Abstract
Viruses are the main cause of respiratory tract infections. Metagenomic next-generation sequencing (mNGS) enables unbiased detection of all potential pathogens. To apply mNGS in viral diagnostics, sensitive and simultaneous detection of RNA and DNA viruses is needed. Herein, were studied the performance of an in-house mNGS protocol for routine diagnostics of viral respiratory infections with potential for automated pan-pathogen detection. The sequencing protocol and bioinformatics analysis were designed and optimized, including exogenous internal controls. Subsequently, the protocol was retrospectively validated using 25 clinical respiratory samples. The developed protocol using Illumina NextSeq 500 sequencing showed high repeatability. Use of the National Center for Biotechnology Information’s RefSeq database as opposed to the National Center for Biotechnology Information’s nucleotide database led to enhanced specificity of classification of viral pathogens. A correlation was established between read counts and PCR cycle threshold value. Sensitivity of mNGS, compared with PCR, varied up to 83%, with specificity of 94%, dependent on the cutoff for defining positive mNGS results. Viral pathogens only detected by mNGS, not present in the routine diagnostic workflow, were influenza C, KI polyomavirus, cytomegalovirus, and enterovirus. Sensitivity and analytical specificity of this mNGS protocol were comparable to PCR and higher when considering off-PCR target viral pathogens. One single test detected all potential viral pathogens and simultaneously obtained detailed information on detected viruses.
Respiratory tract infections pose a great burden on public health, causing extensive morbidity and mortality among patients worldwide.1, 2, 3 Most acute respiratory tract infections are caused by viruses, such as rhinovirus, influenza A and B viruses, metapneumovirus, and respiratory syncytial virus.4 However, in 20% to 62% of the patients, no pathogen is detected.4, 5, 6 This might be the result of diagnostic failures or even infection by unknown pathogens, such as the Middle East respiratory syndrome coronavirus in 2012.7
Rapid identification of the respiratory pathogen is critical to determine downstream decision making, such as isolation measures or treatment, including cessation of antibiotic therapy. Current diagnostic amplification methods, such as real-time quantitative PCR (qPCR), are sensitive and specific, but are only targeting predefined virus species or types. Genetic diversity within the virus genome and the sheer number of potential pathogens in many clinical conditions pose limitations to predefined primer- and probe-based approaches, leading to false-negative results.8 These limitations, combined with the potential emergence of new or unusual pathogens, highlight the need for less restricted approaches that could improve the diagnosis and subsequent outbreak management of infectious diseases.
Metagenomics relates to the study of the complete genomic content in a complex mixture of (micro) organisms.9 Unlike bacteria, viruses do not display a common gene in all virus families, and therefore pan-virus detection relies on catch-all analytic methods. Metagenomic or untargeted next-generation sequencing (mNGS) offers a culture- and nucleotide sequence–independent method that eliminates the need to define the targets for diagnosis beforehand. Besides primary detection, mNGS immediately offers additional information, on virulence markers, epidemiology, genotyping, and evolution of pathogens.7 , 10, 11, 12 Furthermore, quantitative assessment of the presence of virus copies in the sample is enabled by the number of reads.8
Although original mNGS studies typically aim at analysis of (shifts in) population diversity of abundant DNA microbes, detection of viral pathogens in patient samples requires a different technical approach because of the usually low abundance of viral pathogens (<1%) in clinical samples and the requisite of detecting both DNA and RNA viruses. Hence, a low limit of detection for RNA and DNA in one single assay is essential for implementation of mNGS for routine pathogen detection in clinical diagnostic laboratories. Current viral mNGS protocols are optimized for either RNA or DNA detection.11 , 13, 14, 15 Consequently, detection of both RNA and DNA viruses requires parallel workup of both RNA and DNA pretreatment methods. In addition, to increase the relative concentration of viral sequences, viral particle enrichment techniques are often applied.8 , 12 These techniques are laborious and not easily automated for routine clinical diagnostic use. Moreover, during enrichment directed at viral particles, intracellular viral nucleic acids as genomes and mRNAs are being discarded. After sequencing, the bioinformatic classification and interpretation of the results remain a major challenge. Bioinformatic classifiers are often developed for use in either microbiome studies or classification of high abundant reads, whereas extensive validation for clinical diagnostic use in settings of low abundance is limited. After bioinformatics classification, the challenge remains to discriminate between viruses that play a role in disease etiology and nonpathogenic viruses.16 Before considering mNGS in routine diagnostics, there is a need for critical evaluation and validation of every step in the procedure.
In this study, we evaluated a metagenomic protocol for NGS-based pathogen detection with sample pretreatment for DNA and RNA in a single tube. The method was validated using a selection of 25 respiratory pediatric samples from the total 29 positive and 346 negative viral PCR results. The main study objective was to define a sensitive and specific method for mNGS to be used as a broad diagnostic tool for viral respiratory diseases with the potential for automated pan-pathogen detection.
Materials and Methods
Sample Selection
Twenty-five stored clinical respiratory samples (−80°C) from pediatric patients, sent to the microbiological laboratory for routine viral diagnostics in 2016, were selected from the laboratory database (General Laboratory Information Management System; MIPS, Ghent, Belgium) at the Leiden University Medical Center (Leiden, the Netherlands). On the basis of previous PCR test results, a variety of 21 positive and four negative respiratory virus samples with a wide range of quantification cycle (Cq) values were included. The sample types represented routine diagnostic samples from pediatric patients that had been sent to our laboratory: 19 nasopharyngeal washings, two sputa, two bronchoalveolar lavages, one bronchial washing, and one throat swab (in viral transport medium). The patient selection (age range, 1.2 months to 15 years) represented the pediatric population with respiratory diagnostics in our university hospital in terms of (underlying) illness.
Sample Pretreatment
Total nucleic acids were extracted directly from 200 μL of clinical material using the MagNAPure 96 DNA and Viral NA Small Volume Kit (Roche Diagnostics, Almere, the Netherlands) with 100 μL output eluate.
Internal Controls
Clinical material was spiked with equine arteritis virus (EAV) and phocine herpesvirus 1 [PhHV1; kindly provided by Dr. H.G.M. (Bert) Niesters, UMC Groningen, the Netherlands], as internal controls for RNA detection17 and DNA detection, respectively.18 To determine the optimal concentration of the internal controls, a 10-fold dilution series of PhHV1/EAV was added to a mix of two pooled influenza A positive throat swabs (Cq value, 25) and read count and Cq values were compared. Concentration was based on the number of mNGS reads.
Quality Control
Before sequencing, the DNA input concentration was measured with the Qubit (Thermo Fisher Scientific, Waltham, MA), to determine whether there was sufficient DNA in the sample to obtain sequencing results. The range of DNA input for library preparation was 0.5 ng/μL for throat swabs (see reproducibility experiment) up to 300 ng/μL for bronchoalveolar lavages and sputa.
Fragmentation
To compare the effect of different DNA fragmentation techniques, six PCR-positive samples (containing one to three viruses) and three PCR-negative samples were chemically fragmented using zinc (10 minutes) as part of the New England Biolabs Library Prep Kit protocol, as described next in Library Preparation, and physically fragmented using sonication with the Bioruptor pico (Diagenode, Seraing, Belgium; on/off time, 18/30 seconds, 5 cycli).19 Three samples were also tested with the high-intensity settings of the Bioruptor pico (on/off time, 30/40 seconds; 14 cycli).
Library Preparation
Libraries were constructed with 7 μL extracted nucleic acids using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) using single, unique adaptors. This kit has been developed for transcriptome analyses. Several adaptations were made to the manufacturer’s protocol to enable simultaneous detection of both DNA and RNA viruses. The following steps were omitted: poly A mRNA capture isolation (instruction manual New England Biolabs number E7420S/L, version 8.0, chapter 1), rRNA depletion, and DNase step (chapter 2.1 to 2.4, 2.5B, 2.11A).
The size of fragments in the library was 300 to 700 bp. Adaptors were diluted 30-fold given the low RNA/DNA input and 21 PCR cycli were run after adaptor ligation.
Nucleotide Sequence Analysis
Sequencing was performed on Illumina HiSeq 4000 and NextSeq 500 sequencing systems (Illumina, San Diego, CA), obtaining 10 million 150-bp paired-end reads per sample.
Detection Limit
To determine the detection limit of mNGS, serial dilutions (undiluted, 10−1, 10−2, 10−3, and 10−4) of an influenza A–positive sample were tested with both mNGS and laboratory-developed real-time PCR. On the basis of run-off transcript experiments, the typical limit of detection of our real-time RNA PCRs was estimated to be 10 to 50 copies/reaction (data not shown).
Repeatability (Within-Run Precision)
To estimate the reproducibility of metagenomic sequencing, an influenza A–positive clinical sample (throat swab) was divided into four aliquots, nucleic acids were extracted, and library preparation and subsequent sequence analysis on the Illumina HiSeq 4000 were performed in one run.
Bioinformatics
Taxonomic Classification
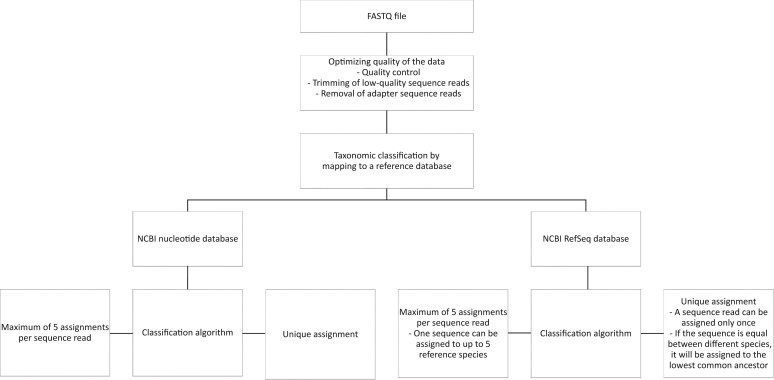
All FASTQ files were processed using the BIOPET Gears pipeline version 0.9.0, developed at the Leiden University Medical Center (http://biopet-docs.readthedocs.io/en/stable, last accessed September 12, 2018). This pipeline performs FASTQ preprocessing (including quality control, quality trimming, and adapter clipping) and taxonomic classification of sequencing reads. In this project, FastQC version 0.11.2 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc, last accessed September 12, 2018) was used for checking the quality of the raw reads. Low-quality read trimming was done using Sickle version 1.33 (https://github.com/najoshi/sickle) with default settings. Adapter clipping was performed using Cutadapt20 version 1.10 with default settings. Taxonomic classification of reads was performed with Centrifuge21 version 1.0.1-beta. The prebuilt nucleotide index, which contains all sequences from the National Center for Biotechnology Information’s (NCBI's) nucleotide database, provided by the Centrifuge developers was used (ftp://ftp.ccb.jhu.edu/pub/infphilo/centrifuge/data/old-indices, last accessed November 16, 2017) as the reference database. An overview of the bioinformatic process is shown in Figure 1 .
Figure 1.
The bioinformatic workflow of the metagenomic next-generation sequencing protocol studied. NCBI, National Center for Biotechnology Information.
In addition, a customized reference centrifuge index with sequence information obtained from the NCBI's RefSeq22 (accessed February 2019) database was built. RefSeq genomic sequences for the domains of bacteria, viruses, archaea, fungi, protozoa, as well as the human reference, along with the taxonomy identifiers, were downloaded with the Centrifuge-download utility and were used as input for Centrifuge-build.
Centrifuge settings were evaluated to increase the sensitivity and specificity. The default setting, with which a read can be assigned to up to five different taxonomic categories, was compared with one unique assignment per read,21 where a read is assigned to a single taxonomic category, corresponding to the lowest common ancestor of all matching species.
Kraken-style reports with taxonomical information were produced by the Centrifuge-kreport utility for all (default) options. Both unique and nonunique assignments can be reported, and these settings were compared. The resulting tree-like structured, Kraken-style reports were visualized with Krona23 version 2.0.
Horizontal coverage (percentage) was determined using GenomeDetective website24 version 1.111 (https://www.genomedetective.com, last accessed May 4, 2019).
In silico simulated EAV reads were analyzed in different databases (NCBI's nucleotide versus RefSeq) and classification algorithms [maximum, five labels per sequence, versus unique, lowest (common ancestor), and reporting (nonunique versus unique)] to determine the most sensitive and specific bioinformatic analyses using Centrifuge.
To determine the amount of reads needed, results of one million reads and 10 million reads were compared. A total of one million reads were randomly selected of the 10 million reads of one FASTQ file and analyzed. The random selection was performed with the FastqSplitter (https://github.com/biopet/biopet/blob/v0.9.0/docs/tools/FastqSplitter.md, last accessed September 12, 2018), which cuts a FASTQ file of 10 million reads into 10 pieces, of which one was selected. Read counts were normalized by the total read count and target virus genome size.
Assembly of PhHV1 Sequences
Because NCBI's databases were lacking a complete PhHV1 genome sequence, PhHV1 was sequenced; and based on the gained sequence reads, the genome was built using SPAdes.25 PhHV1 assembly was done using the biowdl virus-assembly pipeline version 0.1 (https://github.com/biowdl/virus-assembly, last accessed September 12, 2018). The quality control part of the biowdl pipeline determines which adapters need to be clipped by using FastQC version 0.11.7 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc, last accessed September 12, 2018) and cutadapt version 1.16,20 with minimum length setting 1. The resulting reads were down sampled within bowdl to 250,000 reads using seqtk version 1.2 (https://github.com/lh3/seqtk, last accessed September 12, 2018), after which SPADES version 3.11.125 was run to get the first proposed genome contigs.
To retrieve longer assembly contigs, a reiterative assembly approach was used by processing the proposed contigs by the biowdl reAssembly pipeline 0.1. This preassembly pipeline aligns reads to contigs of a previous assembly, then selects the aligned reads, down samples them, and runs a new assembly using SPADES. Subtools used for this consisted of BWA 0.7.1726 for indexing and mapping, SAMtools 1.627 for generating bam files, SAMtools view version 1.7 for filtering out unmapped reads using the setting -G 12, and Picard SamToFastq version 2.18.4 and seqtk for generating FASTQ files with 250,000 reads. The contigs from the reAssembly pipeline were then processed for a second using SPADES, with setting the cov-cutoff to five. The resulting contigs were then processed with the reAssembly pipeline for the third and last time, setting the cov-cutoff in SPADES to 20.
The contigs from the last reAssembly step were then run against the blast nucleotide database using blastn 2.7.128 Of 23 contigs, only five that showed the lowest percentage in identity matches with any other possible non–herpes virus species were selected. The final five contigs contained sequence lengths of 97,893, 8170, 3710, 3294, and 1279 nucleotides; the average coverage was 206, 131, 211, 285, and 154, respectively. The proposed almost complete genome of PhHV1 was added to NCBI's GenBank database (https://www.ncbi.nlm.nih.gov/genbank; accession number MH509440).
Retrospective Validation
Clinical sensitivity was analyzed using the optimized procedure, which in short consisted of total nucleic acid extraction, including internal controls (1:100 dilution); the adapted New England Biolabs Next library preparation protocol, including fragmentation with zinc, for combined RNA and DNA detection (see Library Preparation); and sequencing of 10 million reads (Illumina NextSeq 500). Bioinformatic analyses were performed using Centrifuge with NCBI's RefSeq database and unique assignment of the sequence reads.
Sensitivity and specificity of the metagenomic NGS procedure were compared with a published updated version of our laboratory-developed multiplex qPCR.29 The routine multiplex PCR panel consisted of 15 respiratory target pathogens: influenza A/B viruses, respiratory syncytial virus, metapneumovirus, adenovirus, human bocavirus, parainfluenza viruses 1/2/3/4, rhinovirus, and the coronaviruses HKU1, NL63, 227E, and OC43. Thus, in total, 375 PCR results were available (15 targets × 25 samples), of which 29 were PCR positive and 346 were PCR negative for comparison with mNGS.
Ethical Approval of Patient Studies
The study design was approved by the medical ethics review committee of the Leiden University Medical Center (reference B16.004).
Results
Internal Controls
Serial dilutions of EAV and PhHV1 were added to an influenza A PCR-positive sample. Serial dilution 1:10,000 detected EAV with a substantial read count in the presence of a viral infection and without a significant decline in target virus family reads (Table 1 ). On the basis of these results, the concentration of internal controls was determined for further experiments.
Table 1.
Internal Controls EAV/PhHV-1: Serial Dilutions against a Clinical Sample Background and Within-Run Precision (INFA)
| Sample EAV/PhHV-1 dilution | Cq value |
Centrifuge reads (log) |
||||
|---|---|---|---|---|---|---|
| INFA | EAV | PhHV-1 | INFA | EAV | PhHV-1 | |
| 1:100 | 24.52 | 21.59 | 23.52 | 4438 (3.6) | 12,925 (4.1) | 347 (2.5) |
| 1:1000 | 24.67 | 24.91 | 26.83 | 3742 (3.6) | 1202 (3.1) | 49 (1.7) |
| 1:10,000 | 24.76 | 28.45 | 30.33 | 4628 (3.7) | 95 (2.0) | 14 (1.1) |
| 1:100,000 | 24.79 | 30.85 | 32.55 | 4093 (3.6) | 18 (1.3) | 14 (1.1) |
Cq, quantification cycle; EAV, equine arteritis virus; INFA, influenza A virus; PhHV-1, phocine herpesvirus 1.
The EAV Cq value of the dilutions correlated with the number of EAV reads from the Centrifuge analysis.
Fragmentation
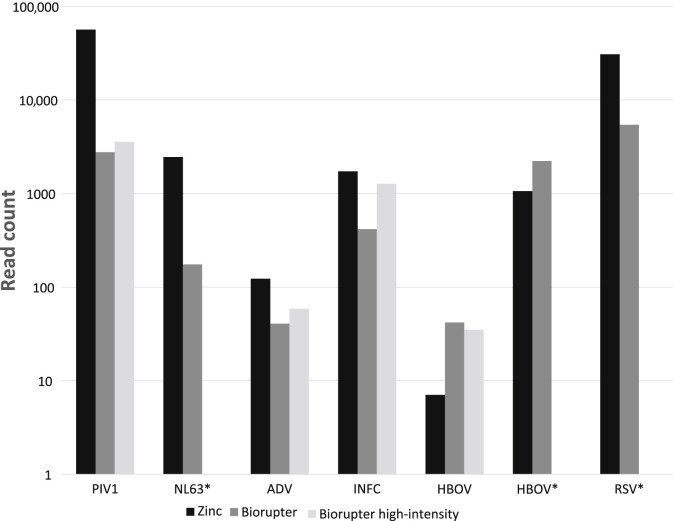
The comparison of fragmentation methods was done using a selection of samples with relevant target reads and performed on the Illumina NextSeq 500. The total reads were comparable among the three protocols (Figure 2). The protocol with zinc fragmentation had higher yield in target virus reads for all RNA viruses tested and adenovirus.
Figure 2.
Comparison of fragmentation methods on target reads (species level, log scale). Asterisks indicate not tested with Bioruptor setting high intensity. ADV, adenovirus; HBOV, human bocavirus; INFC, influenza C virus; NL63, coronavirus NL63; PIV, parainfluenza virus; RSV, respiratory syncytial virus.
Detection Limit
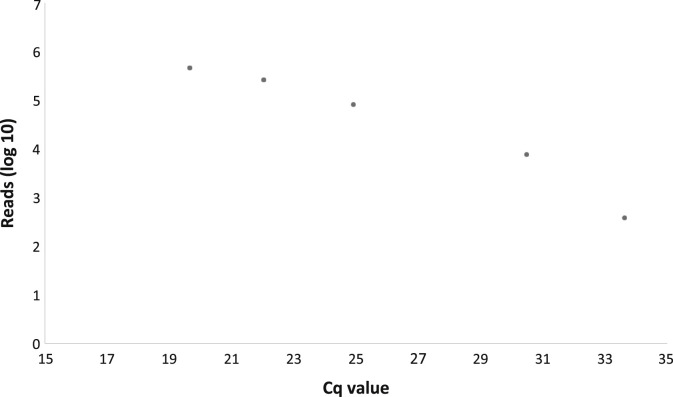
The detection threshold of our NGS limit, deduced from serial dilutions of influenza A (Figure 3 ) and EAV (Table 1), was comparable with a real-time PCR Cq value of >35, corresponding to approximately <50 to 250 copies/reaction.
Figure 3.
Serial dilutions of an influenza A–positive clinical sample. Cq, quantification cycle.
Repeatability: Within-Run Precision
The mNGS results of an influenza A–positive sample tested in quadruple could be reproduced with only minor differences (Table 1): CV of 1.1%: 0.04 log SD/3.6 log average.
Bioinformatics: Taxonomic Classification
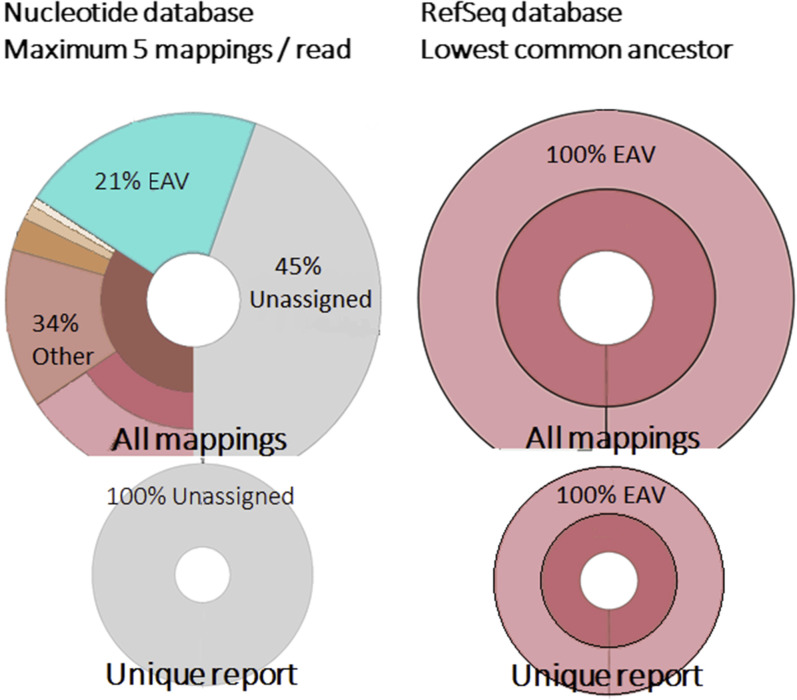
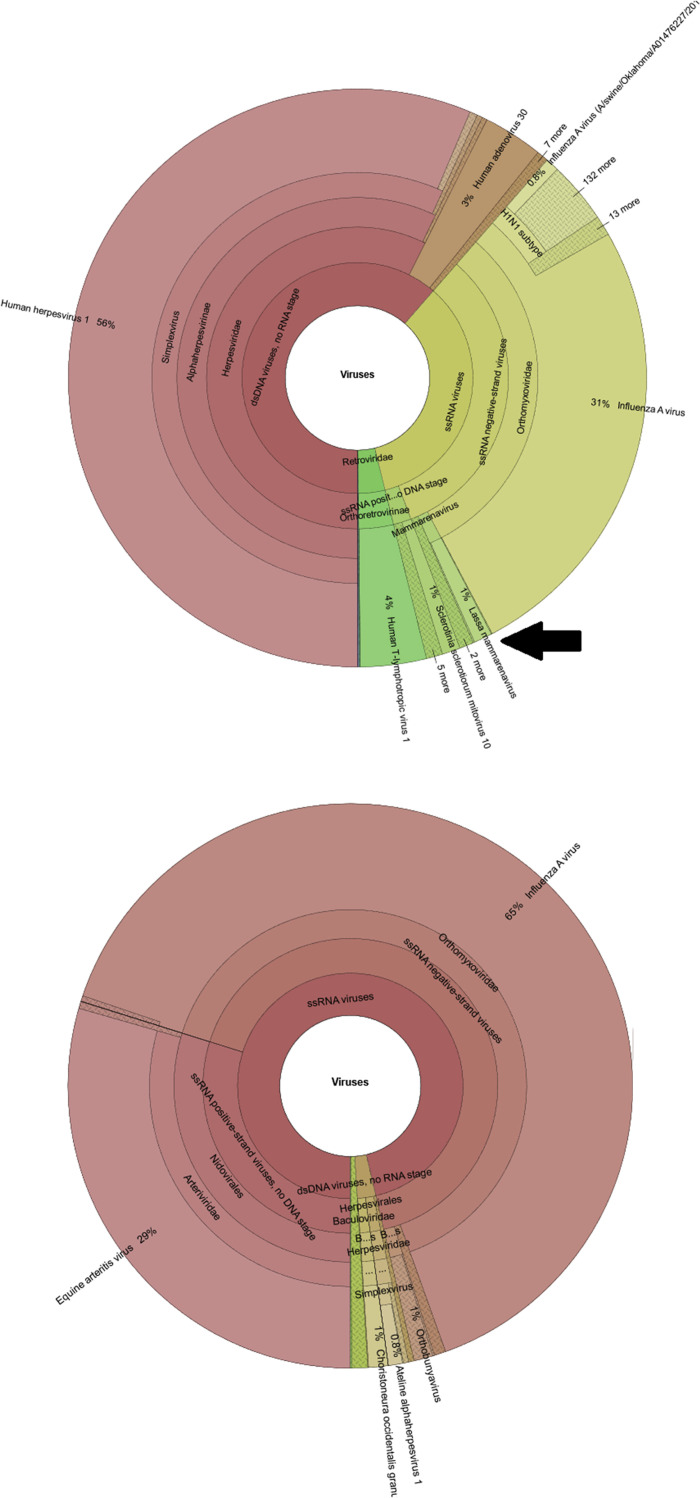
The Centrifuge default settings, with NCBI's nucleotide database and assignment of sequence reads to a maximum of five labels per sequence, resulted in various spurious classifications (Figure 4 ) [eg, Lassa virus (Figure 5 ), evidently highly unlikely to be present in patient samples from the Netherlands with respiratory complaints]. The specificity could be increased by using NCBI's RefSeq database instead of NCBI's nucleotide database. The classification was further improved by changing the Centrifuge tool settings to limit the assignment of homologous reads to the lowest common ancestor (maximum, one label per sequence).
Figure 4.
Analysis of in silico simulated equine arteritis virus (EAV) reads with the different bioinformatic settings of the Centrifuge pipeline.
Figure 5.
Spurious Lassa virus reads detected using the National Center for Biotechnology Information’s (NCBI's) nucleotide database (top), versus NCBI's RefSeq database (bottom). Black arrow points to the spurious Lassa virus reads. dsDNA, double-stranded DNA; ssRNA, single-stranded RNA.
The Centrifuge reporting of shared sequences between different organisms/subtypes differs, dependent of the classification and reporting algorithm. The default classification will assign a shared read to a maximum of five organisms (one read will be assigned five times); and with the lowest common ancestor classification setting, this read will only be assigned once (namely, to the lowest ancestor these organisms/subtypes have in common). Classification with a maximum of five labels per read resulted in two different outcomes using the report with all mappings and the report with unique mappings, with the latter not reporting the reads assigned to multiple organisms.
Comparison of classification using these different settings shows the highest sensitivity and specificity using NCBI's RefSeq database with one label (lowest common ancestor) assignment, with both in silico prepared data sets containing solely EAV sequence fragments (Figure 4) and clinical data sets (with highly abundant background) (Figure 5).
To determine the effect of the total number of sequencing reads obtained per sample on sensitivity, 1 million and 10 million total reads were compared by in silico analysis (Table 2 ). One million total reads resulted in an approximate 10-fold decrease in target virus read count compared with 10 million total reads, implicating a reduction of sensitivity.
Table 2.
Comparison of Analysis of 1 Million versus 10 Million Reads
| Virus | Virus family | Cq value | 10 million reads |
1 million reads |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total reads | Virus family reads | % of total | % of viral | Total reads | Virus family reads | % of total | % of viral | |||
| RV | Picornaviridae | 37.7 | 8,203,894 | 8941 | 0.06 | 84.37 | 822,218 | 889 | 0.07 | 86.11 |
| PIV4 | Paramyxoviridae | 24.9 | 10,886,798 | 2136 | 0.04 | 41.90 | 1,088,067 | 199 | 0.08 | 40.73 |
| CMV | Herpesviridae | 34.5 | 15,889,428 | 22 | 00.01 | 10.88 | 1,588,922 | 2 | 0.04 | 11.87 |
| ADV | Adenoviridae | 30.2 | 11,146,488 | 0 | 0 | 0 | 1,115,135 | 0 | 0.03 | 0 |
| RSV | Pneumoviridae | 27.3 | 10,191,995 | 1477 | 0.02 | 53.29 | 1,019,415 | 163 | 0.04 | 59.25 |
| INFB | Orthomyxoviridae | 30 | 8,535,672 | 652 | 0.01 | 48.67 | 853,149 | 61 | 0.02 | 46.58 |
| NL63 | Coronaviridae | 36.2 | 10,386,928 | 0 | 0 | 0 | 1,038,469 | 0 | 0.02 | 0 |
| INFA | Orthomyxoviridae | 27.5 | 10,981,601 | 8403 | 0.11 | 70.28 | 1,097,872 | 855 | 0.17 | 69.84 |
| MPV | Pneumoviridae | 34.1 | 12,972,626 | 2 | 0 | 0.10 | 1,297,151 | 0 | 0.02 | 0 |
| HBOV | Parvoviridae | 32.2 | 11,819,805 | 0 | 0 | 0 | 1,181,738 | 0 | 0 | 0 |
| RV | Picornaviridae | 23.1 | 11,819,805 | 58,695 | 0.42 | 84.27 | 1,183,738 | 5754 | 0.49 | 84.25 |
% of total, percentage of total reads; % of viral, percentage of all viral reads; ADV, adenovirus; CMV, cytomegalovirus; Cq, quantification cycle; HBOV, human bocavirus; INFA, influenza A virus; INFB, influenza B virus; MPV, metapneumovirus; NL63, coronavirus NL63; PIV4, parainfluenza virus 4; RSV, respiratory syncytial virus; RV, rhinovirus.
Retrospective Validation
Clinical Sensitivity Based on PCR Target Pathogens
Clinical sensitivity was analyzed using the optimized mNGS procedure. The sample collection consisted of 21 clinical specimens positive for at least one of the following PCR target viruses: rhinovirus, influenza A and B, parainfluenza viruses 1 and 4, metapneumovirus, respiratory syncytial virus, coronaviruses NL63 and HKU1, human bocavirus, and adenovirus. Fourteen samples were positive for one virus, six samples were positive for two viruses, and one sample was positive for three viruses with the laboratory-developed respiratory multiplex qPCR. Cq values ranged from Cq 17 to Cq 35, with a median of 23.
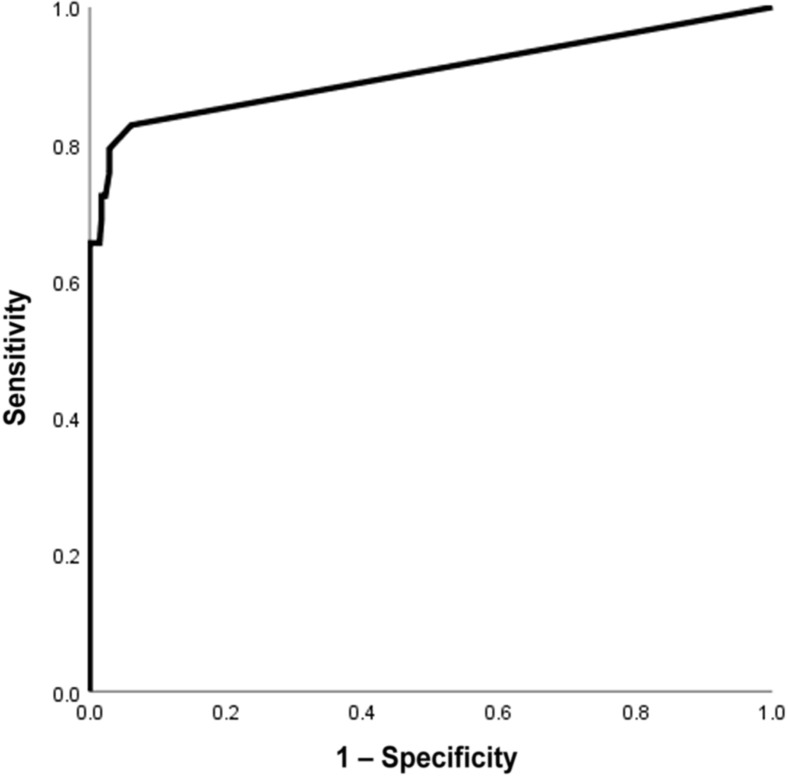
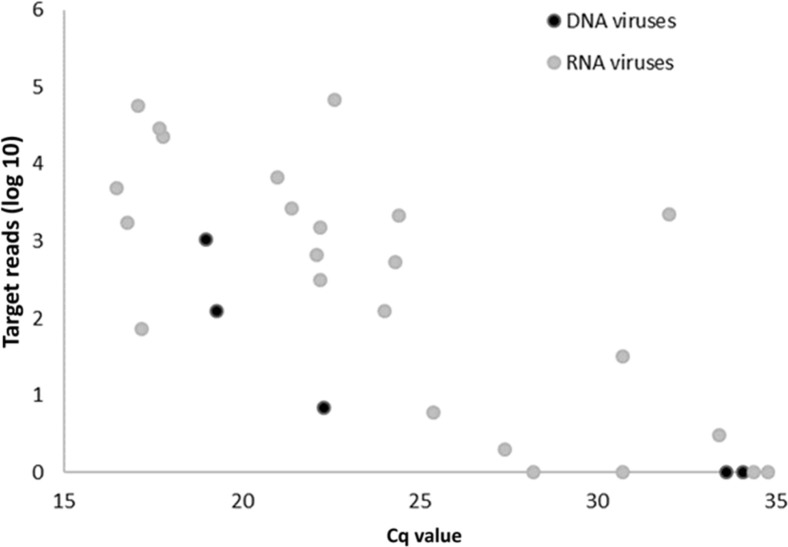
With mNGS, 24 of the 29 viruses demonstrated in routine diagnostics were detected (Table 3 ), resulting in a sensitivity of 83% for PCR targets. If a cutoff of 15 reads was handled, sensitivity declined to 66% (19/29) (Table 4 ). A receiver-operating characteristic curve for mNGS detection of PCR target viruses, depending on the cutoff level of the number of mapped sequence reads for defining a positive result, is shown in Figure 6 ; mNGS target read count (log value) showed a correlation (Pearson correlation coefficient, −0.582; P = 0.003) with the Cq values of the qPCR (Figure 7 ).
Table 3.
Detection of qPCR Virus Positive Respiratory Samples with mNGS
| Material | Routine diagnostics |
mNGS |
||||
|---|---|---|---|---|---|---|
| PCR positive | Cq value | Virus genus | Genus reads∗ | Virus species | Species reads∗ | |
| NP wash | RV | 30.7 | Enterovirus | 0 | Rhinovirus | 0 |
| PIV1 | 17.1 | Respirovirus | 58,619 | Human respirovirus 1 | 56,407 | |
| ADV | 33.6 | Mastadenovirus | 0 | Human mastadenovirus C | 0 | |
| NP wash | MPV | 24 | Metapneumovirus | 127 | Human metapneumovirus | 123 |
| BAL | NL63 | 24.4 | Alphacoronavirus | 1999 | Human coronavirus NL63 | 2176 |
| HKU1 | 28.2 | Betacoronavirus | 1 | Human coronavirus HKU1 | 1 | |
| Sputum | RV | 32 | Enterovirus | 2326 | Rhinovirus C | 2204 |
| NP wash | INFA | 22.2 | Alphainfluenzavirus | 1490 | Influenza A virus (A/California/07/2009 (H1N1)) | 1490 |
| NP wash | MPV | 33.4 | Metapneumovirus | 1 | Human metapneumovirus | 3 |
| ADV | 19.3 | Mastadenovirus | 125 | Human mastadenovirus C | 123 | |
| Sputum | PIV4 | 21 | Orthorubulavirus | 7729 | Human rubulavirus virus 4 (subtype a) | 6798 |
| NP wash | HBOV | 22.3 | Bocaparvovirus | 7 | Human bocavirus | 7 |
| NP wash | MPV | 22.2 | Metapneumovirus | 139 | Human metapneumovirus | 312 |
| NP wash | INFB | 16.5 | Betainfluenzavirus | 4971 | Influenza B virus (B/Lee/1940) | 4971 |
| NP wash | RV | 25.4 | Enterovirus | 8 | Rhinovirus A | 6 |
| RSV | 30.7 | Orthopneumovirus | 32 | Human orthopneumovirus | 32 | |
| NP wash | INFB | 21.4 | Betainfluenzavirus | 2686 | Influenza B virus (B/Lee/1940) | 2686 |
| NP wash | RSV | 17.8 | Orthopneumovirus | 29,900 | Human orthopneumovirus | 22,483 |
| NP wash | RV | 34.4 | Enterovirus | 0 | Rhinovirus | 0 |
| INFB | 22.6 | Betainfluenzavirus | 68,972 | Influenza B virus (B/Lee/1940) | 68,972 | |
| BAL | INFB | 34.8 | Betainfluenzavirus | 0 | Influenza B virus | 0 |
| HBOV | 34.1 | Bocaparvovirus | 0 | Human bocavirus | 0 | |
| NP wash | HKU1 | 24.3 | Betacoronavirus | 534 | Human coronavirus HKU1 | 535 |
| NP wash | RV | 16.8 | Enterovirus | 3877 | Rhinovirus A | 1721 |
| NP wash | RV | 27.4 | Enterovirus | 1 | Rhinovirus B | 2 |
| HBOV | 19 | Bocaparvovirus | 1014 | Human bocavirus | 1064 | |
| NP wash | INFA | 22.1 | Alphainfluenzavirus | 657 | Influenza A virus (A/California/07/2009 (H1N1)) | 657 |
| NP wash | RSV | 17.2 | Orthopneumovirus | 31,179 | Human orthopneumovirus | 72 |
| NP wash | RV | 17.7 | Enterovirus | 50,642 | Rhinovirus A | 29,293 |
ADV, adenovirus; BAL, bronchoalveolar lavage; Cq, quantification cycle; HBOV, human bocavirus; HKU1, coronavirus HKU1; INFA, influenza A virus; INFB, influenza B virus; mNGS, metagenomic next-generation sequencing; MPV, metapneumovirus; NL63, coronavirus NL63; NP, nasopharyngeal; PIV, parainfluenza virus; qPCR, real-time quantitative PCR; RSV, respiratory syncytial virus; RV, rhinovirus.
Number of reads assigned to the genus or species of the target virus.
Table 4.
Sensitivity and Specificity of the mNGS Protocol Tested, Based on PCR Target Viruses, with Different Sequence Read Cutoff Levels for Defining a Positive Result
| Variable | All reads | ≥15 sequence reads | ≥50 sequence reads |
|---|---|---|---|
| Sensitivity | 83 (24/29) | 66 (19/29) | 62 (18/29) |
| Specificity | 94 (325/346) | 100 (345/346) | 100 (346/346) |
Data are given as percentage (number/total).
mNGS, metagenomic next-generation sequencing.
Figure 6.
Receiver-operating characteristic curve for metagenomic next-generation sequencing detection of PCR target viruses, depending on the cutoff level of the number of mapped sequence reads for defining a positive result.
Figure 7.
Semiquantification of the metagenomic next-generation sequencing assay for target virus detection in clinical samples with real-time quantitative PCR confirms human respiratory viruses. Cq, quantification cycle.
Detection of Additional Viral Pathogens by mNGS: Off-PCR Target Viruses
Next to the viral pathogens tested by PCR, mNGS also detected other pathogenic viruses, indicating additional viral sequences uncovered by mNGS but not included in the routine diagnostics, with influenza C virus being the most prominent. A high amount, 2221 reads (99% horizontal coverage), of influenza C virus reads (58% of all viral reads and 0.02 of the total reads) was found in one sample; confirmatory PCR was not routinely available. Other potential respiratory pathogens detected by mNGS and not included in PCR analysis were KI polyomavirus [two samples: 262 and 46 reads; retrospective in-house PCR Cq 25 (1:10 dilution) and 26, respectively], cytomegalovirus (human betaherpesvirus 5; 55 and 3 reads; retrospective in-house PCR Cq 22 and 27, respectively), and enterovirus (10,073 reads; retrospective in-house PCR rhinovirus/enterovirus Cq 18). All these viruses are not included routinely in the diagnostic multiplex qPCRs.
Internal Controls
The spiked-in internal controls were detected by mNGS in all samples. EAV sequence reads ranged from 14 to 19,894 (median, 362), and PhHV1 sequence reads ranged from 41 to 1206 (median, 121).
Analytical Specificity Based on PCR Target Viruses
In total, 25 pediatric respiratory samples were available to evaluate the analytical specificity of mNGS: four samples were negative for all 15 viral pathogens in the multiplex PCR panel (influenza A/B, respiratory syncytial virus, human metapneumovirus, adenovirus, human bocavirus, parainfluenza viruses 1/2/3/4, rhinovirus, HKU1, NL63, 227E, and OC43), and 21 samples were negative for 12 to 14 of these PCR target pathogens.
Out of a total 346 negative target PCR results from these 25 samples, 325 results corresponded with the finding of 0 target-specific reads by mNGS. If a cutoff of 15 reads was used, 345 of the 346 negative PCR targets were negative with mNGS. The sample positive by mNGS and negative by PCR was human parainfluenza virus 3 (18 reads). Although no conclusive proof for either true- or false-positive mNGS results could be found, specificity of mNGS was 94% (325/346) when encountering all reads and ≥99% (345/346) with a 15-read cutoff (Table 4 and receiver-operating characteristic curve in Figure 6).
Antiviral Susceptibility
In addition to subtyping (Table 3), using the metagenomic sequence data, the nucleotide positions that conferred resistance to either oseltamivir or zanamivir were analyzed. Sequence data of amino acids I117, E119, D198, I222, H274, R292, N294, and I314 showed susceptibility to oseltamivir; and sequence data of amino acids V116, R118, E119, Q136, D151, R152, R224, E276, R292, and R371 revealed susceptibility to zanamivir.30 , 31
Data Access
The raw sequence data of the samples, after removal of human reads, have been deposited to the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra; accession numbers SRX6715205 to SRX6715229).
Discussion
Metagenomic sequencing has not yet been implemented as a routine tool in clinical diagnostics of viral infections. Such application would require the careful definition and validation of several parameters to enable the accurate assessment of a clinical sample with regard to the presence or absence of a pathogen, to fulfill current accreditation guidelines. Therefore, this study has initiated the optimization of several steps throughout the presequencing and post-sequencing workflow, which are considered essential for sensitive and specific mNGS-based virus detection. Many virus discovery or virus diagnostic protocols have focused on the enrichment of viral particles32 with the intention to increase the relative amount of virus reads. However, these methods are laborious and intrinsically exclude viral nucleic acid located in host cells. Herein, a sample pretreatment protocol was designed with potential for: i) automation, ii) pan-pathogen detection, and iii) detection of intracellular viral nucleic acids. Consequently, any type of viral enrichment was excluded (filtration, centrifugation, nucleases, and rRNA removal). The current protocol enabled high-throughput sample pretreatment by means of automated nucleic acid extraction and without depletion of bacterial or human genome, with potential for pan-pathogen detection. Several adaptations in the bioinformatic script resulted in more accurate reporting of the classification output.
Addition of an internal control to a PCR is commonly used for quality control in qPCR.33 Although the addition of internal controls in mNGS is not yet an accepted standard procedure, EAV and PhHV1 were used as an RNA and a DNA control, respectively, to monitor the workflow in this diagnostic application. The amount of internal control reads and target virus reads has been reported to be dependent on the amount of background reads (negative correlation).34 In our protocol, the internal controls were used as qualitative controls but may be used as indicator of the amount of background. PhHV1 showed less linearity in the dilution series, compared with EAV, which may be indicative for a potential relative difference in efficiency of amplification of PhHV1 viral sequences. Because NCBI's databases were lacking a complete PhHV1 genome, the Centrifuge index building and classification was limited to classification on a higher taxonomic rank. To achieve classification of PhHV1 at the species level, the whole genome of PhHV1 was sequenced; and based on the gained sequence reads, the genome was built.25 The proposed nearly complete genome of PhHV1 was submitted to NCBI's GenBank database.
Sensitivity of the mNGS protocol was maximum 83% based on PCR target viruses and depended on the cutoff level of reads for defining a positive result. Five viruses, which were not recovered by mNGS, had high Cq values, >30 (ie, a relatively low viral load). This may be a drawback of the retrospective nature of this clinical evaluation as RNA viruses may be degraded because of storage and freeze-thaw steps, resulting in lower sensitivity of mNGS. A correlation was found between read counts and PCR Cq value, demonstrating the quantitative nature of viral detection by mNGS. Discrepancies between the Cq values and the number of mNGS reads may be explained by unrepresentative Cq values (eg, by primer mismatch for highly divergent viruses, like rhinoviruses/enteroviruses and differences in sensitivity of mNGS for several groups of viruses, as has been reported by others).35 In addition, viral pathogens were detected that were not targeted by the routine PCR assays, including influenza C virus, which is typical of the unbiased nature of the method. In addition, although not within the scope of this study, bacterial pathogens, including Bordetella pertussis (qPCR confirmed), were also detected. In the current study, only viruses were targeted because these could be well compared with qPCR results; bacterial targets remain to be studied in clinical sample types as sputum or bronchoalveolar lavages that are more suitable for bacterial detection. The analytical specificity of mNGS appeared to be high, especially with a cutoff of 15 reads. However, the clinical specificity, the relevance of the lower read numbers, still needs further investigation in clinical studies.
Sequencing using Illumina HiSeq 4000 with single, unique indexes resulted in rhinovirus-C sequences (55 to 909 reads) in all samples run on one lane, which appeared to be identical sequences. Retesting of the samples with Illumina NextSeq 500 resulted in disappearance of these reads. This problem could be attributed to index hopping (index misassignment), as described earlier.36 Because of the chemistry, essential for the increased speed, the HiSeq 4000 is more prone to index hopping between neighboring samples. Although the percentage of reads that contributed to the index hopping was low, this is critical for clinical viral diagnostics, as this is aimed specifically at low abundance targets.36 , 37
Bioinformatics classification of metagenomic sequence data with the pipeline Centrifuge required identification of the optimal parameters to minimize misclassified and unclassified reads. Default settings of this pipeline resulted in higher rates of both false-positive and false-negative results. NCBI's nucleotide database includes a wide variety of unannotated viral sequences, such as partial sequences and (chimeric) constructs, in contrast to the curated and well-annotated sequences in NCBI's RefSeq database, which resulted in a higher specificity. In addition to the database, settings for the assignment algorithm were adapted as well. The assignment settings were adjusted to unique assignment in the case of homology to the lowest common ancestor. This modification resulted in higher sensitivity and specificity than the default settings; however, the ability to further subtyping diminished. This is likely to be attributed to the limited representation/availability of strain types within NCBI's RefSeq database. In consequence, this leads to a more accurate estimation of the common ancestor for particular viruses, but limited typing results in case of highly variable ones. To obtain optimal typing results, additional annotated sequences may be added or a new database should be built, with a high variety of well-defined and frequently updated virus strain types.
To conclude, this study contributes to the increasing evidence that metagenomic NGS can effectively be used for a wide variety of diagnostic assays in virology, such as unbiased virus detection, resistance mutations, virulence markers, and epidemiology, as shown by the ability to detect single-nucleotide polymorphisms in influenza virus.
These findings support the feasibility of moving this promising field forward to a role in the routine detection of pathogens by the use of mNGS. Further optimization should include the parallel evaluation of adult samples, the inclusion of additional annotated strain sequences to the database, and further elaboration of the classification algorithm and reporting for clinical diagnostics. The importance of both negative nontemplate control samples38 and healthy control cases may support the critical discrimination of contaminants and viral colonization from clinically relevant pathogens.
Conclusions
Optimal sample preparation and bioinformatics analysis are essential for sensitive and specific mNGS-based virus detection.
Using a high-throughput genome extraction method without viral enrichment, both RNA and DNA viruses could be detected with a sensitivity comparable to PCR.
Using mNGS, all potential pathogens can be detected in one single test, while simultaneously obtaining additional detailed information on detected viruses. Interpretation of clinical relevance is an important issue but essentially not different from the use of PCR-based assays and supported by the available information on typing and relative quantities. These findings support the feasibility of a role of mNGS in the routine detection of pathogens.
Acknowledgments
We thank our project partners Floyd Wittink, Wouter Suring (Hogeschool Leiden), Danny Duijsings (BaseClear), and Christiaan Henkel (Leiden University); Tom Vreeswijk, Lopje Höcker, and Mario van Bussel [Clinical Microbiology Laboratory, Leiden University Medical Center (LUMC)] for help with the presequencing experiments; and Jeroen Laros (Human Genetics, LUMC) for help with the bioinformatics.
Footnotes
Supported in part by GENERADE Centre of Expertise Genomics (Leiden, the Netherlands) grant 2016_001 (J.J.C.d.V.).
S.v.B. and A.L.v.R. contributed equally to this work.
A.C.M.K. and J.J.C.d.V. contributed equally to this work as senior authors.
Disclosures: None declared.
Current address of S.v.B., Department of Clinical Virology, Erasmus Medical Center, Rotterdam, the Netherlands; of N.P., Theoretical Biology and Bioinformatics, Department of Biology, Science for Life, Utrecht University, Utrecht, the Netherlands.
Author Contributions
S.v.B., A.L.v.R., E.C.J.C., A.C.M.K., and J.J.C.d.V. participated in the study design; S.v.B. performed the prelibrary preparation experiments; S.v.B., N.P., E.C.C., R.H.P.V., I.S., P.J.v.H., and H.M. performed bioinformatic analyses; S.v.B., A.L.v.R., and E.C.C. analyzed the data; S.v.B. and A.L.v.R. wrote the first version of the manuscript; and all authors contributed and revised the manuscript and approved the final manuscript.
References
- 1.Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nair H., Simoes E.A., Rudan I., Gessner B.D., Azziz-Baumgartner E., Zhang J.S. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet. 2013;381:1380–1390. doi: 10.1016/S0140-6736(12)61901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates M., Mudenda V., Mwaba P., Zumla A. Deaths due to respiratory tract infections in Africa: a review of autopsy studies. Curr Opin Pulm Med. 2013;19:229–237. doi: 10.1097/MCP.0b013e32835f4fe4. [DOI] [PubMed] [Google Scholar]
- 4.Jain S., Self W.H., Wunderink R.G., Fakhran S., Balk R., Bramley A.M., Reed C., Grijalva C.G., Anderson E.J., Courtney D.M., Chappell J.D., Qi C., Hart E.M., Carroll F., Trabue C., Donnelly H.K., Williams D.J., Zhu Y., Arnold S.R., Ampofo K., Waterer G.W., Levine M., Lindstrom S., Winchell J.M., Katz J.M., Erdman D., Schneider E., Hicks L.A., McCullers J.A., Pavia A.T., Edwards K.M., Finelli L., CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heikkinen T., Jarvinen A. The common cold. Lancet. 2003;361:51–59. doi: 10.1016/S0140-6736(03)12162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ieven M., Coenen S., Loens K., Lammens C., Coenjaerts F., Vanderstraeten A., Henriques-Normark B., Crook D., Huygen K., Butler C.C., Verheij T.J.M., Little P., Zlateva K., van Loon A., Claas E.C.J., Goossens H. Aetiology of lower respiratory tract infection in adults in primary care: a prospective study in 11 European countries. Clin Microbiol Infect. 2018;24:1158–1163. doi: 10.1016/j.cmi.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 8.Prachayangprecha S., Schapendonk C.M., Koopmans M.P., Osterhaus A.D., Schurch A.C., Pas S.D., van der Eijk A.A., Poovorawan Y., Haagmans B.L., Smits S.L. Exploring the potential of next-generation sequencing in detection of respiratory viruses. J Clin Microbiol. 2014;52:3722–3730. doi: 10.1128/JCM.01641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wooley J.C., Godzik A., Friedberg I. A primer on metagenomics. PLoS Comput Biol. 2010;6:e1000667. doi: 10.1371/journal.pcbi.1000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann B., Scheuch M., Hoper D., Jungblut R., Holsteg M., Schirrmeier H., Eschbaumer M., Goller K.V., Wernike K., Fischer M., Breithaupt A., Mettenleiter T.C., Beer M. Novel orthobunyavirus in cattle, Europe, 2011. Emerg Infect Dis. 2012;18:469–472. doi: 10.3201/eid1803.111905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mongkolrattanothai K., Naccache S.N., Bender J.M., Samayoa E., Pham E., Yu G., Dien Bard J., Miller S., Aldrovandi G., Chiu C.Y. Neurobrucellosis: unexpected answer from metagenomic next-generation sequencing. J Pediatr Infect Dis Soc. 2017;6:393–398. doi: 10.1093/jpids/piw066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., Osterhaus A.D., Haagmans B.L., Gorbalenya A.E., Snijder E.J., Fouchier R.A. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6) doi: 10.1128/mBio.00473-12. e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohl C., Brinkmann A., Dabrowski P.W., Radonic A., Nitsche A., Kurth A. Protocol for metagenomic virus detection in clinical specimens. Emerg Infect Dis. 2015;21:48–57. doi: 10.3201/eid2101.140766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker J., Chen J. Application of next generation sequencing for the detection of human viral pathogens in clinical specimens. J Clin Virol. 2017;86:20–26. doi: 10.1016/j.jcv.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou X., Tang G., Zhao X., Huang Y., Chen T., Lei M., Chen W., Yang L., Zhu W., Zhuang L., Yang J., Feng Z., Wang D., Wang D., Shu Y. Simultaneous virus identification and characterization of severe unexplained pneumonia cases using a metagenomics sequencing technique. Sci China Life Sci. 2017;60:279–286. doi: 10.1007/s11427-016-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wylie K.M., Mihindukulasuriya K.A., Sodergren E., Weinstock G.M., Storch G.A. Sequence analysis of the human virome in febrile and afebrile children. PLoS One. 2012;7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheltinga S.A., Templeton K.E., Beersma M.F., Claas E.C. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J Clin Virol. 2005;33:306–311. doi: 10.1016/j.jcv.2004.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalpoe J.S., Kroes A.C., de Jong M.D., Schinkel J., de Brouwer C.S., Beersma M.F., Claas E.C. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J Clin Microbiol. 2004;42:1498–1504. doi: 10.1128/JCM.42.4.1498-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wery M., Descrimes M., Thermes C., Gautheret D., Morillon A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods. 2013;63:25–31. doi: 10.1016/j.ymeth.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 20.Martin M. Cutadept removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. [Google Scholar]
- 21.Kim D., Song L., Breitwieser F.P., Salzberg S.L. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilsker M., Moosa Y., Nooij S., Fonseca V., Ghysens Y., Dumon K., Pauwels R., Alcantara L.C., Vanden Eynden E., Vandamme A.M., Deforche K., de Oliveira T. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics. 2019;35:871–873. doi: 10.1093/bioinformatics/bty695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H. Aligning sequence reads, clone sequences and assembly contigs with. BWA-MEM. arXiv, 2013 1303.3997v2 [q-bio.GN] [Google Scholar]
- 27.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Loens K., van Loon A.M., Coenjaerts F., van Aarle Y., Goossens H., Wallace P., Claas E.J., Ieven M. Performance of different mono- and multiplex nucleic acid amplification tests on a multipathogen external quality assessment panel. J Clin Microbiol. 2012;50:977–987. doi: 10.1128/JCM.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orozovic G., Orozovic K., Lennerstrand J., Olsen B. Detection of resistance mutations to antivirals oseltamivir and zanamivir in avian influenza A viruses isolated from wild birds. PLoS One. 2011;6:e16028. doi: 10.1371/journal.pone.0016028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh N.H., Lin Y.J., Yang Y.F., Liao C.M. Assessing the oseltamivir-induced resistance risk and implications for influenza infection control strategies. Infect Drug Resist. 2017;10:215–226. doi: 10.2147/IDR.S138317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasan M.R., Rawat A., Tang P., Jithesh P.V., Thomas E., Tan R., Tilley P. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol. 2016;54:919–927. doi: 10.1128/JCM.03050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ninove L., Nougairede A., Gazin C., Thirion L., Delogu I., Zandotti C., Charrel R.N., De Lamballerie X. RNA and DNA bacteriophages as molecular diagnosis controls in clinical virology: a comprehensive study of more than 45,000 routine PCR tests. PLoS One. 2011;6:e16142. doi: 10.1371/journal.pone.0016142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlaberg R., Chiu C.Y., Miller S., Procop G.W., Weinstock G. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch Pathol Lab Med. 2017;141:776–786. doi: 10.5858/arpa.2016-0539-RA. [DOI] [PubMed] [Google Scholar]
- 35.Bal A., Pichon M., Picard C., Casalegno J.S., Valette M., Schuffenecker I., Billard L., Vallet S., Vilchez G., Cheynet V., Oriol G., Trouillet-Assant S., Gillet Y., Lina B., Brengel-Pesce K., Morfin F., Josset L. Quality control implementation for universal characterization of DNA and RNA viruses in clinical respiratory samples using single metagenomic next-generation sequencing workflow. BMC Infect Dis. 2018;18:537. doi: 10.1186/s12879-018-3446-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinha R., Stanley G., Gulati G.S., Ezran C., Travaglini K.J., Wei E., Chan C.K.F., Nabhan A.N., Su T., Morganti R.M., Conley S.D., Chaib H., Red-Horse K., Longaker M.T., Snyder M.P., Krasnow M.A., Weissman I.L. Index switching causes “spreading-of-signal” among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. bioRxiv, 2017 [Epub] https://doi.org/10.1101/125724. [Google Scholar]
- 37.van der Valk T., Vezzi F., Ormestad M., Dalen L., Guschanski K. Estimating the rate of index hopping on the Illumina HiSeq X platform. Mol Ecol Resour. 2018 doi: 10.1111/1755-0998.13009. [Epub] https://doi.org/10.1111/1755-0998.13009. [DOI] [PubMed] [Google Scholar]
- 38.Naccache S.N., Hackett J., Jr., Delwart E.L., Chiu C.Y. Concerns over the origin of NIH-CQV, a novel virus discovered in Chinese patients with seronegative hepatitis. Proc Natl Acad Sci U S A. 2014;111:E976. doi: 10.1073/pnas.1317064111. [DOI] [PMC free article] [PubMed] [Google Scholar]