalence of pslA gene. The presence of pslA gene had proven itself a good marker of biofilm formation in Pseudomonas aeruginosa isolates reported in previous findings by Overhage et al. (2005[22]), Ma et al. (2006[19]) and Hou et al. (2012[11]), but in some studies the pslA was also detected in non biofilm producing isolates (Heydari and Eftekhar, 2015[9]). It might be due to the fact that pslA plays an essential role in initial biofilm formation. So, in addition to pslA, there might be some other factors which play their role in biofilm formation (Colvin et al., 2011[3]).
Conclusion
In conclusion, all the MDR isolates of P. aeruginosa showed biofilm forming potential and 90 % of them were positive for pslA gene. This presence of MDR strains along with biofilm formation is depicting a lethal combination of bacterial armory that poses a serious threat for public health.
Acknowledgements
This study was partially funded by National Research Program for Universities (NRPU) of higher education Commission (HEC), Pakistan (5680/Punjab/NRPU/ R&D/HEC/2016).
Disclosure
The authors declare that they have no conflict of interest.
References
- 1.Abootaleb M, Zolfaghari MR, Soleimani NA, Ghorbanmehr N, Yazdian MR. Biofilm formation with Microtiter plate 96 and pslA detection of Pseudomonas aeruginosa isolates from clinical samples in Iran. Int J Adv Biol Biomed Res. 2020;8:58–66. [Google Scholar]
- 2.Bukholm G, Tannaes T, Kjelsberg AB, Smith-Erichsen N. An outbreak of multidrug-resistant Pseudomonas aeruginosa associated with increased risk of patient death in an intensive care unit. Infect Control Hosp Epidemiol. 2002;23:441–446. doi: 10.1086/502082. [DOI] [PubMed] [Google Scholar]
- 3.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douraghi M, Ghasemi F, Dallal MMS, Rahbar M, Rahimiforoushani A. Molecular identification of Pseudomonas aeruginosa recovered from cystic fibrosis patients. J Prev Med Hyg. 2014;55:1–3. [PMC free article] [PubMed] [Google Scholar]
- 5.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghadaksaz A, Fooladi AAI, Hosseini HM, Amin M. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J Appl Biomed. 2014;13:61–68. [Google Scholar]
- 7.Ghafoor A, Hay ID, Rehm BHA. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil-Perotin S, Ramirez P, Marti V, Sahuquillo JM, Gonzalez E, Calleja I, et al. Implications of endotracheal tube biofilm in ventilator-associated pneumonia response: a state of concept. Crit Care. 2012;16:2–8. doi: 10.1186/cc11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heydari S, Eftekhar F. Biofilm formation and β-lactamase production in burn isolates of Pseudomonas aeruginosa. Jundishapur J Microbiol. 2015;8:1–5. doi: 10.5812/jjm.15514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10:441–451. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou W, Sun X, Wang Z, Zhang Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus and Pseudomonas aeruginosa from ocular infections. Invest Ophthalmol Vis Sci. 2012;53:5624–5631. doi: 10.1167/iovs.11-9114. [DOI] [PubMed] [Google Scholar]
- 12.Hussain M, Munir S, Fatima M, Rahim K, Ahmed I, Basit A, et al. Antimicrobial susceptibility patterns and CTX-M β-lactamase producing clinical isolates from burn patients in Islamabad, Pakistan. Asian Pac J Trop Dis. 2017;7:486–490. [Google Scholar]
- 13.Ijaz M, Siddique AB, Rasool MH, Shafique M. Frequency of multi drug resistant Pseudomonas aeruginosa in different wound types of hospitalized patients. Pak J Pharm Sci. 2019;32:865–870. [PubMed] [Google Scholar]
- 14.Ilyas S, Qamar MU, Rasool MH, Abdulhaq N, Nawaz Z. Multidrug-resistant pathogens isolated from ready-to-eat salads available at a local market in Pakistan. Brit Food J. 2016;118:2076–2091. [Google Scholar]
- 15.Khalid F, Siddique AB, Nawaz Z, Shafique M, Zahoor MA, Nisar MA, et al. Efficacy of bacteriophage against multidrug resistant Pseudomonas aeruginosa isolates. Southeast Asian J Trop Med Public Health. 2017;48:1056–1062. [Google Scholar]
- 16.Khan F, Khan A, Kazmi SU. Prevalence and susceptibility pattern of multi drug resistant clinical isolates of Pseudomonas aeruginosa in Karachi. Pak J Med Sci. 2014;30:951–954. doi: 10.12669/pjms.305.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan HA, Ahmad A, Mehboob R. Nosocomial infections and their control strategies. Asian Pac J Trop Biomed. 2015;5:509–514. [Google Scholar]
- 18.Lima JLDC, Alves LR, Paz JNPD, Rabelo MA, Maciel MAV, Morais MMC. Analysis of biofilm production by clinical isolates of Pseudomonas aeruginosa from patients with ventilator-associated pneumonia. Rev Bras Ter Intensiva. 2017;29:310–316. doi: 10.5935/0103-507X.20170039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure post attachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O'Toole GA. A genetic basis of Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- 21.Maita P, Boonbumrung K. Association between biofilm formation of Pseudomonas aeruginosa clinical isolates versus antibiotic resistance and genes involved with biofilm. J Chem Pharm. 2014;6:1022–1028. [Google Scholar]
- 22.Overhage J, Schemionek M, Webb JS, Rehm BH. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl Environ Microbiol. 2005;71:4407–4413. doi: 10.1128/AEM.71.8.4407-4413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagani L, Mantengoli E, Migliavacca R, Nucleo E, Pollini S, Spalla M, et al. Multifocal detection of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum β-lactamase in northern Italy. J Clin Microbiol. 2004;42:2523–9. doi: 10.1128/JCM.42.6.2523-2529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SC, Park Y, Hahm KS. The role of antimicrobial peptides in preventing multidrug-resistant bacterial infections and biofilm formation. Int J Mol Sci. 2011;12:5971–5992. doi: 10.3390/ijms12095971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Procop GW, Church D, Hall G, et al. Koneman's color atlas and textbook of diagnostic microbiology (Color Atlas & Textbook of Diagnostic Microbiology) Philadelphia, PA: Lippincott, Williams & Wilkins; 2017. [Google Scholar]
- 26.Qureshi R, Shafique M, Shahid M, Rasool MH, Muzammil S. Molecular detection of blaVIM Metallo-β-lactamase producing clinically isolated Pseudomonas aeruginosa from tertiary care hospital, Faisalabad. Pak J Pharm Sci. 2018;31:2673–2677. [PubMed] [Google Scholar]
- 27.Rodriguez-Martinez JM, Poirel L, Nordmann P. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53:4783–4788. doi: 10.1128/AAC.00574-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–8. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samad A, Ahmed T, Rahim A, Khalil A, Ali I. Antimicrobial susceptibility patterns of clinical isolates of Pseudomonas aeruginosa isolated from patients of respiratory tract infections in a Tertiary Care Hospital, Peshawar. Pak J Med Sci. 2017;33:670–674. doi: 10.12669/pjms.333.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samad A, Khan AA, Sajid M, Zahra R. Assessment of biofilm formation by Pseudomonas aeruginosa and hydrodynamic evaluation of microtiter plate assay. J Pak Med Assoc. 2019;69:666–671. [PubMed] [Google Scholar]
- 31.Schaber JA, Triffo WJ, Suh SJ, Oliver JW, Hastert MC, Griswold JA, et al. Pseudomonas aeruginosa forms biofilms in acute infection independent of cell-to-cell signaling. Infect Immun. 2007;75:3715–21. doi: 10.1128/IAI.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stepanovic S, Vukovic D, Dakic I, Savic B, Svabic-Vlahovic M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J Microbiol Meth. 2000;40:175–9. doi: 10.1016/s0167-7012(00)00122-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Zhou JY, QU TT, Shen P, Wei ZQ, Yu YS, et al. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Chinese hospitals. Int J Antimicrob Agents. 2010;35:486–491. doi: 10.1016/j.ijantimicag.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O’Toole GA, et al. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Nat Acad Sci U S A. 2003;100:7907–12. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]