Abstract
Infectious bronchitis virus (Gammacoronavirus, Coronaviridae) is a genetically variable RNA virus that causes one of the most persistent respiratory diseases in poultry. The virus is classified in genotypes and lineages with different epidemiological relevance. Two lineages of the GI genotype (11 and 16) have been widely circulating for decades in South America. GI-11 is an exclusive South American lineage while the GI-16 lineage is distributed in Asia, Europe and South America. Here, we obtained the whole genome of two Uruguayan strains of the GI-11 and GI-16 lineages using Illumina high-throughput sequencing. The strains here sequenced are the first obtained in South America for the infectious bronchitis virus and provide new insights into the origin, spreading and evolution of viral variants. The complete genome of the GI-11 and GI-16 strains have 27,621 and 27,638 nucleotides, respectively, and possess the same genomic organization. Phylogenetic incongruence analysis reveals that both strains have a mosaic genome that arose by recombination between Euro Asiatic strains of the GI-16 lineage and ancestral South American GI-11 viruses. The recombination occurred in South America and produced two viral variants that have retained the full-length S1 sequences of the parental lineages but are extremely similar in the rest of their genomes. These recombinant virus have been extraordinary successful, persisting in the continent for several years with a notorious wide geographic distribution. Our findings reveal a singular viral dynamics and emphasize the importance of complete genomic characterization to understand the emergence and evolutionary history of viral variants.
Keywords: Infectious bronchitis virus, Genomic evolution, Lineage, South America, High-throughput sequencing
Highlights
-
•
Genomic analysis was performed in two main lineages of Infectious bronchitis virus.
-
•
Lineages differ in their S1 sequences but are similar in the rest of the genome.
-
•
Genomic similarity between both lineages arise by inter-lineage recombination.
-
•
Inter-lineage recombination occurred in South America between European/Asiatic and local strain.
-
•
Recombinant forms have persisted in the continent for several years with wide geographic distribution.
1. Introduction
Infectious bronchitis (IB) is still one of the most persistent respiratory diseases of chickens despite intensive control and research efforts made since its first description in 1931 (Shalk and Hawn, 1931). In regions where there is no highly pathogenic avian influenza virus or velogenic Newcastle disease virus, infectious bronchitis virus (IBV) is the cause of the major economic losses to the poultry industry (Cook et al., 2012).
IBV belongs to the genus Gammacoronavirus within the Coronaviridae family (de Groot, 2012). The positive-sense single-stranded RNA viral genome (27.6 kb) contains six genes and at least ten open reading frames (ORFs) in the order 1a-1b-S-3a-3b-E-M-5a-5b-N. The 1a and 1b ORFs code for the 1ab polyprotein using a ribosomal frameshift event at the end of ORF la; the large 1ab polyprotein is then post-translationally cleaved into 15 non-structural polypeptides, which are required for RNA replication and transcription. ORFs S, E, M and N encode four major structural proteins: the spike (S) glycoprotein, the small envelope (E) protein, the membrane (M) glycoprotein and the nucleocapsid (N) protein, respectively. ORFs 3a, 3b, 5a and 5b encode small non-structural proteins, which are not essential to viral replication (Cavanagh, 2007).
The IBV genome has high rates of mutation and recombination, leading to the continuous emergence of novel genetic and antigenic variants worldwide (Gough et al., 1992, Liu and Kong, 2004). Strain classification and evolutionary analysis of IBV variants are usually accomplished by the phylogenetic analysis of the complete coding region of the S1 subunit, a highly variable region of the S protein. The S1 subunit carries most of the virus-neutralizing epitopes and has a direct implication in protective immunity (Cavanagh and Davis, 1986, Cavanagh et al., 1992, Mockett et al., 1984). The most recent comprehensive classification identified six main genotypes (GI–GVI), 32 viral lineages (1 − 32) and a number of inter-lineage recombinants in global strains (Valastro et al., 2016).
Two lineages of the GI genotype (11 and 16), also called South America I (SAI) and Asia/South America II (A/SAII) genotypes, have been widely circulating for decades in South America (Alvarado et al., 2005, Marandino et al., 2015, Rimondi et al., 2009). These lineages are responsible for most outbreaks in commercial flocks and cause major economic losses in countries with high production levels, such as Brazil and Argentina, which are among the world's top producers of broiler meat (USDA, 2014). GI-11 is an exclusive South American lineage that emerged in the 1960s and is now apparently restricted to Argentina, Brazil and Uruguay (Chacon et al., 2011, Felippe et al., 2010, Marandino et al., 2015, Villarreal et al., 2007). The GI-16 lineage contains viruses collected in South America (Argentina, Chile, Colombia, Peru and Uruguay), Asia (China and Taiwan) and Europe (Italy, Russia and Slovenia) (Alvarado et al., 2005, Han et al., 2011, Huang et al., 2004, Liu et al., 2009, Marandino et al., 2015, Sesti et al., 2014, Tataje-Lavanda et al., 2016, Toffan et al., 2013, Valastro et al., 2016). Strains of this lineage were first reported in China in 1996, but an Italian strain from 1986 was later described (Valastro et al., 2016).
In order to understand IBV biology, in addition to the analysis of the S1 region, it is desirable to obtain complete genome sequences of different viral population. Genomic studies provide information about the origin and spread of IBV genetic variants (Naguib et al., 2016) and help to identify the genetic changes associated with inter and intra-continental spreading. So far, there are not genomes available for any South American strains of IBV. In the present study, we obtained full-length genome of two South American strains of the GI-11 and GI-16 lineages, and performed phylogenetic analyses to determine the evolutionary relationships between these strains and worldwide field strains.
2. Materials and methods
2.1. Strains
The UY/11/CA/18 and UY/09/CA/01 strains were isolated from IBV outbreaks in commercial broilers during 2009 and 2011 from Uruguay. These strains were previously classified within the GI-11 and GI-16 lineage by full-length S1 analysis using Sanger sequencing (Marandino et al., 2015, Valastro et al., 2016).
2.2. Propagation and purification of IBV particles
The strains were propagated in 10-day-old embryonated chicken eggs by allantoic route inoculation to obtain large quantity of IBV virions. Forty mL of allantoic fluid was harvested 72 h post inoculation and then concentrated through a 20% (wt/vol) sucrose cushion. Viral pellets were re-suspended in 1200 μl of phosphate buffered saline (PBS) and the virus suspension was layered over a continuous gradient of 30 to 55% (wt/vol) sucrose in PBS. The gradient was centrifuged at 100,000g for 4 h at 4 °C.
2.3. RNA extraction and Illumina sequencing
RNA extraction was performed using the Quick-RNA™ MiniPrep kit (Zymo Research, Irvine, CA, USA) and 200 μl of purified viral particles. The extracted RNA was eluted in 35 μl of RNase-free water. Reverse transcription was carried out using the Maxima H Minus Double-Stranded cDNA Synthesis kit (Fermentas Life Sciences Inc., Hanover, MD, USA) and 13 μl of extracted RNA. Viral RNA enrichment was determined by quantitative PCR (Callison et al., 2006). Nextera XT DNA Sample Preparation kit (Illumina, USA) was used from 1 ng of double strand cDNA. After 12 PCR cycles, the final library was purified with AMPure XP (Benchman, USA) and quantified with the Qubit dsDNA HS assay kit (Invitrogen, USA). Quality and length of the library were assessed with the Agilent high-sensitivity DNA kit (Agilent, USA) using an Angilent 2100 Bioanalyzer (Agilent, USA). Sequencing was performed on an Illumina MiSeq (Illumina, USA) platform at the Institute Pasteur, Montevideo, Uruguay.
2.4. Genome assembly and annotation
The raw reads generated by the Illumina sequencer were analyzed using the Galaxy web-based platform (Goecks et al., 2010). Paired-end reads were converted to FASTQ Sanger format using FASTQ Groomer (Blankenberg et al., 2010). The read quality was assessed by FastQC (Andrews, 2010) and adapter sequences were trimmed and low-quality reads were removed from the raw reads using Trim Galore (Krueger, 2012). Reads were mapped to the closest genomic reference IBV strains (accession numbers: EU418976 and KP780179) using Burrows–Wheeler Alignment tools with Galaxy's default settings. Alignments were visually inspected and manually optimized using Integrative Genomics Viewer (IGV) (Robinson et al., 2011). The sequences were deposited in the GenBank database under the accession numbers MF421319 and MF421320. The ORF prediction was carried out in http://covdb.microbiology.hku.hk and http://www.jcvi.org/vigor/ (Huang et al., 2008, Wang et al., 2010).
2.5. Genome sequence analysis
Sequences were aligned using MUSCLE algorithm implemented in MEGA 5.0 (Tamura et al., 2011). The best-fit model of nucleotide substitution was selected under the Akaike information criterion and Bayesian information criterion as implemented in jModelTest.
Identification of potential recombinant and parental sequences and localization of possible recombinant breakpoints were performed using the RDP4 program, which implements seven distinct algorithms for characterization of recombinant sequences (Martin et al., 2015).
Maximum-likelihood trees, with approximate likelihood ratio tests for internal nodes support, were inferred using PhyML. Phylogenetic trees were visualized and edited with Figtree (Rambaut, 2012).
3. Results
3.1. Purification of IBV particles and genome sequencing
Purified virions were obtained from the UY/11/CA/18 and UY/09/CA/01 strains by gradient centrifugation; viral particles were observed as a single clear band and obtained by fractionation of the gradient. The virion-extracted RNA had a concentration of 9.83 × 106 RNA copies/μl for the UY/11/CA/18 strain and 1.35 × 106 RNA copies/μl for the UY/09/CA/01. The Illumina sequencer for these strains generated 2.3 × 106 and 3.7 × 106 reads, respectively; a low percentage of these reads matched with chicken genome (2.15 and 3.83%), while 86.4 and 86.8% of them matched with a reference IBV strain (quality control statistics is available as Supplementary material). The reads were assembled and the genome sequence of both strains were obtained; the sequences included the coding genome, the UTRs, and the poly-A tail. The complete genome sequence of UY/11/CA/18 and UY/09/CA/01 strains have a size of 27,621 and 27,638 nucleotides respectively, excluding the poly(A)-tail. Both South America strains have thirteen ORFs (1a-1b-S-3a-3b-E-M-4b-4c-5a-5b-N-6b). The ORFs have the same length, except for the ORFs 1a and S that differ in 24 and 36 nucleotides, respectively.
3.2. Strain classification
A full-length genome dataset was built using complete IBV genomes available in the GenBank database (n = 158).
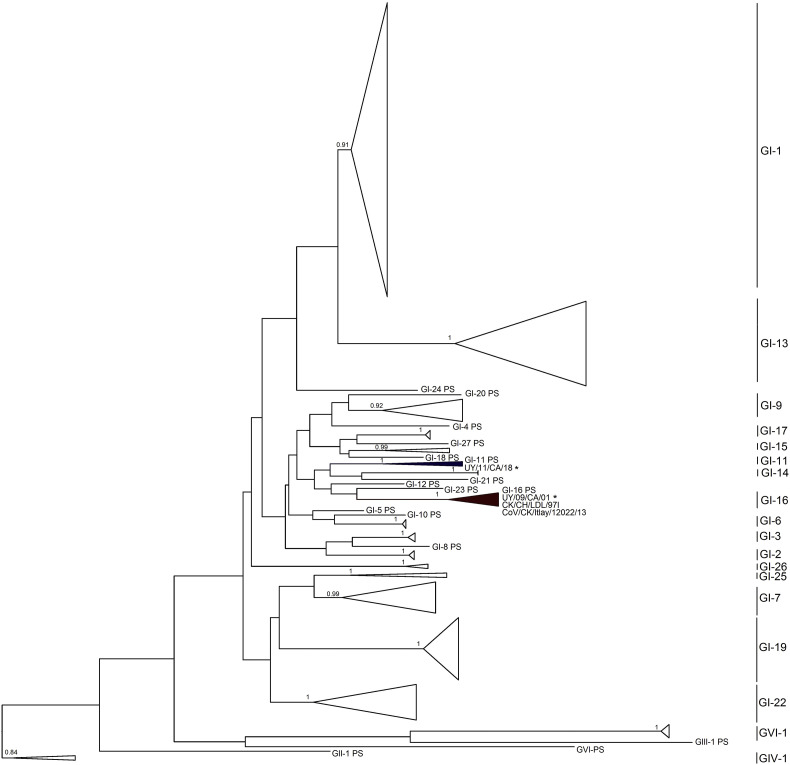
Strain classification in genotypes and lineages was performed by S1 phylogenetic analysis with non-recombinant S1 sequences retrieved from the complete genomes and prototype IBV strains following Valastro et al. (2016) (Supplementary Fig. 1). The UY/11/CA/18 and UY/09/CA/01 strains clustered within the GI-11 and GI-16 lineages, respectively.
3.3. Phylogenetic incongruence and recombination
The South American strains were analyzed using incongruence between phylogenetic trees derived from the essential ORFs present in all IBV strains (1a, 1b, S, 3a, 3b, E, M, 5a, 5b and N). Contiguous ORFs with the same phylogenetic relationship were considered as single units for the analysis, and denoted as regions I to V starting from the 5′ end.
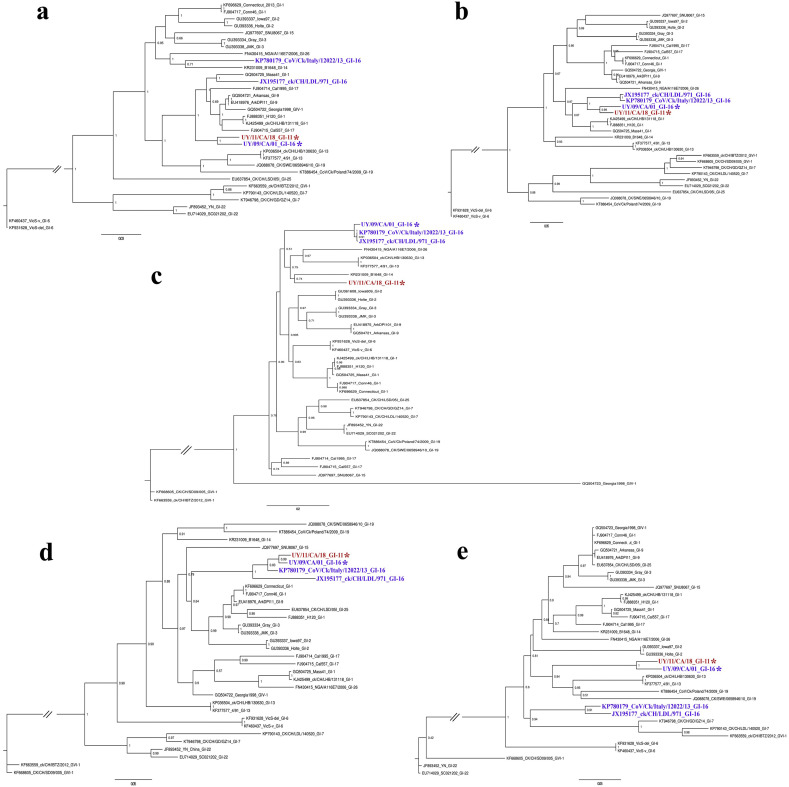
In the ORF1a phylogeny (region I), the South American strain UY/09/CA/01 (lineage GI-16) clusters with the South American strain UY/11/CA/18 (lineage GI-11) with an average nucleotide identity of 96% (Fig. 1a). Other strains of the GI-16 lineage, the Asiatic strain CK/CH/LDL/97I (accession number: JX195177) and the European strain CoV/Ck/Italy/I2022/13 (accession number: KP780179), fall in different positions in the tree and have an average nucleotide identity ranging from 90 to 92%.
Fig. 1.
Phylogenetic trees obtained with the maximum-likelihood method and JC model (a), GTR model with gamma distribution and invariant sites (b and c), and HKY model with gamma distribution and invariant sites (d and e). Phylogenetic reconstruction was carried out using the ORF1a (a), ORF1b (b), S1 sequence (c), S2 sequence and ORFs 3a, 3b, E, M, 5a and 5b (d) and ORF N (e). Mapping uncertainties for internal nodes are shown as approximate likelihood ratio test values. Strains of GI-16 and GI-11 lineages are labeled with an asterisk. This analysis was performed using a sub-sampled non-recombinant data set with representatives of the IBV variability.
In the ORF1b phylogeny (region II), strains of the GI-16 and GI-11 lineages form a monophyletic group (Fig. 1b). The nucleotide identity of this group ranges from 97 to 98%.
In the phylogenetic tree based on the S1 coding region of ORF S (region III), the UY/09/CA/01 strain clusters with Asiatic and European strains of the GI-16 lineage, while the UY/11/CA/18 strain appears separated as the only complete-genome representative of the GI-11 lineage (Fig. 1c).
The phylogenetic trees based on the S2 coding region and ORFs 3a, 3b, E, M, 5a and 5b (region IV) have the same topology for the GI-11 and GI-16 lineages and were then concatenated to produce a single phylogeny. As observed for the ORF1b phylogeny, the GI-11 and GI-16 strains cluster together in a monophyletic group with 94˗98% nucleotide identity (Fig. 1d).
The phylogenetic tree based on the ORF N (region V), encoding the N protein, showed that the South American strains form a monophyletic group, with a nucleotide identity of 97%. The Asiatic and European strains of GI-16 lineage form another monophyletic group clearly separated from the South American strains (Fig. 1e).
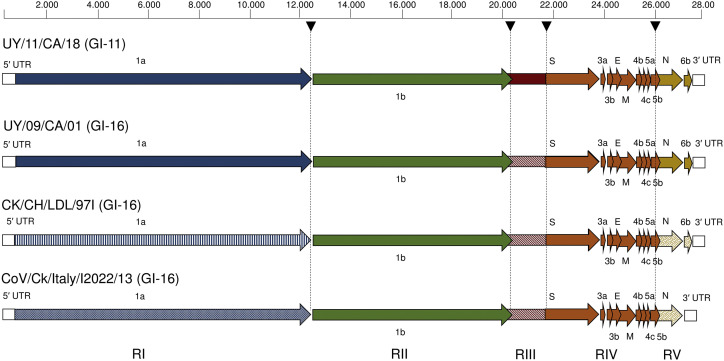
The full-length sequence of the UY/11/CA/18, UY/09/CA/01, CK/CH/LDL/97I and CoV/Ck/Italy/I2022/13 strains were further examined for the detection of recombinant events. Six RDP4 algorithms detected four recombinant breakpoints in South American strains, located at the end of ORF1a (nt 12,319), at both ends of the S1 coding region (nt 20,330 and 21,854) and the beginning of the ORF N (nt 25,900) (Fig. 2 ).
Fig. 2.
Genomic organization of the GI-16 and GI-11 strains. Essential (1a, 1b, S, 3a. 3b, E, M, 5a, 5b and N) and non-essential ORFs (4b, 4c and 6b) are indicated. European strain does not have ORF6b. Recombinant points involved in the emergence of the South American strains are labeled with a head arrow. The five regions of the South American mosaic genomes are delimited by recombinant points (indicated with different colors). Divergence of the sequences are indicated by different shade of the same color.
4. Discussion
The traditional sequencing methodology for IBV genomes requires the producing of several overlapping PCR fragments followed by Sanger sequencing (Abro et al., 2012, Mondal and Cardona, 2007). This approach is particularly laborious because coronaviruses have the largest known RNA genomes (Lai and Cavanagh, 1997). High-throughput or deep sequencing approaches drastically reduced time and cost requirements for genomic sequencing. Additional advantages of this methodology are the significant increase in sequence coverage and depth that permit to study co-infection, recombination, and quasispecies diversity (Isakov et al., 2015, Pérez et al., 2014).
Relatively few studies have used high-throughput sequencing to obtain IBV genomes (Abolnik, 2015, Naguib et al., 2016, Quinteros et al., 2015, Reddy et al., 2015). All methods use viral enrichment in allantoic cavity of embryonated chicken eggs and different protocols to purify IBV particles, including RNase treatment of allantoic fluid, sucrose gradient or genome amplification with commercial kits. The purification of IBV particles with sucrose cushion and gradient along with Illumina sequencing technology were here used with good effectivity and extremely high coverage values. This standardized protocol allowed the straightforward obtainment of the full-length genome of the UY/11/CA/18 and UY/09/CA/01 strains, including the coding genome and UTR regions. These strains were classified as belonging to the GI-11 and GI-16 lineages, supporting previous analysis using S1 analysis with Sanger sequencing (Marandino et al., 2015).
The GI-11 and GI-16 lineages are good examples of successful genetic variants that have been widely circulating for decades in South America (Marandino et al., 2015). GI-16 is also one of the four most distributed lineages of IBV, together with GI-1 (Massachusetts), GI-13 (793B) and GI-19 (QX) (Valastro et al., 2016). Despite their wide distribution, extended field persistence, and economical relevance, the genomic evolution of these lineages is poorly understood. The strains here sequenced are the first obtained in South America and provide meaningful contributions toward our understanding of IBV variability in the continent.
South American strains have a genome with thirteen ORFs (Fig. 2), including the ORFs 4b, 4c and 6b which are quite variable in both presence and absence, and in their nucleotide sequence. The same organization was observed in the Asiatic strain of the GI-16 lineage but the European strain of this lineage lacks the ORF for the 6b putative protein. The role of ORF6b in IBV remain uncertain but the ORF6b homologue of SARS coronavirus was identified as an endoplasmic reticulum/Golgi membrane-localized protein that induces apoptosis (Ye et al., 2010). This protein accelerated the replication of murine coronavirus, increasing the virulence of the original attenuated virus (Tangudu et al., 2007).
The different clustering observed in the phylogenetic analysis shows that the UY/11/CA/18 and UY/09/CA/01 strains have a mosaic genome formed by regions with different evolutionary relationships. The region III (S1 sequence) is clearly divergent between them and lead to the classification of the strains in different lineages (GI-11 and GI-16) (Fig. 1c). Region II (ORF1b) and region IV (S2 sequence and ORFs 3a, 3b, E, M, 5a, 5b) of the South American genomes examined in this study are closely related with Euro-Asiatic strains of GI-16 lineage (Fig. 1b and d). Region I (ORF1a) and region V (ORF N) differ from other IBV strains and probably came from the ancestral South American GI-11 virus (Fig. 1a and e).
Our findings suggest that the mosaic genome of South American strains here analyzed arose by recombination between the GI-11 and GI-16 lineages. Recombination is frequently described in IBV field strains (Dolz et al., 2008, Kottier et al., 1995, Lee and Jackwood, 2000) as a result of the large genome size, a replication machinery that dissociates and reassociates from the template RNA (site-assisted copy choice recombination), and the availability of full-length and subgenomic-length strands for template switching (Fu and Baric, 1992). Most recombination studies in IBV have been conducted using the S1 coding region, but relatively few analysis have been performed in a genomic context (Ammayappan and Vakharia, 2009, Thor et al., 2011, Xu et al., 2016).
The recombination in these South American strains involved four recombinant points that delimit the five genomic regions and were then involved in the generation of the mosaic genomes (Fig. 2). Notable, these breakpoints do not disrupt protein coding regions and two of them are located within genes in inter-ORF regions. Intragenic breakpoints are not very frequent in coronaviruses as recombination generally occurs at intergenic regions by specific template-switching events during the discontinuous transcription of subgenomic mRNAs (Simon-Loriere and Holmes, 2011).
We propose the following evolutionary scenario to explain the pattern of variability in the strains of the GI-16 and GI-11 lineages. This proposal supports the emergence and spreading of the GI-16 lineage in Eurasia before being introduced in South America (Marandino et al., 2015, Yu et al., 2001). This Euro Asiatic spreading of the GI-16 lineage involved sequence divergence (Fig. 1a) (Franzo et al., 2015), and changes in the structure of the genome as evidenced by the differences in the presence of the 6b ORF between the Chinese and Italian strains (Fig. 2).
In South America, the strain of the GI-16 lineage would have experienced extensive recombination with GI-11 ancestral strains to produce two recombinant lineages that have retained the S1 sequences of the parental viruses. Notable, both strains remain extremely similar in the rest of their chimeric genome that is comprised by the ORF1b, S2, 3a, 3b, E, M, 4b, 4c, 5a and 5b of the GI-16 Euro-Asiatic strains, and the 1a, N and 6b of a South American ancestral strain of the GI-11 lineage.
Our findings underscore the fact that different S1 lineages can exist with virtually identical genomes originated by recombination between ancestral strains. It is notable that the S1 protein had persisted as a single unit without the detection of recombination within the S1 coding region in all the strain already analyzed (Marandino et al., 2015). The fact that recombinant breakpoints occur outside the S1 region suggests that the S1 coding region is positively selected and transmitted as an intact unit to avoid disrupting favorable intra-protein interactions (Lefeuvre et al., 2007). S1 is a predominant determinant of antigenicity of IBV and its variability could be related to the evasion of the immune response produced by the vaccine used for IBV control. Most South American countries apply vaccines with the Massachusetts serotype (GI-1 lineage) that is highly divergent from the field strains here analyzed (Fig. 1) (Marandino et al., 2015). In this context, the selection exerted by the continuous use of the Massachusetts vaccine, together with gene flow and local differentiation, have been driven forces for the successful persistence of the GI-11 and GI-16 lineages in South America.
Together, our results reveals that IBV dynamics in South America is singular as it involves two main lineages that have emerged by the transferring of genomic regions between viruses of different origin without disrupting the S1 coding region. These recombinant virus have been extraordinary successful, persisting in the continent for several years with a notorious wide geographic distribution. Our results reveal an interesting recombination pattern that emphasizes the importance of complete genomic characterization to understand viral emergence and evolutionary history.
The following are the supplementary data related to this article.
Fig. S1.
Phylogenetic tree obtained with the maximum-likelihood method and GTR model with gamma distribution and invariant sites. Phylogenetic reconstruction was carried out using all non-recombinant S1 sequences (n = 147) retrieved from the completely sequenced genome strains and prototype strains (PS) as described by Valastro et al. (2016). Mapping uncertainties for internal nodes are shown as approximate likelihood ratio test values. South American strains are labeled with an asterisk. Complete genome strains grouped in 18 of the 32 recognized lineages.
Quality control of alignment sequencing data obtained with Qualimap 2 (Okonechnikov et al., 2016).
Acknowledgments
This study was supported by grants from the Instituto Nacional de Investigación Agropecuaria – INIA Uruguay – under project number 319 of the Fondo de Promoción de Tecnología Agropecuaria (FPTA), the Comisión Sectorial de Investigación Científica (Grant: CSIC I +D 594, 2012), Programa de Desarrollo de las Ciencias Básicas, and the Agencia Nacional de Investigación e Innovación (Grant: POS_NAC_2013_1_11865). We thank Granjas Hepa Ltda. for collaboration.
References
- Abolnik C. Genomic and single nucleotide polymorphism analysis of infectious bronchitis coronavirus. Infect. Genet. Evol. 2015;32:416–424. doi: 10.1016/j.meegid.2015.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abro S.H., Renström L.H.M., Ullman K., Isaksson M., Zohari S., Jansson D.S. Emergence of novel strains of avian infectious bronchitis virus in Sweden. Vet. Microbiol. 2012;155(2–4):237–246. doi: 10.1016/j.vetmic.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado A.I.R., Villegas P., Mossos N., Jackwood M.W. Molecular characterization of avian infectious bronchitis virus strains isolated in Colombia during 2003. Avian Dis. 2005;49(4):494–499. doi: 10.1637/7202-050304R.1. [DOI] [PubMed] [Google Scholar]
- Ammayappan A., Vakharia V.N. Complete nucleotide analysis of the structural genome of the infectious bronchitis virus strain Md27 reveals its mosaic nature. Viruses. 2009;1(3):1166–1177. doi: 10.3390/v1031166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
- Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenko A. Manipulation of FASTQ data with Galaxy. Bioinformatics. 2010;26(14):1783–1785. doi: 10.1093/bioinformatics/btq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callison S., Hilt D., Boynton T.O., Sample B.F., Robison R., Swayne D.E. Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J. Virol. Methods. 2006;138(1–2):60–65. doi: 10.1016/j.jviromet.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38(2):281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J. Coronavirus IBV: removal of spike glycopolypeptide S1 by urea abolishes infectivity and haemagglutination but not attachment to cells. J. Gen. Virol. 1986;67(7):1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K., Li D., Kant A., Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21(1):33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Chacon J.L., Rodrigues J.N., Assayag M.S., Peloso C., Pedroso A.C., Ferreira A. Epidemiological survey and molecular characterization of avian infectious bronchitis virus in Brazil between 2003 and 2009. Avian Pathol. 2011;40(2):153–162. doi: 10.1080/03079457.2010.544641. [DOI] [PubMed] [Google Scholar]
- Cook J.K.A., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41(3):239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- Dolz R., Pujols J., Ordóñez G., Porta R., Majó N. Molecular epidemiology and evolution of avian infectious bronchitis virus in Spain over a fourteen-year period. Virology. 2008;374(1):50–59. doi: 10.1016/j.virol.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippe A., Silva L., Santos M., Spilki F.R., Arns C.W. Genetic diversity of avian infectious bronchitis virus isolated from domestic chicken flocks and coronaviruses from feral pigeons in Brazil between 2003 and 2009. Avian Dis. 2010;54(4):1191–1196. doi: 10.1637/9371-041510-Reg.1. [DOI] [PubMed] [Google Scholar]
- Franzo G., Listorti V., Naylor C.J., Lupini C., Laconi A., Felice V. Molecular investigation of a full-length genome of a Q1-like IBV strain isolated in Italy in 2013. Virus Res. 2015;210:77–80. doi: 10.1016/j.virusres.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu K., Baric R.S. Evidence for variable rates of recombination in the MHV genome. Virology. 1992;189:88–102. doi: 10.1016/0042-6822(92)90684-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J., Nekrutenko A., Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough R.E., Randall C.J., Dagless M., Alexander D.J., Cox W.J., Pearson D. A “new” strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet. Rec. 1992;130(22):493–494. doi: 10.1136/vr.130.22.493. [DOI] [PubMed] [Google Scholar]
- de Groot . Family Coronaviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy, 9th Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, CA: 2012. pp. 806–828. [Google Scholar]
- Han Z., Sun C., Yan B., Zhang X., Wang Y., Li C. A 15-year analysis of molecular epidemiology of avian infectious bronchitis coronavirus in China. Infect. Genet. Evol. 2011;11(1):190–200. doi: 10.1016/j.meegid.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.P., Lee H.C., Cheng M.C., Wang C.H. S1 and N gene analysis of avian infectious bronchitis viruses in Taiwan. Avian Dis. 2004;48(3):581–589. doi: 10.1637/7186-033004R. [DOI] [PubMed] [Google Scholar]
- Huang Y., Lau S., Woo P., Yuen K. CoVDB: a comprehensive database for comparative analysis of coronavirus genes and genomes. Nucleic Acids Res. 2008;36:504–511. doi: 10.1093/nar/gkm754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakov O., Bordería A.V., Golan D., Hamenahem A., Celniker G., Yoffe L. Deep sequencing analysis of viral infection and evolution allows rapid and detailed characterization of viral mutant spectrum. Bioinformatics. 2015;31(13):2141–2150. doi: 10.1093/bioinformatics/btv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottier S., Cavanagh D., Britton P. Experimental evidence of recombination in coronavirus infectious bronchitis virus. Virology. 1995;213(2):569–580. doi: 10.1006/viro.1995.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F. Trim Galore. 2012. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ Babraham Bioinformatics.
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000;145(10):2135–2148. doi: 10.1007/s007050070044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P., Lett J.M., Reynaud B., Martin D.P. Avoidance of protein fold disruption in natural virus recombinants. PLoS Pathog. 2007;3(11):e181. doi: 10.1371/journal.ppat.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004;33(3):321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang X., Wang Y., Li C., Han Z., Shao Y. Molecular characterization and pathogenicity of infectious bronchitis coronaviruses: complicated evolution and epidemiology in china caused by cocirculation of multiple types of infectious bronchitis coronaviruses. Intervirology. 2009;52(4):223–234. doi: 10.1159/000227134. [DOI] [PubMed] [Google Scholar]
- Marandino A., Pereda A., Tomás G., Hernández M., Iraola G. Phylodynamic analysis of avian infectious bronchitis virus in South America. J. Gen. Virol. 2015;96(6):1340–1346. doi: 10.1099/vir.0.000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1(1):vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockett A.P., Cavanagh D., Brown T.D. Monoclonal antibodies to the S1 spike and membrane proteins of avian infectious bronchitis coronavirus strain Massachusetts M41. J. Gen. Virol. 1984;65(12):2281–2286. doi: 10.1099/0022-1317-65-12-2281. [DOI] [PubMed] [Google Scholar]
- Mondal S.P., Cardona C.J. Genotypic and phenotypic characterization of the California 99 (Cal99) variant of infectious bronchitis virus. Virus Genes. 2007;34(3):327–341. doi: 10.1007/s11262-006-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguib M.M., Hoper D., Arafa A.S., Setta A.M., Abed M., Monne I. Full virus from Sudan reveals distinct spots of recombination. Infect. Genet. Evol. 2016;46:42–49. doi: 10.1016/j.meegid.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez R., Calleros L., Marandino A., Sarute N., Iraola G., Grecco S. Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PLoS One. 2014;9(11) doi: 10.1371/journal.pone.0111779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinteros J.A., Markham P.F., Lee S.W., Hewson K.A., Hartley C.A., Legione A.R. Analysis of the complete genomic sequences of two virus subpopulations of the Australian infectious bronchitis virus vaccine VicS. Avian Pathol. 2015;44(3):182–191. doi: 10.1080/03079457.2015.1022857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. FigTree version 1.4.0. 2012. http://tree.bio.ed.ac.uk/software/figtree/
- Reddy V., Theuns S., Roukaerts I., Zeller M., Matthijnssens J., Nauwynck H.J. Genetic characterization of the Belgian nephropathogenic infectious bronchitis virus (NIBV) reference strain B1648. Viruses. 2015;7(8):4488–4506. doi: 10.3390/v7082827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimondi A., Craig M.I., Vagnozzi A., König G., Delamer M., Pereda A. Molecular characterization of avian infectious bronchitis virus strains from outbreaks in Argentina (2001–2008) Avian Pathol. 2009;38(2):149–153. doi: 10.1080/03079450902737821. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G. Integrative genomics viewer. Nat. Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesti L., Sara L., Alvarado L., Mató T., Palya V., de Wit J.J. Proceedings of the VIII International Symposium on Avian Corona- and Pneumoviruses and Complicating Pathogens, Rauischholzhausen, Germany. 2014. Diagnostic, epidemiology and control of the Q1 infectious bronchitis virus (IBV) variant strain in Peru, Colombia, Argentina and Chile; pp. 56–64. [Google Scholar]
- Shalk A., Hawn M. An apparently new respiratory disease of baby chicks. J. Am. Vet. Med. Assoc. 1931;78:413. [Google Scholar]
- Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011;9(8):617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81(3):1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataje-Lavanda L., Montalván Á., Bueno C., Requena D., Fernández-Díaz M. First evidence of detection of Asia/South America II (A/SAII) infectious bronchitis virus in a commercial broiler flock in Peru. Vet. Rec. Case Rep. 2016;4 [Google Scholar]
- Thor S.W., Hilt D.A., Kissinger J.C., Paterson A.H., Jackwood M.W. Recombination in avian gamma-coronavirus infectious bronchitis virus. Viruses. 2011;3(9):1777–1799. doi: 10.3390/v3091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffan A., Bonci M., Bano L., Valastro V., Vascellari M., Capua I. Diagnostic and clinical observation on the infectious bronchitis virus strain Q1 in Italy. Vet. Ital. 2013;49(4):347–355. doi: 10.12834/VetIt.1303.01. [DOI] [PubMed] [Google Scholar]
- USDA Livestock and poultry: world markets and trade. 2014. http://www.fas.usda.gov/data/livestock-and-poultry-world-markets-and-trade
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal A., Brandão P.E., Chacón J.L., Saidenberg A., Assayag M.S., Ferreira A. Molecular characterization of infectious bronchitis virus strains isolated from the enteric contents of Brazilian laying hens and broilers. Avian Dis. 2007;51(4):974–978. doi: 10.1637/7983-041307.1. [DOI] [PubMed] [Google Scholar]
- Wang S., Sundaram J.P., Spiro D. VIGOR, an annotation program for small viral genomes. BMC Bioinf. 2010;11:451. doi: 10.1186/1471-2105-11-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Liu X.Y., Zhao Y., Chen Y., Zhao J., Zhang G.Z. Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2018. Virol. J. 2016;13:40. doi: 10.1186/s12985-016-0497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.D., Wong C.K., Li P., Xie Y. The role of SARS-CoV protein, ORF-6, in the induction of host cell death. Hong Kong Med. J. 2010;16(4):22–26. [PubMed] [Google Scholar]
- Yu L., Jiang Y., Low S., Wang Z., Nam S.J., Liu W. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 2001;45(2):416–424. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quality control of alignment sequencing data obtained with Qualimap 2 (Okonechnikov et al., 2016).