Highlights
► Canine coronavirus prevalence in 4 kennels and shelters varied from 5.3% to 33.3%. ► Transmission pathways were demonstrated by quantitative PCR and sequencing. ► A small number of persistently shedding dogs may contribute to maintenance of infection. ► CECoV may be a marker for hygiene and biosecurity.
Keywords: Canine enteric coronavirus, Shelters, Kennelled dogs, Transmission, Biosecurity, Molecular epidemiology
Abstract
Previous studies have suggested that kennelled dogs are more likely to test positive for CECoV than household pets. Here we describe both cross sectional and longitudinal studies in two rescue kennels and two boarding kennels, together with molecular diagnostics, to provide a new insight into the epidemiology of CECoV. Prevalence of CECoV in the cross sectional studies tended to be higher in the rescue kennels (13.8% and 33.3%) than the boarding kennels (5.3% and 13.5%). In each kennel, type I CECoV was more prevalent than type 2 CECoV. The mean quantity of type I detected was equivalent to 6.3 × 108 gc/gm (range = 5 × 106, 8.5 × 1011), compared to 1.3 × 108 gc/gm (range = 3 × 106, 2.4 × 1010) for type II. In one rescue shelter where dogs were followed longitudinally, infection was significantly associated with accommodation block as well as the length of stay (increased risk of CECoV per week in residence of ×1.9). Of those animals sampled on two or more occasions, none tested positive on arrival, and 54.5% later shed CECoV, suggesting that infection may have been acquired within the kennel. Shedding patterns and sequence analysis suggested both types I and II CECoV were maintained in this population by a combination of introductions into the shelter and within-shelter transmission. The findings suggest that some kennel environments may be important in maintaining CECoV infection in the population. We also propose that the diversity of viruses like CECoV in these populations may provide a novel surrogate marker for the success of biosecurity.
1. Introduction
Canine enteric coronavirus (CECoV) belongs to the genus Alpacoronavirus of the Coronaviridae (Decaro and Buonavoglia, 2011). Field infections are typically associated with mild and self-limiting diarrhoea, (Binn et al., 1974, Decaro and Buonavoglia, 2008, Pratelli, 2005), although there are also reports of more severe haemorrhagic disease with high mortality (Buonavoglia et al., 2006, Evermann et al., 2005, Tennant et al., 1993), sometimes in combination with other pathogens (Evermann et al., 1980, Pratelli et al., 2001, Pratelli et al., 1999b). Both mild and severe disease have been recreated experimentally (Decaro et al., 2008, Decaro et al., 2010a), suggesting the more severe disease phenotype may be associated with the genetic evolution of more virulent strains (Decaro et al., 2008, Evermann et al., 2005). Although CECoV infection is generally considered to be short-lived, occasionally virus shedding has been detected for weeks to months (Decaro et al., 2005, Pratelli et al., 2002, Tennant et al., 1991). The epidemiological significance of these long-term shedders is unknown.
Canine enteric coronavirus exists as at least two genetically distinct genotypes, the more recently identified strain designated type I, and the original strain designated type II (Decaro et al., 2010b, Pratelli et al., 2003). The strains are named in this way due to their respective homologies to types I and II feline coronavirus (FCoV) (Decaro et al., 2010a, Herrewegh et al., 1998). Genotype II has recently been subdivided into CECoV IIa and CECoV IIb, with the latter thought to have formed via a recombination with transmissible gastroenteritis virus (TGEV) (Decaro et al., 2009, Decaro et al., 2010b). There is conflicting evidence on the relative prevalence of CECoV I and II, with some studies suggesting type I is more common (Decaro et al., 2005, Stavisky et al., 2010), and others suggesting type II (Decaro et al., 2011, Soma et al., 2011), but these findings will depend on a number of factors including the nature of the sample population. Dual infection with both types has been reported to occur, both naturally and experimentally (Decaro et al., 2011, Decaro et al., 2005, Soma et al., 2011). There is some evidence to show that type I may be shed at a greater titre than type II CECoV (Decaro et al., 2011, Decaro et al., 2005, Soma et al., 2011).
Overall prevalence estimates of CECoV vary substantially, and are complicated by a number of factors including the different detection methods used (electron microscopy, virus isolation, RT-PCR), whether both types of CECoV are being considered, the clinical status of the animals and the characteristics of the sample population. In general, CECoV prevalence is lowest in healthy pet dogs, ranging from 0% to 17.5% (Schulz et al., 2008, Tennant et al., 1993), and somewhat greater in pet dogs with diarrhoea, ranging from 7.9% to 66.3% (Godsall et al., 2010, Soma et al., 2011). CECoV shedding is also common in dogs in rescue kennels, with a reported prevalence of 43–59% and 73% in non-diarrhoeic dogs and diarrhoeic dogs, respectively (Sokolow et al., 2005, Tennant et al., 1993). In contrast, no CECoV was isolated from 26 dogs sampled in a boarding kennel (Tennant et al., 1993). Sporadic outbreaks of CECoV-related disease in kennels has led to the suggestion that dogs living in these environments may be more susceptible to CECoV (Benetka et al., 2006, Binn et al., 1974, Ntafis et al., 2010, Tennant et al., 1993).
The aims of this study were to investigate the prevalence and molecular epidemiology of CECoV in different kennel environments, including the role of long term shedders in maintaining and transmitting CECoV, using quantitative real-time PCR and phylogenetic analysis of sequence taken from both the conserved M gene region and the variable S gene region of the CECoV genome. Such environments also provide a valuable opportunity to study the shedding patterns associated with type I and II CECoV infection of individual, naturally infected dogs.
2. Materials and methods
2.1. Kennels
Four kennels were selected to participate, on a convenience basis; two boarding kennels designated as B1 (Northern England) and B2 (West Midlands), and two rescue shelters designated R1 (Northern England) and R2 (South-Eastern England).
2.1.1. Cross-sectional sampling
A cross-sectional study was performed for all four kennels, whereby the kennel was visited early in the morning, and a voided faecal sample was collected for each dog before cleaning commenced. For kennel R1, which was large and consisted of three separate blocks, cross-sectional sampling was conducted for each of the blocks on separate days within the same week (week 0). The three kennel blocks at R1 each had a distinct intended function. On entry, dogs were usually housed in block 1 (92 kennels) for at least 7 days, and could remain there for the duration of their stay. Block 2 (20 kennels) was used for those dogs deemed by staff to be the most easily re-homed. A dog normally only entered block 2 after its 7-day holding period (a statutory requirement for any stray dog). A small number of dogs which were healthy and had been relinquished by their owners rather than admitted as strays were housed from entry in block 2. Block 3 (32 kennels) was used as a quarantine facility, most frequently for dogs exhibiting signs of upper respiratory tract disease, and also for bitches in season. Most dogs were housed singly, with the exception of bonded pairs relinquished together. Dogs in blocks 1 and 2 were put in individual runs in daytime, whereas dogs in block 3 sometimes shared the use of the outdoor runs, at staff discretion.
2.1.2. Longitudinal sampling
Longitudinal sampling was also carried out at kennel R1. Following the initial cross-sectional sampling (week 0), the kennel was subsequently visited one day a week for a further 8 weeks (weeks 1–8). During this period, every dog admitted within the previous 24 h had a faecal sample taken, giving each dog’s infection status on entry. Recruited dogs were then sampled weekly until the conclusion of the study, or until they were lost to follow-up when rehomed, reclaimed, euthanased or died. Dogs sampled during the cross-sectional phase of the study (week 0) were also included in the longitudinal phase, if their cross-sectional sample was obtained within 24-h of their admission to the shelter. Data were also collected on the within-kennel location and length of stay of each dog, to identify potential risk factors for CECoV infection.
2.2. Sample analysis
Samples were stored and genetic material extracted as previously described (Stavisky et al., 2010). Fluorogenic RT-PCR for CECoV was carried out according to a protocol developed by Decaro et al (2005) with a modified internal positive control and quantification standard as previously described (Stavisky et al., 2011).
Conventional PCR was carried out on all samples found to be positive by real-time RT-PCR, using the primers CCOV1/CCOV2 (Pratelli et al., 1999a), which detect both types I and II CECoV and are targeted at the conserved M gene region of the genome. Additionally, type-specific primers (Table 1 ) were designed from alignments of the antigenically important spike (S) gene from sequences available on Genbank. The two pairs of S gene primers were designed to differentiate between type I and type II CECoV and could therefore be used in samples with mixed infections. Cycling conditions consisted of DNA denaturation at 94 °C for 10 min, followed by 34 cycles of 94 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min, with a final extension at 72 °C for 7 min.
Table 1.
Type-specific S-gene primers used for conventional PCR.
| Primer ID | Type specificity | Sequence | Polarity | Amplicon size |
|---|---|---|---|---|
| S1For | Type I CECoV | 5′-CTA GTG GAC TTG GCA CTG TTG ATG AAG AC-3′ | + | 779 |
| S1RevR | 5′-TCA CCT CTT CCC ATT CGG TTG GAA GC-3′ | − | ||
| S2ForR | Type II CECoV | 5′-GCT TTT TGA TAA GGT TGT AAC ATC-3′ | + | 796 |
| S2RevAR | 5′-GTT TCA TAA GCT GTT GGT AAT AGC-3′ | − | ||
2.3. Sequencing and sequence analysis
Products from conventional PCR on samples collected from R1 were sequenced bi-directionally using the Applied Biosystems Genetic Analyzer with Bigdye terminator. Consensus sequences were produced using ChromasPro version 1.32 (Technelysium Ptl. Ltd.). Sequence alignments, nucleotide distance calculations (Jukes–Cantor), and phylogenetic analysis were performed using MEGA version 4.0 (Tamura et al., 2007). Support for individual nodes was sought by bootstrap analysis using 1000 repetitions.
2.4. Statistical analysis
Statistical analysis was carried out to identify factors associated with for CECoV positivity in kennel R1. Descriptive statistics were calculated using SPSS for Windows (Rel. 16.0 2007. Chicago SPSS Inc.). Chi squared tests were conducted to investigate the association between factors such as length of stay (when categorised) and CECoV status. A generalised additive model was constructed to examine the relationship between CECoV status and length of stay as a continuous variable (S-Plus). For continuous data (titre), the data were log-transformed for normality and a Student’s t-test applied. A cutoff p-value of <0.05 was applied to determine significance.
Logistic regression analysis was performed using Egret (Cytel Software Corporation). Univariable and multivariable evaluation of the effect of variables on the outcome of CECoV was performed on the data from R1 using a logistic regression model with a random effect to allow for clustering within dog due to the repeated measures at the different time points. All factors with a p ⩽ 0.3 were considered for inclusion in the multivariable logistic regression model, which was built using backwards elimination. Factors with a likelihood ratio test statistic p-value of >0.05 were excluded step-wise from the model. All eliminated factors were then rechecked against the final model.
3. Results
3.1. Cross-sectional kennel sampling
Overall, 18.7% (41/219) of the dogs sampled during the cross-sectional phase of the study tested positive for CECoV by fluorogenic RT-PCR (95% CI 14.1, 24.4) (Table 2 ). The prevalence of CECoV in dogs at individual kennels ranged from 5.3% to 33.3%. The boarding kennels had a lower prevalence (5.3% and 13.5%) than the two rescue shelters (13.8% and 33.3%). Of the 41 samples that tested positive for CECoV, type I alone was found in 23 (10.5%), type II alone in 6 (2.7%), and there were 12 mixed infections (5.5%). Type I CECoV was found in all the four kennels, and type II in only two of them.
Table 2.
Results of cross-sectional sampling. Percentage of dogs which were positive for CECoV during their stay in two boarding kennels (B1 and B2) and two rescue shelters (R1 and R2), as determined by real-time RT–PCR. ∗ For shelter R1, a total of 87 samples were obtained from the 81 dogs.
| Kennel | No. of dogs | Total +ve | % +ve (95% CI) | Type I (%) | Type II (%) | Mixed (%) |
|---|---|---|---|---|---|---|
| B1 | 57 | 3 | 5.3 (0.1, 11.1) | 2 (3.5) | 1 (1.8) | 0 |
| B2 | 52 | 7 | 13.5 (4.2, 22.7) | 7 (13.5) | 0 | 0 |
| R1 | 81∗ | 27 | 33.3 (24.0, 44.2) | 10 (12.3) | 5 (6.2) | 12 (14.8) |
| R2 | 29 | 4 | 13.8 (1.2, 26.3) | 4 (13.8) | 0 | 0 |
| Total | 219 | 41 | 18.7 (14.1, 24.4) | 23 (10.5) | 6 (2.7) | 12 (5.5) |
3.2. Quantity of CECoV shed
The quantity of CECoV detected in individual samples varied considerably, from 3 × 106 to 8.5 × 1011 genome copies per gram of faeces (gc/gm) (data not presented). The mean quantity of type I CECoV detected was equivalent to 6.3. × 108 gc/gm (range = 5 × 106, 8.5 × 1011), compared to 1.3 × 108 gc/gm (range = 3 × 106, 2.4 × 1010) for type II, with no significant difference between types (p = 0.06).
3.3. Sampling at R1
3.3.1. Shedding patterns at R1
In total, 184 samples (87 cross-sectional samples and 97 from the longitudinal phase) were collected from kennel R1 (Table 3 ). Forty-three (23%) tested positive for CECoV (16 type I, 10 type II and 17 mixed infections). The samples were obtained from 140 dogs, of which 40 (29%) tested positive at some point during their stay in the kennel. One hundred and fourteen dogs were sampled only once, and 26 dogs had two or more samples taken. For 22 of these 26 dogs, a sample from within 24 h of arrival was available, and these all tested negative. In contrast, twelve of these dogs (54.5%) later tested positive suggestive of possible kennel-acquired infection.
Table 3.
Results of real-time RT–PCR on faecal samples from kennel R1. Blank = not sampled; 0 = CECoV negative; I = positive for just CECoV I; II = positive for just CECoV II; M = mixed type I and II CECoV. ∗ Indicates the two individuals further detailed in Fig. 1. N = denotes a number of dogs sharing the same sampling and shedding pattern.
| Dog reference number (if referred to in the text) or number of dogs | Date of sampling |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 22/5 |
24/5 |
29/5 |
5/6 |
12/6 |
19/6 |
26/6 |
3/7 |
10/7 |
17/7 |
24/7 |
|
| Cross sectional sampling | Longitudinal sampling | ||||||||||
| 13 | 0 | I | |||||||||
| 19 | 0 | 0 | 0 | ||||||||
| 37 | 0 | 0 | |||||||||
| 129 | 0 | M | |||||||||
| N = 40 | 0 | ||||||||||
| 3 | II | ||||||||||
| 57 | II | ||||||||||
| 135 | I | ||||||||||
| N = 7 | 0 | ||||||||||
| 53 | II | ||||||||||
| 54, 56, 124 | M | ||||||||||
| 60, 121, 122, 123, 140 | I | ||||||||||
| 119 | I | ||||||||||
| 4, 49, 51, 58, 126, 130, 134, 139 | M | ||||||||||
| N = 3 | 0 | ||||||||||
| 52, 128 | I | ||||||||||
| 48, 133 | II | ||||||||||
| 16∗ | 0 | 0 | 0 | 0 | 0 | I | I | I | I | ||
| 17 | 0 | 0 | 0 | 0 | II | II | |||||
| N = 6 | 0 | ||||||||||
| 69 | 0 | 0 | |||||||||
| N = 2 | 0 | 0 | |||||||||
| 71∗ | 0 | 0 | II | M | M | M | I | ||||
| 72 | 0 | 0 | 0 | ||||||||
| 77 | 0 | 0 | 0 | ||||||||
| 76 | 0 | 0 | II | II | |||||||
| 62 | 0 | M | |||||||||
| N = 4 | 0 | ||||||||||
| 66 | I | ||||||||||
| N = 2 | 0 | ||||||||||
| 96 | 0 | II | 0 | 0 | |||||||
| 61 | I | ||||||||||
| N = 10 | 0 | ||||||||||
| 80 | 0 | 0 | 0 | I | |||||||
| 85 | 0 | I | 0 | 0 | |||||||
| 88 | 0 | 0 | |||||||||
| 118 | M | ||||||||||
| 78 | 0 | ||||||||||
| 97 | 0 | 0 | 0 | 0 | |||||||
| 115 | 0 | 0 | II | ||||||||
| N = 5 | 0 | ||||||||||
| N = 4 | 0 | 0 | |||||||||
| N = 4 | 0 | ||||||||||
| 116 | 0 | I | |||||||||
| 108 | 0 | 0 | |||||||||
| N = 4 | 0 | ||||||||||
Dogs shed CECoV for a variable period of time. Two dogs (85 and 96) only had CECoV detected in a single sample, despite being sampled over several weeks, implying a duration of shedding of <14 days (Table 3). In contrast, two other dogs (16 and 71) shed virus for prolonged periods (four or more weeks), and still had high titres in their final sample, suggesting they might shed virus for a much longer period. On entry to the kennel, dog 16 tested negative for CECoV (Table 3 and Fig. 1 a). After 5 weeks, it started shedding type I CECoV, with the viral load rising over a week from zero to 2.8 × 1010 genome copies per gram of faeces. It then continued to shed type I CECoV for the remainder of the sampling period, but never shed type II CECoV. Fig. 1b shows the shedding pattern of dog 71. This dog also initially tested negative for CECoV shedding, then shed type II CECoV during its second week in the shelter, and then, a week later, shed both type I and type II CECoV. Three weeks after its first positive sample, the dog ceased shedding type II, but continued to shed type I until it was lost to follow up. The peak titre of type I CECoV shed by dog 71 (2.1 × 1010) was slightly higher than the peak type II CECoV titre (4.2 × 109), in keeping with the overall trend noted in this study.
Fig. 1.
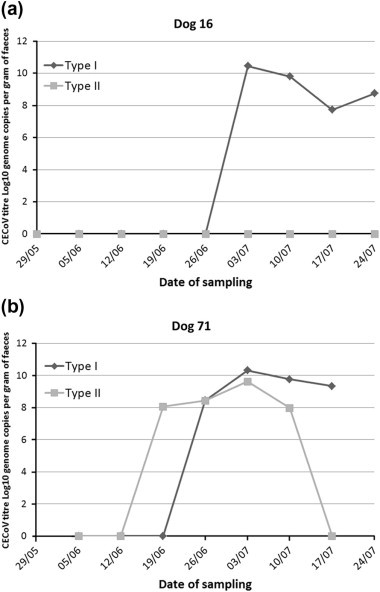
Graph of log10 RNA copy number of type I and type II CECoV per gram of faeces in (a) dog 16 and (b) dog 71. Dates relate to the weeks of sampling, and are the same as those used in Fig. 3.
3.3.2. Risk factors for shedding at R1
Kennel block and length of stay were considered for inclusion into the multivariable model. Other factors such as use of medication, occurrence of diarrhoea, age and breed were also recorded where available, but examination of the kennel records revealed that a substantial quantity of this information was either estimated or not reliably recorded, and was therefore unsuitable for inclusion in the analysis.
Length of stay was correlated with an increased risk of CECoV. Of the 54 dogs sampled within one day of entry, two (3.7%) shed CECoV; in contrast, of the 15 dogs sampled after being in the kennels for 56 days or more, 12 (80%) were CECoV positive (data not presented), a significant difference (Fisher’s Exact Test, p < 0.0001).
A generalised additive model was used to investigate the relationship between duration of stay in kennel R1 and probability of testing CECoV positive (Fig. 2 ). This showed that the probability of testing CECoV positive increased until approximately 50 days, at which point it appeared to level; therefore, length of stay was considered piecewise to 30, 35, 40, 45 and 50 days. The minimum deviance (indicating best fit) was obtained when a limit of 40 days was used, and this was therefore included in the multivariable model.
Fig. 2.
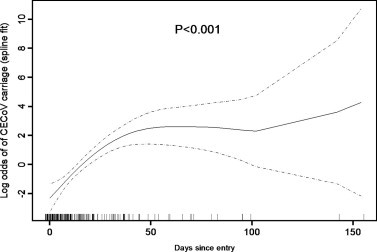
Generalised additive model to explore the relationship between days since entry (x axis) and log odds of CECoV carriage (y axis) (fitted using spline smoother). Dashed lines show 95% confidence intervals. The rug plot (vertical lines on x-axis) represents the number of individual data points.
As well as the effect of length of stay, the location of accommodation also had a considerable impact on CECoV shedding, with those dogs housed in block 2 (OR 7.2, 95% CI 2.3, 22) and block 3 (OR 18.6, 95% CI 7.1, 49.8) being at considerably greater risk of shedding CECoV than those housed in block 1.
In the multivariable model, block 2 was no longer a significant risk factor for CECoV shedding (OR 0.93, 95% CI 0.22, 3.82), after allowing for length of stay. However, block 3 remained a significant risk factor (OR 6.01, 95% CI 2.18, 16.52), as did the length of stay. The increased risk of CECoV per day in residence was 1.1 (95% CI 1.05, 1.1) and corresponds to an increased odds ratio of 1.9 (1.17) per week (up to 40 days).
3.4. Sequencing
Overall, clear partial M gene sequence was obtained for 23 samples (Fig. 3 a), and partial S gene sequence for 26 samples (Fig. 3b). In all cases, the sequence typing was concordant with the real-time PCR typing. Additionally, in the nine samples for which both M gene and S gene sequence was generated, sequences grouped similarly in both regions of the genome, suggesting recombination was not a strong feature in this data set. In those dogs (dogs number 16 and 71) for which repeated M or S gene sequences were obtained, the sequences for either type I or type II in each dog were identical, suggesting they were infected with one strain, and not undergoing cycles of reinfection. Unfortunately, no clear sequence was obtained from kennels B1, B2 or R2; most of these samples were of a relatively low titre (data not shown), which might have affected the success of the sequencing.
Fig. 3.
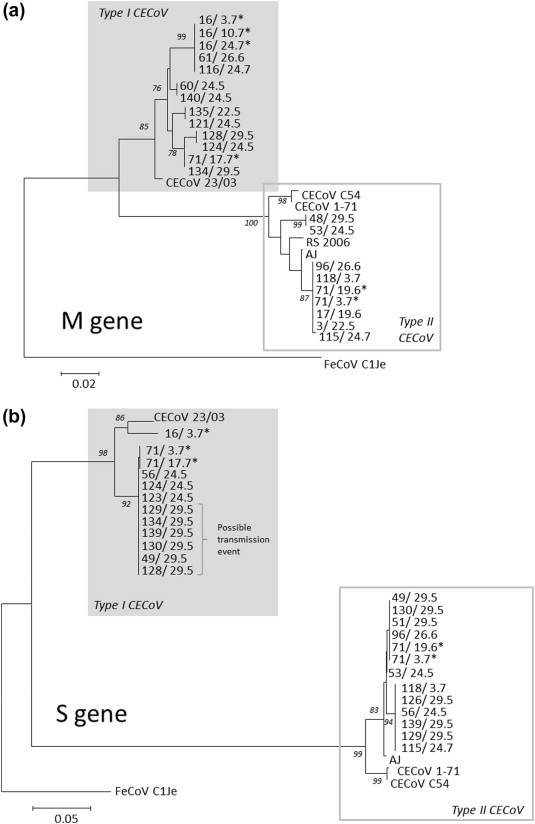
Neighbour-joining tree of partial (a) M gene (ORF 3a), and (b) S gene (ORF 7b) sequences generated in this study and published reference canine and feline coronaviruses. All branch lengths are proportional to distances established using the Jukes–Cantor method. Bootstraps are expressed as percentages, and only included where greater than 75%. Each sample is identified by a combination of unique dog identification number and the date of sampling. Reference strains CECoV 23/03 (AY548235), CECoV 1-71 (AY664662) and FcoVC1Je (DQ848678); additionally CECoV C54 (Tennant et al., 1991), RS 2006 (sequence from a sample collected at R1 in 2006) and AJ (a diagnostic sample received in 2009) are included in the trees. Accession numbers for sequences obtained in this study JX035812–JX035862. ∗ Samples from dogs 16 and 71, whose titres are shown in Fig. 1. The potential transmission event referred to in the text is indicated.
In total over the 9 weeks of sampling, 8 and 5 distinct sequences were seen in the kennel based on M and S genes, respectively, suggesting that in this kennel, the high CECoV prevalence could not be attributed entirely to within-kennel transmission. However, several putative transmission events were suggested by the phylogeny; for example six dogs housed in block 3 (129, 134, 139, 130, 49 and 128) on the same date (29.05), shed virus with identical spike gene sequence (Fig. 3b). In addition, both of the long-term shedders (dogs 16 and 71), tested negative on arrival in the shelter, and subsequently began shedding viruses, with dog 71 shedding sequence identical to one already present within the shelter.
4. Discussion
Previous studies have suggested that kennelled dogs are more likely to test positive for CECoV (Naylor et al., 2001, Tennant et al., 1993), although the mechanisms for this are largely unknown. Such environments may be important for maintaining this infection in the general population. In this study, we have for the first time used both cross sectional and longitudinal study designs, together with molecular diagnostics, to provide a unique insight into the epidemiology of this coronavirus in high density, high turnover populations of kennelled dogs. The prevalence of CECoV in the four kennels ranged from 5% to 33%, with type I being both more common. The prevalence in newly housed animals was relatively low, in line with that reported previously in the general dog population (Stavisky et al., 2010, Tennant et al., 1993). During housing, both length of stay and kennel block were significantly associated with infection risk. Shedding patterns and sequence analysis in one of the shelters strongly suggested the viruses were maintained in this population by a combination of introductions into the shelter and within-shelter transmission. This is important, since CECoV may be associated with diarrhoea, and it has been shown that animals with diarrhoea or ill-health have either a delay in being rehomed (Litster et al., 2011), or a greater chance of being returned to the shelter (Wells and Hepper, 1999). As well as its significance to the health of the animals, such studies also provide a marker that other diseases may be readily transmitted in such environments.
Although the way in which such kennels operate makes detailed epidemiological study difficult, our study design allowed us to identify some within-kennel risk factors for infection. Longitudinal sampling clearly showed the risk of infection increased with the length of stay in kennel R1, from 3.7% within the first week of entry to 80% after 8 weeks or more. This suggests some exposure to cumulative risk factors (for example infectious burden of CECoV, concurrent infections, stress) likely impact on infection risk. This is similar to studies in feline rescue shelters with the related feline enteric coronavirus, where the prevalence of infection rose from 33% on relinquishment to 60% within one week, with all adults cats that tested positive on both occasions showing a rise in viral titre of 101–106 (Pedersen et al., 2004). In our study most animals tested negative on arrival, and for the three animals that did test positive on arrival, repeat samples were not available so whether there was a similar rise in titre could not be assessed.
As well as an association with length of time within kennel R1, dogs housed in block 3 were significantly more likely to shed CECoV than those housed elsewhere, regardless of their length of stay. Husbandry factors specific to this block may at least in part explain this risk, since dogs were sometimes mixed in outdoor runs or put into runs which had not been cleaned of faeces and urine from other dogs. This was not common practice in blocks 1 and 2 and, therefore, enhanced opportunity for transmission in the dogs in block 3 may partially explain the different infection rates. Respiratory infection was the most common reason for moving a dog to block 3, and immunocompromise, from concurrent ill-health, may have led to an increased susceptibility to CECoV infection. However, other unmeasured factors could also have contributed to this difference in risk between the three blocks. Using sequence analysis, we showed several dogs contemporaneously housed in this block all shedding the same virus, providing strong evidence for within-kennel transmission. Data such as this, where a risk factor is clearly linked to accommodation, can allow limited resources to be efficiently targeted to those areas where disease transmission is most problematic.
Despite the epidemiological and molecular evidence of transmission, sequence analysis also clearly showed multiple strains present within shelter R1. Such strain complexity suggests that new strains of CECoV are regularly introduced into this shelter, presumably with those occasional new residents that are shedding CECoV on admission. The observed strain complexity present within these high density, high turnover populations is clearly a balance between the introduction of new distinct strains in new animals, and the transmission of strains within the colony. The level of strain diversity within these populations can, therefore, provide an objective assessment of the biosecurity in such high density populations, identify practices that could be improved (Beersma et al., 2009, Coyne et al., 2007, Radford et al., 2001), and enable the effectiveness of interventions to be monitored.
The use of boarding and rescue kennels in our study provided an opportunity to contrast the natural history of CECoV in different environments. The overall prevalence of CECoV appeared higher in the two rescue kennels (13.8% and 33.3%) than the boarding kennels (5.3% and 13.5%), consistent with the pattern seen in previous studies using virus isolation (which only detects type II CECoV), where prevalence rates of 23–86% and 0% were seen in rescue and boarding kennels, respectively (Tennant et al., 1993). Numerous reasons could underlie the variations in prevalence shown. The boarding kennels mainly housed owned dogs for relatively short periods. We have shown time is a risk factor for CECoV faecal shedding and many may leave the kennel before they can become infected and/or start shedding. In addition, such dogs are likely to have better health, nutritional status and veterinary care. In contrast, many animals in rescue shelters are young (New et al., 2000), a known risk factor for infection (Tennant et al., 1993, Soma et al., 2011). Unfortunately, reliable data on both demographics and whether the dogs had a history of diarrhoea were not available, due to constraints of limited record keeping (rescue shelters) and data protection (private boarding kennels). Additionally, the success of the longitudinal sampling was impaired by the high turnover of dogs in R1. Further studies in different environments, such as working or hunt kennels, could circumvent these problems and provide valuable additional information.
The prevalence of CECoV infection in the rescue shelters studied here was lower than that reported previously in two similar populations (Sokolow et al., 2005, Tennant et al., 1993). Both of these latter kennels reported ongoing problems with diarrhoea. However, infection was also prevalent in non-diarrhoeic dogs suggesting disease alone was not driving the reported high prevalence. It seems likely that many other factors will influence the overall prevalence of CECoV infection in a rescue shelter environment, such as population demographics, temporal fluctuations, and shelter management, and clearly this is an area that would benefit from future research. Additionally, in the present study, only four kennels were investigated, and only one of these was studied longitudinally. The diversity of these environments is such that further work in other similar types of kennels would be beneficial, in order to enable clearer understanding of the drivers for infection, transmission and disease.
In our study, the prevalence of type I CECoV was consistently higher than that of type II, with all four kennels shedding type I, whereas only two kennels shedding type II. There is some uncertainty about which type of CECoV is most prevalent in the population, and most recently it has been suggested that the significance of each type may be different in different countries (Decaro et al., 2011). In this latter study, only eight samples were available from the UK, one of which tested positive for CECoV II. These authors commented that their findings may be compromised by the low number of samples available from the UK. Our findings reported here are consistent with our previous work (Stavisky et al., 2010, Stavisky et al., 2011), which together suggest type I CECoV may be the predominant variant in the UK. The samples here underwent PCR and sequencing in fragments of the variable S gene and conserved M gene in order to allow detection of recombination. No recombination was detected in this study; however sequencing was not successful in both regions for all samples, possibly due to some of the samples being of lower titre. The regions selected, whilst useful in determining transmission patterns and potential recombination events, did not allow differentiation of the CECoV type IIa and IIb (TGEV-like) strains. These ‘TGEV-like’ strains have been previously recognised in the UK and elsewhere (Decaro et al., 2009, Decaro et al., 2010b, Erles and Brownlie, 2009) and it has been suggested that they may show an enhanced ability to spread systemically, as compared with other strains (Ntafis et al., 2011). In future work it would be to useful to examine both the prevalence of these subtypes in rescue and other kennel populations, and their transmission characteristics.
In these naturally infected dogs, shedding occurred for a variable period. In some dogs, CECoV was only detected on a single sampling occasion, whilst others shed for 4–5 weeks. In these dogs, sequentially sequenced samples proved to be genetically identical, suggesting that these dogs were true chronic shedders, rather than undergoing cycles of re-infection with different viral strains. Similar periods of more long-term shedding have also been reported following natural (Tennant et al., 1991) and experimental (Decaro et al., 2005) CECoV infection, and similar patterns of shedding have also been described for the related feline coronavirus (Addie et al., 2003). Interestingly in our study, both of the animals that shed virus for the longest tested negative on arrival, suggesting they were newly infected within the shelter or were undergoing some stress induced recrudescence. Evidence for the former comes from sequence analysis which clearly identified identical viruses already in the shelter when these two carrier animals arrived. Although further work is needed to understand the mechanism of coronavirus persistence, clearly such long term shedders are likely to play an important part in the ongoing epidemiology of infection.
The titres of CECoV shed were very variable. For those dogs which shed CECoV for several weeks during the sampling period, peak titres were seen at different points of the infectious period. Therefore, the observed variability of the titres we detected may depend in part on when in the infectious cycle a dog was sampled. There may also be a component of both the immunity of the dog, and the virulence of the strain involved. The mean type I titre detected was slightly higher than type II; this difference was not significantly different, but the same tendency has been observed previously (Decaro et al., 2005). The titres obtained must be considered as semi-quantitative rather than fully quantitative when used as a measure of viral shedding since both the consistency of the faeces and other factors will affect the amount of virus shed.
5. Conclusion
This study has shown that CECoV can be shed at a high prevalence in kennelled dog populations. Longitudinal sampling suggested most dogs in R1 were negative for CECoV on entry, but had a high risk of acquiring CECoV. Shedding occurred for a variable period, which in some cases continued for at least five weeks. The study design used may help identify interventions to manage both between- and within-kennel risk factors for transmission such as biosecurity and stress, particularly in populations of dogs with uncertain health status, and high density and turnover.
Acknowledgements
This project was funded by MSD Animal Health. The authors would like to also thank Ruth Ryvar and Bryony Parsons for technical assistance, and the workers at the kennels for their support.
References
- Addie D.D., Schaap I.A.T., Nicolson L., Jarrett O. Persistence and transmission of natural type I feline coronavirus infection. Journal of General Virology. 2003;84:2735–2744. doi: 10.1099/vir.0.19129-0. [DOI] [PubMed] [Google Scholar]
- Beersma M.F.C., Schutten M., Vennema H., Hartwig N.G., Mes T.H.M., Osterhaus A.D.M.E., van Doornum G.J.J., Koopmans M. Norovirus in a Dutch tertiary care hospital (2002–2007): frequent nosocomial transmission and dominance of GIIb strains in young children. Journal of Hospital Infection. 2009;71:199–205. doi: 10.1016/j.jhin.2008.11.018. [DOI] [PubMed] [Google Scholar]
- Benetka V., Kolodziejek J., Walk K., Rennhofer M., Mostl K. M gene analysis of atypical strains of feline and canine coronavirus circulating in an Austrian animal shelter. Veterinary Record. 2006;159:170–174. doi: 10.1136/vr.159.6.170. [DOI] [PubMed] [Google Scholar]
- Binn, L.N., Lazar, E.C., Keenan, K.P., Huxsoll, D.L., Marchwicki, R.H., Strano, A.J., 1974. Recovery and characterization of a coronavirus from military dogs with diarrhea. In: Proceedings, Annual Meeting of the United States Animal Health Association, p. 359. [PubMed]
- Buonavoglia C., Decaro N., Martella V., Elia G., Campolo M., Desario C., Castagnaro M., Tempesta M. Canine coronavirus highly pathogenic for dogs. Emerging Infectious Diseases. 2006;12:492–494. doi: 10.3201/eid1203.050839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne K.P., Edwards D., Radford A.D., Cripps P., Jones D., Wood J.L.N., Gaskell R.M., Dawson S. Longitudinal molecular epidemiological analysis of feline calicivirus infection in an animal shelter: a model for investigating calicivirus transmission within high-density, high-turnover populations. Journal of Clinical Microbiology. 2007;45:3239–3244. doi: 10.1128/JCM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Veterinary Microbiology. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. Canine coronavirus: not only an enteric pathogen. Veterinary Clinics of North America: Small Animal Practice. 2011;41:1121–1132. doi: 10.1016/j.cvsm.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Campolo M., Lorusso A., Desario C., Mari V., Colaianni M.L., Elia G., Martella V., Buonavoglia C. Experimental infection of dogs with a novel strain of canine coronavirus causing systemic disease and lymphopenia. Veterinary Microbiology. 2008;128:253–260. doi: 10.1016/j.vetmic.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Desario C., Billi M., Mari V., Elia G., Cavalli A., Martella V., Buonavoglia C. Western European epidemiological survey for parvovirus and coronavirus infections in dogs. Veterinary Journal. 2011;187:195–199. doi: 10.1016/j.tvjl.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Elia G., Martella V., Campolo M., Mari V., Desario C., Lucente M.S., Lorusso E., Kanellos T., Gibbons R.H., Buonavoglia C. Immunity after natural exposure to enteric canine coronavirus does not provide complete protection against infection with the new pantropic CB/05 strain. Vaccine. 2010;28:724–729. doi: 10.1016/j.vaccine.2009.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Campolo M., Lorusso A., Camero M., Elia G., Martella V., Cordioli P., Enjuanes L., Buonavoglia C. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. Journal of Virology. 2009;83:1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Elia G., Addie D.D., Camero M., Lucente M.S., Martella V., Buonavoglia C. Recombinant canine coronaviruses in dogs, Europe. Emerging Infectious Diseases. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M., Cavaliere N., Di Trani L., Tempesta M., Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for the detection and quantitation of canine coronavirus type I and type II RNA in faecal samples of dogs. Journal of Virology Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erles K., Brownlie J. Sequence analysis of divergent canine coronavirus strains present in a UK dog population. Virus Research. 2009;141:21–25. doi: 10.1016/j.virusres.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evermann J.F., Abbott J.R., Han S. Canine coronavirus-associated puppy mortality without evidence of concurrent canine parvovirus infection. Journal of Veterinary Diagnostic Investigation. 2005;17:610–614. doi: 10.1177/104063870501700618. [DOI] [PubMed] [Google Scholar]
- Evermann J.F., Foreyt W., Maag-Miller L., Leathers C.W., McKeirnan A.J., LeaMaster B. Acute haemorrhagic enteritis associated with canine coronavirus and parvovirus infections in a captive coyote population. Journal of the American Veterinary Medicine Association. 1980;177:784–786. [PubMed] [Google Scholar]
- Godsall S.A., Clegg S.R., Stavisky J.H., Radford A.D., Pinchbeck G. Epidemiology of canine parvovirus and coronavirus in dogs presented with severe diarrhoea to PDSA PetAid hospitals. Veterinary Record. 2010;167:196–201. doi: 10.1136/vr.c3095. [DOI] [PubMed] [Google Scholar]
- Herrewegh A.A.P.M., Smeenk I., Horzinek M.C., Rottier P.J.M., Groot R.J.d. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. Journal of Virology. 1998;72:4508–4514. doi: 10.1128/jvi.72.5.4508-4514.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litster A., Allen J., Mohamed A., He S. Risk factors for delays between intake and veterinary approval for adoption on medical grounds in shelter puppies and kittens. Preventive Veterinary Medicine. 2011;101:107–112. doi: 10.1016/j.prevetmed.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor M.J., Monckton R.P., Lehrbach P.R., Deane E.M. Canine coronavirus in Australian dogs. Australian Veterinary Journal. 2001;79:116–119. doi: 10.1111/j.1751-0813.2001.tb10718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New J.C., Jr., Salman M.D., King M., Scarlett J.M., Kass P.H., Hutchison J.M. Characteristics of shelter-relinquished animals and their owners compared with animals and their owners in US pet-owning households. Journal of Applied Animal Welfare Science. 2000;3:179–201. [Google Scholar]
- Ntafis V., Mari V., Danika S., Fragkiadaki E., Buonavoglia C. An outbreak of Canine coronavirus in puppies in a Greek kennel. Journal of Veterinary Diagnostic Investigation. 2010;22:320–323. doi: 10.1177/104063871002200231. [DOI] [PubMed] [Google Scholar]
- Ntafis V., Mari V., Decaro N., Papanastassopoulou M., Papaioannou N., Mpatziou R., Buonavoglia C., Xylouri E. Isolation, tissue distribution and molecular characterization of two recombinant canine coronavirus strains. Veterinary Microbiology. 2011;151:238–244. doi: 10.1016/j.vetmic.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.C., Sato R., Foley J.E., Poland A.M. Common virus infections in cats, before and after being placed in shelters, with emphasis on feline enteric coronavirus. Journal of Feline Medicine and Surgery. 2004;6:83–88. doi: 10.1016/j.jfms.2003.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli, A., 2005. Canine Coronavirus Infection. In: Carmichael L. (Ed.), Recent Advances in Canine Infectious Diseases, International Veterinary Information Service.
- Pratelli A., Elia G., Martella V., Tinelli A., Decaro N., Marsilio F., Buonavoglia D., Tempesta M., Buonavoglia C. M gene evolution of canine coronavirus in naturally infected dogs. Veterinary Record. 2002;151:758–761. [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Tempesta M., Guarda F., Capucchio M.T., Carmichael L.E., Buonavoglia C. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. Journal of Veterinary Medicine: Series B. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Pistello M., Elia G., Decaro N., Buonavoglia D., Camero M., Tempesta M., Buonavoglia C. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. Journal of Virological Methods. 2003;107:213–222. doi: 10.1016/S0166-0934(02)00246-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Tempesta M., Greco G., Martella V., Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. Journal of Virological Methods. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Tempesta M., Roperto F.P., Sagazio P., Carmichael L., Buonavoglia C. Fatal coronavirus infection in puppies following canine parvovirus 2b infection. Journal of Veterinary Diagnostic Investigation. 1999;11:550–553. doi: 10.1177/104063879901100615. [DOI] [PubMed] [Google Scholar]
- Radford A.D., Dawson S., Kerins A.M., Sommerville L.M., Ryvar R., Gaskell R.M. Molecular analysis of isolates of feline calicivirus from a population of cats in a rescue shelter. Veterinary Record. 2001;149:477–481. doi: 10.1136/vr.149.16.477. [DOI] [PubMed] [Google Scholar]
- Schulz B.S., Strauch C., Mueller R.S., Eichhorn W., Hartmann K. Comparison of the prevalence of enteric viruses in healthy dogs and those with acute haemorrhagic diarrhoea by electron microscopy. Journal of Small Animal Practice. 2008;49:84–88. doi: 10.1111/j.1748-5827.2007.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolow S.H., Rand C., Marks S.L., Drazenovich N.L., Kather E.J., Foley J.E. Epidemiologic evaluation of diarrhea in dogs in an animal shelter. American Journal of Veterinary Research. 2005;66:1018–1024. doi: 10.2460/ajvr.2005.66.1018. [DOI] [PubMed] [Google Scholar]
- Soma T., Ohinata T., Ishii H., Takahashi T., Taharaguchi S., Hara M. Detection and genotyping of canine coronavirus RNA in diarrheic dogs in Japan. Research in Veterinary Science. 2011;90:205–207. doi: 10.1016/j.rvsc.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavisky J., Pinchbeck G.L., German A.J., Dawson S., Gaskell R.M., Ryvar R., Radford A.D. Prevalence of canine enteric coronavirus in a cross-sectional survey of dogs presenting at veterinary practices. Veterinary Microbiology. 2010;140:18–24. doi: 10.1016/j.vetmic.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavisky J., Radford A.D., Gaskell R., Dawson S., German A., Parsons B., Clegg S., Newman J., Pinchbeck G. A case-control study of pathogen and lifestyle risk factors for diarrhoea in dogs. Preventive Veterinary Medicine. 2011;99:185–192. doi: 10.1016/j.prevetmed.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tennant B., Gaskell R.M., Kelly D.F., Carter S.D., Gaskell C.J. Canine coronavirus infection in the dog following oronasal inoculation. Research in Veterinary Science. 1991:11–18. doi: 10.1016/0034-5288(91)90023-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant B.J., Gaskell R.M., Jones R.C., Gaskell C.J. Studies on the epizootiology of canine coronavirus. Veterinary Record. 1993;132:7–11. doi: 10.1136/vr.132.1.7. [DOI] [PubMed] [Google Scholar]
- Wells D.L., Hepper P.G. Prevalence of disease in dogs purchased from an animal rescue shelter. Veterinary Record. 1999;144:35–38. doi: 10.1136/vr.144.2.35. [DOI] [PubMed] [Google Scholar]


