Abstract
Feline calicivirus (FCV) is a highly prevalent pathogen of the domestic cat that causes acute infections of the oral and upper respiratory tract. The E region of the ORF2 protein is responsible for the induction of virus-neutralizing antibodies, thus it is important to understand the codon usage of this gene. Here, analysed 90 coding sequences of ORF2 and show that it undergoes a low codon usage bias. In addition, although mutational bias is one of the factors shaping the codon usage bias of this gene, natural selection plays a more significant role. Our results reveal part of the mechanisms driving FCV evolution, which will lay foundation for the further research of FCV.
Keywords: Feline calicivirus, Open reading frame 2 (ORF2), Codon usage, Mutation pressure, Natural selection, Neutrality analysis
Highlights
-
•
The codon usage of the ORF2 gene of FCV was first analysed in this study.
-
•
The ORF2 gene of FCV shows a low codon usage bias.
1. Introduction
Feline calicivirus (FCV) is a highly prevalent pathogen of the domestic cat, with widespread distribution across the world (Radford et al., 2007). It has been suggested that almost all of the members of Felidae, such as cats, tigers and cheetahs are susceptible to the virus. FCV has also been isolated from the faeces of dogs (Gabriel et al., 1996). FCV strains have been shown to exhibit interspecific circulation among different animal species.
FCV belongs to the genus vesivirus of the Caliciviridae. It has a small, non-enveloped, positive-sense, single-stranded RNA genome of approximately 7700 nucleotides of which the 5′ end is linked covalently to the VPg protein and the 3′ end is linked to poly(A). The genome contains three open reading frames (ORFs) referred to as ORF1, ORF2 and ORF3 (Prikhodko et al., 2014). ORF2 encodes the capsid precursor protein, which is processed by the viral protease to release a small 124 amino acid protein called the leader of the capsid (LC) and the mature capsid protein (VP1). Comparative analysis of ORF2 sequences has been used to elucidate phylogenetic relationships among different FCV isolates (Prikhodko et al., 2014).
Codons that encode the same amino acid are referred to as synonymous codons. Although synonymous codons encode for the same amino acid, their corresponding tRNAs may differ in relative abundance in the cell as well as the ribosome recognition speed, thus affecting the codon usage. The usage of synonymous codons is a non-random process with some codons being used more often than others (Marín et al., 1989). This phenomenon which called ‘codon usage bias’, can be found in numerous species such as prokaryotes, eukaryotes and viruses (Liu et al., 2011). Codon usage is influenced by two major factors, natural selection and mutation bias (Gu et al., 2004). The codon usage between the virus and the host will affect the overall survival of the virus, the ability to evade the host immune system and evolution (Moratorio et al., 2013). Thus, understanding the codon usage of viruses can provide information about viral evolution and expand our understanding of the regulation of viral gene expression based on codon adoption. This can aid rational vaccine design to achieve efficient viral protein expression to induce long-lasting immunity.
Because different FCV strains cause disease with a wide range of clinical signs, it is important to characterize the genetic variation, evolution and the codon usage pattern of FCV to understand how these viral strains cause disease. The aim of this study was to describe the genetic features of the ORF2 gene of FCV. To this end, we analysed in detail the genetic evolution, the codon usage pattern and the evolutionary characterization of the codon usage pattern of FCV.
2. Materials and methods
2.1. Sequence data
90 coding sequences (CDS) of ORF2 of FCV strains were included in this study, which were retrieved from the National Center for Biotechnology (NCBI) GenBank database (https://www.ncbi.nlm.nih.gov/nucleotide/). The details of the sequences analysed including accession number, time of collection and geographical distribution are shown in Table S1.
2.2. Nucleotide composition analysis
The frequency of each nucleotide (A%, U%, G% and C%) was calculated using BioEdit. The nucleotide composition of the third synonymous codon position of each codon (A3s, T3 s, G3 s, C3s) was calculated using the Codon W package. The G + C at the first (GC1s), second (GC2s) and third codon positions (GC3s) were calculated using the CodonW program. Additionally, the G + C at the first and the second positions (GC12s) were calculated with the same program.
2.3. Synonymous codon usage analysis
2.3.1. Relative synonymous codon usage (RSCU)
The RSCU value of each codon (except for Met, Trp and termination codons and excluding the influence of amino acid composition and sequence length) was calculated to directly reflect the usage characteristics as first proposed in by sharp et al. (Sharp and Li, 1986). The RSCU value of a codon is the ratio of its observed frequency to its expected frequency assuming that all codons for a particular amino acid are used evenly (Peden, 1999) and it was calculated using the following equation:
It is essential to note that gij represents the observed number of the i thcodon for the j th amino acid which has ni kinds of synonymous codons (Nasrullah et al., 2015). Normally, it is considered that a high RSCU value reflects a strong codon usage bias. Codon usages with RSCU values of < 1.0, 1.0, > 1.0 stand for negative codon usage bias, no bias and positive codon usage bias respectively (Chen et al., 2014a).
2.3.2. Effective number of codons (ENC)
The ENC is considered the magnitude of the codon usage bias of a single gene (Wright, 1990). The ENC value is not influenced by the amino acid or the gene length (Morla et al., 2016) . The ENC value was calculated using the formula given below:
where the s value stands for the GC3s composition of each codon (Chen et al., 2014a). The ENC value ranges from 20 (only one of the possible synonymous codons is used for the corresponding amino acid) to 61 (all possible synonymous codons are used equally for the corresponding amino acid (Wright, 1990). In contrast to the RSCU value, the smaller the ENC value, the greater the extent of codon usage bias. A ENC value equal or < 35 is considered to be a sign of strong codon bias (Comeron and Aguadé, 1998).
2.3.3. ENC-plot
ENC plots were drawn to determining factors (especially mutation pressure) that influence the codon usage bias (Wright, 1990) taking the GC3s values in the x axes and the ENC values in the y axes. Under the null model, if codon usage is only constrained by G + C, the predicted ENC values would sit on or around the standard curve (Jiang et al., 2007). Otherwise, if predicted ENC values sit far lower than the standard curve, other factors such as natural selection play a major role in shaping codon usage bias.
2.3.4. Principal component analysis (PCA)
PCA, a widely used multivariate statistical approach to determine the major trends in codon usage variation of genes was performed using GraphPad Prism 6.0 (Gupta and Ghosh, 2001). The RSCU value of each gene, excluding Met, Trp and termination codons, is explained by a 59-dimensional space vector and transformed into a smaller number of unrelated factors (Lu et al., 2013).
2.3.5. General average GRAVY and AROMA indices
To unravel if natural selection shapes codon usage bias, the Gravy and Aroma score were determined. Both indices were obtained from CodonW, which reveals the frequencies of hydrophobic and aromatic amino acids respectively (Kyte and Doolittle, 1982). A higher Gravy or Aroma value suggests a more hydrophobic or aromatic amino acid product.
2.3.6. Neutrality analysis
Neutrality analysis was used to determine the role of mutational bias and natural pressure shaping codon usage bias. In neutrality plots, GC12s are drawn against GC3s each dot representing an independent FCV strain. It is essential to demonstrate that the slope of the line near to zero is an indication of only natural selection constrains the codon usage bias, while the near to one represents complete neutrality (Sueoka 1988).
2.4. Statistical analysis
Correlation analysis was performed using GraphPad Prism 6.0.
3. Results
3.1. Nucleotide composition of the FCV ORF2 gene
The nucleotide compositions of 90 coding sequences of FCV ORF2 were calculated. The mean values of A%, C%, G% and U% were 26.48%, 21.23%, 23.20% and 29.10%, with standard deviations (SD) of 0.53, 0.73, 0.44 and 0.62 respectively. This indicates that U and A were more abundant than C and G, while U was the most preferred nucleotide. The codon compositions at the third position (A3, U3, G3, C3, GC3) revealed that the mean U3% (43.57%) was the highest among the four nucleotides, which is consist with the nucleotide content of FCV ORF2 gene (Table S2). The GC3 values raged from 32.8% to 44.3% (mean 38.3%.), indicating that A/U terminated codons are preferred over G/C terminated codons.
3.2. Relative synonymous codon usage (RSCU) and effective number of codons (ENC) of the FCV ORF2 gene
The RSCU values of all 61 codons were calculated and are displayed in Table 1 . Among the 18 most frequently employed synonymous codons, 17 optional codons ended with U (GCU for Ala, UGU for Cys, GAU for Asp, UUU for Phe, GGU for Gly, AUU for Ile, CUU for Leu, CCU for Pro, UCU for Ser, ACU for Thr, GUU for Val, UAU for Tyr), 3 preferred codons terminated with A (GAA for Glu, AAA for Lys, CAA for Gln), 2 codons ended with C (CAC for His, AAC for Asn) and one codon terminated with G (AGG for Arg). It is interesting to note that codons ending in U were the most frequently used. This is in accordance with the fact that U was the most abundantly used nucleotide, demonstrating that codon usage is influenced by compositional constraints.
Table 1.
RSCU of 90 sequences of the ORF2 gene of FCV.
| Amino acid | Codon | RSCU/number | Amino acid | Codon | RSCU/number |
|---|---|---|---|---|---|
| Ala | GCA | 1.1088/971 | Met | ATG | 1/1264 |
| GCC | 0.8655/758 | Asn | AAC | 1.0364/1522 | |
| GCG | 0.2170/190 | AAT | 0.9636/1415 | ||
| GCT | 1.8088/1584 | Pro | CCA | 1.4004/1372 | |
| Cys | TGC | 0.8711/331 | CCC | 0.8359/819 | |
| TGT | 1.1289/429 | CCG | 0.2715/266 | ||
| Asp | GAC | 0.8030/1645 | CCT | 1.4922/1462 | |
| GAT | 1.1970/2452 | Gln | CAA | 1.4014/1290 | |
| Glu | GAA | 1.2405/1372 | CAG | 0.5986/551 | |
| GAG | 0.7595/840 | Arg | AGA | 1.6088/514 | |
| Phe | TTC | 0.9181/1328 | AGG | 1.6651/532 | |
| TTT | 1.0819/1565 | CGA | 0.6291/201 | ||
| Gly | GGA | 1.2784/1473 | CGC | 0.7418/237 | |
| GGC | 0.7021/809 | CGG | 0.6354/203 | ||
| GGG | 0.6735/776 | CGT | 0.7199/230 | ||
| GGT | 1.3461/1551 | Ser | AGC | 0.5226/482 | |
| His | CAC | 1.1190/823 | AGT | 0.8403/775 | |
| CAT | 0.8810/648 | TCA | 1.2089/1115 | ||
| Ile | ATA | 0.4546/726 | TCC | 0.9389/866 | |
| ATC | 0.9305/1486 | TCG | 0.2906/268 | ||
| ATT | 1.6149/2579 | TCT | 2.1988/2028 | ||
| Lys | AAA | 1.0958/1275 | Thr | ACA | 1.2033/1234 |
| AAG | 0.9042/1052 | ACC | 1.0054/1031 | ||
| Leu | CTA | 0.7986/630 | ACG | 0.3023/310 | |
| CTC | 0.9394/741 | ACT | 1.4890/1527 | ||
| CTG | 0.6554/517 | Val | GTA | 0.5403/548 | |
| CTT | 1.9294/1522 | GTC | 0.8233/835 | ||
| TTA | 0.6377/503 | GTG | 1.1171/1133 | ||
| TTG | 1.0395/820 | GTT | 1.5193/1541 | ||
| Trp | TGG | 1/1259 | Tyr | TAC | 0.9414/900 |
| TAT | 1.0586/1012 |
The values of the ENC analysis ranged from 49.38 to 57.55 (average ± SD of 53.70 ± 1.639) indicating fluctuation among the 90 FCV strains. The high ENC values (ENC > 45) indicate a low codon usage bias.
3.3. The role of mutational bias in shaping the codon usage of the FCV ORF2 gene
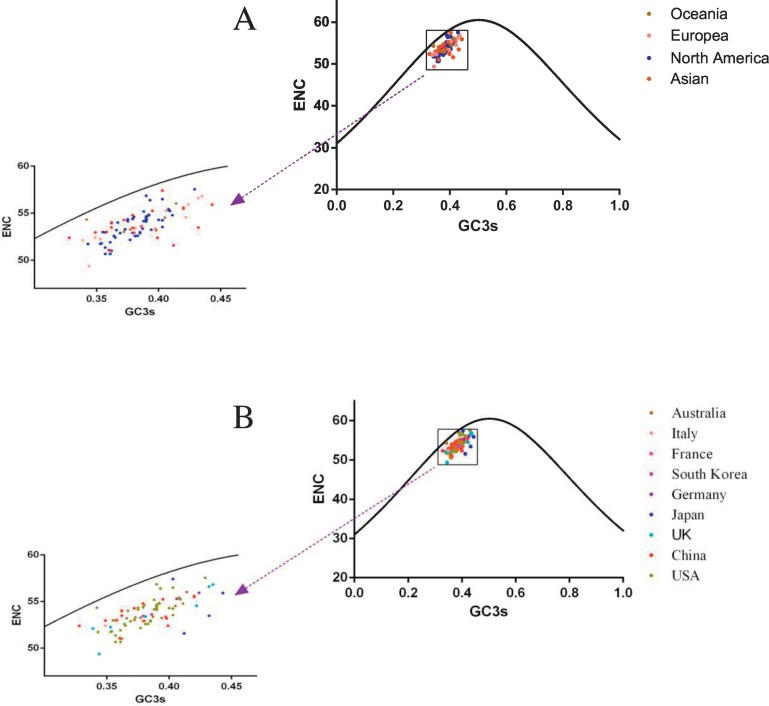
ENC-plots were drawn with ENC values plotted against GC3s values according to the geographical distribution of the strains used in this study (Fig. 1 ). All the strains, represented in different colours for each continent, located below the theoretical curve (Fig. 1A). Additionally, strains isolated from the same continents did not cluster together. In particular, all 90 strains collected from 9 countries were distributed widely (Fig. 1B). This indicates that mutational pressure combined with other factors contributes to codon usage bias of the FCV ORF2 gene (Sueoka 1988).
Fig. 1.
ENC plots depicting the relationship between ENC and the GC content at the third codon position (GC3s) according to geographical distribution of each strain by (A) continents or (B) country.
In addition, there was a correlation between the nucleotide composition (A%, U%, G%, C%, GC%) and the codon contents (A3s, T3 s, C3s, G3 s, GC3s) (p < 0.05), except between the relationship of A3s and T. Furthermore, there was a significant correlation between the ENC values and the nucleotide compositions (p < 0.01), which indicates that mutational bias influences the synonymous codon usage pattern of the ORF2 gene of FCV.
3.4. Principal component analysis (PCA)
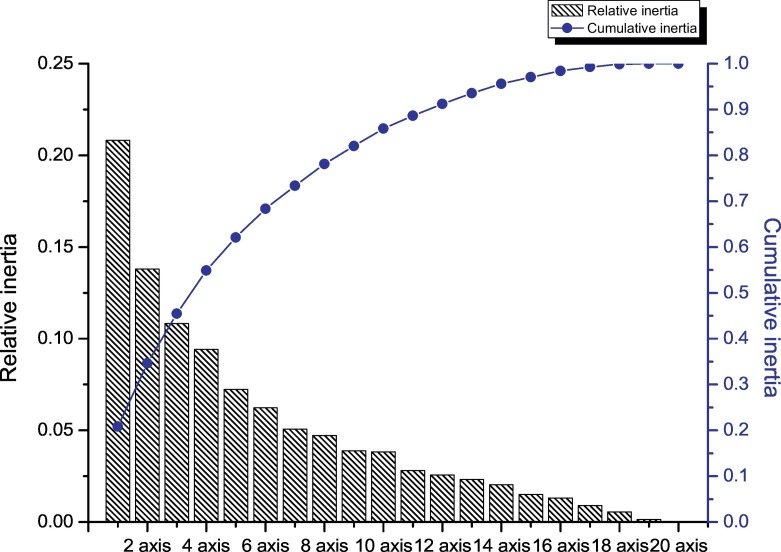
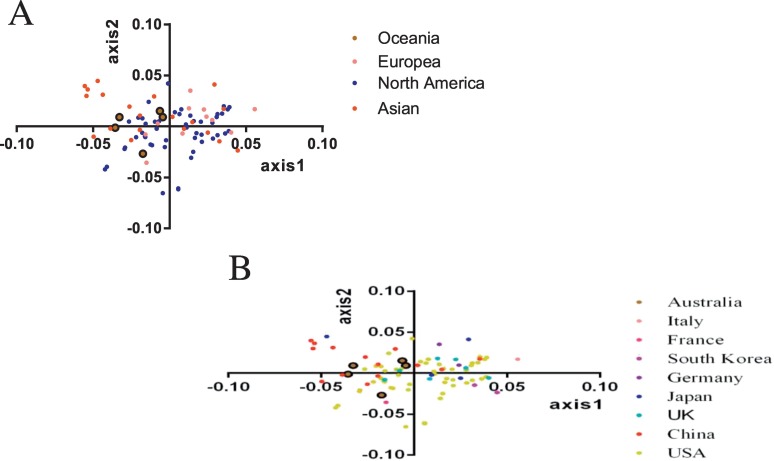
PCA analysis, a multivariable method, was employed to unravel the variation of the synonymous codon usage (Singh et al. 2016). We found that the first four principal axes accounted for 54.86% of the total variation with the first, second, third and fourth principal axis accounting for 20.81%, 13.8%, 10.83% and 9.42% respectively (Fig. 2 ). This suggests that the first and second axis contributed to the variation of RSCU of synonymous codons. PCA analysis was performed based on the continent and country of isolation (Fig. 3 ). Based on the distribution of different strains on the first two axes, we found that the distribution of Asian strains, especially strains collected from China, was more widespread than the distribution of strains isolated from Oceania, Europe and North America. Moreover, most of the strains isolated from North America located near the origin, indicating that mutational pressure contributed to codon usage of the FCV ORF2 gene.
Fig. 2.
Principal component analysis of the amino acid usage frequencies of the ORF2 gene. The relative and cumulative inertia of the first 20 factors are shown based on principal component analysis.
Fig. 3.
Principal component analysis based on geographical distribution according to: (A) continent or (B) country. Different geographical distributions are represented by different colours.
3.5. The role of natural selection in shaping the codon usage pattern of the ORF2 gene of FCV
It is normally considered that natural selection contributes to some extent to codon usage bias, therefore we evaluated the correlation between the Gravy and Aroma values and the codon contents (A3s, G3 s, C3s, U3 s and GC3s) (Table 2 ). We found a correlation between the Aroma values and U3 s, G3 s and GC3s (p < 0.05), confirming that natural selection influences the codon usage bias of the FCV ORF2 gene.
Table 2.
Correlation analysis of the nucleotide composition, Axis1, Axis2, Gravy, Aroma, nucleotide at the third position and ENC.
| A | C | G | T | GC | Gravy | Aromo | Axis1 | Axis2 | |
|---|---|---|---|---|---|---|---|---|---|
| Nc | − 0.4652** | 0.6024** | 0.4801** | − 0.6511** | 0.7161** | − 0.2012 | − 0.1371 | 0.1103 | 0.3489** |
| GC3s | − 0.6888** | 0.7532** | 0.6418** | − 0.7512** | 0.9126** | − 0.0802 | − 0.2666* | 0.1408 | 0.4044** |
| T3 s | 0.3443** | − 0.76** | − 0.46** | 0.9259** | − 0.83** | 0.1378 | 0.2722** | − 0.2728** | − 0.2962** |
| C3s | − 0.5442** | 0.8967** | 0.2532* | − 0.7673** | 0.841** | − 0.1449 | − 0.2608* | 0.1534 | 0.3926** |
| A3s | 0.8373** | − 0.377** | − 0.54** | 0.1082 | − 0.561** | − 0.0879 | 0.1313 | 0.015 | − 0.375** |
| G3 s | − 0.5055** | 0.1586 | 0.8325** | − 0.3455** | 0.5283** | 0.0371 | − 0.1316 | − 0.0318 | 0.1925 |
means p < 0.05.
means p < 0.01.
3.6. Natural selection plays a more important role than mutation pressure in shaping the codon usage of FCV
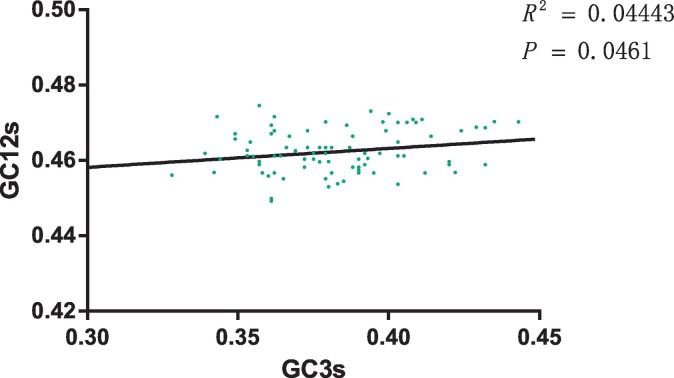
We found that both mutation pressure and natural selection contribute to the codon usage bias of the ORF2 gene of FCV. Thus, to understand which one plays a more important role, the GC12s values (the mean value of GC1s and GC2s) were plotted against the GC3s values (Fig. 4 ). We found a correlation between GC12s and GC3s (p < 0.05) with a correlation coefficient of 0.22, indicating that relative neutrality was 22% or, conversely, natural selection was 78%. Thus, natural selection plays a major role in shaping the codon usage bias of ORF2 gene of FCV compared to mutational pressure.
Fig. 4.
Neutrality analysis in relation to GC3s and GC12s.
4. Discussion
RNA zoonotic viruses; such as influenza viruses and coronaviruses which have highly susceptible to recombination and cross species transmission (Su et al., 2015, Su et al., 2016). FCV is a RNA virus and as such it has experienced a high evolution rate since its emergence. Previous studies on FCV have mostly focused on infectivity (Pesavento et al. 2004) and prevalence (Knowles et al. 1989). However, there are no studies on codon usage bias of FCV. ORF2 is one of three ORF of FCV genome and encodes is the major capsid protein VP1. Therefore, the codon usage of ORF2 of FCV was first studied.
Previous studies on codon usage bias of other RNA viruses showed high codon usage bias. For example, analysis of the G gene of Rabies virus (RABV) showed ENC values ranging from 44.40% to 51.40% (Zhao et al. 2016); Foot and Mouth Disease Virus (FMDV) displayed ENC values of 51.42% (Zhou et al. 2013); Porcine Epidemic Diarrhea Virus (PEDV) of 47.91% (Chen et al. 2014b); and Severe Acute Respiratory Syndrome (SARS) of 48.99% (Zhao et al. 2008). However, the mean ENC value of ORF2 of FCV reported here was 53.70% (SD ± 1.639), thus in comparison with the above viruses, the degree of codon usage bias of FCV is lower.
Codon usage bias is mainly influenced by natural selection (Romero et al. 2003) and mutation pressure (Jenkins et al. 2001). Here, we used ENC-Plots (Fig. 1) and PCA(Fig. 3) analysis according to the geographical distribution to investigate the major factors shaping the codon usage bias of the FCV ORF2 gene . We found only one strain isolated from USA, one from Japan and one from Australia to sit near the standard values, suggesting that mutation pressure contributed to the codon usage bias of these three strains. PCA analysis revealed that mutation pressure is the dominant force shaping the codon usage of sequences isolated from North America (Fig. 3). Furthermore, analysis of the relationships between nucleotide composition and the codon contents at the third base positions suggested that mutation pressure is one of the factors in shaping the codon usage of FCV ORF2. Correlation analysis of Gravy, Aroma values and codon content (A3s, G3 s, C3s, U3 s and GC3s) showed that there is a correlation between Aroma and U3 s, G3 s and GC3s, confirming that natural selection contributes to the codon usage of the ORF2 gene of FCV. Since both mutation pressure and natural selection are important in driving the codon usage of ORF2, we performed neutrality analysis to understand which of the two forces has a bigger impact. We found that natural selection is the major force driving the codon usage of ORF2.
This is the first study analysing the codon usage of the FCV ORF2 gene and describing the forces that drive FCV evolution. In the future, more epidemiological surveys and sequence analysis are required to examine the factors that resulted in FCV evolution.
Acknowledgments
This paper was supported in part by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.meegid.2017.06.013.
Appendix A. Supplementary data
Supplementary tables
References
- Chen Y., Shi Y., Deng H., Gu T., Xu J., Ou J., Jiang Z., Jiao Y., Zou T., Wang C. Characterization of the porcine epidemic diarrhea virus codon usage bias. Infect. Genet. Evol. 2014;28:95–100. doi: 10.1016/j.meegid.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shi Y., Deng H., Gu T., Xu J., Ou J., Jiang Z., Jiao Y., Zou T., Wang C. Characterization of the porcine epidemic diarrhea virus codon usage bias. Infect. Genet. Evol. Journal of Molecular Epidemiology & Evolutionary Genetics in Infectious Diseases. 2014;28:95–100. doi: 10.1016/j.meegid.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J.M., Aguadé M. An evaluation of measures of synonymous codon usage bias. J. Mol. Evol. 1998;47:268–274. doi: 10.1007/pl00006384. [DOI] [PubMed] [Google Scholar]
- Gabriel S.S., Tohya Y., Mochizuki M. Isolation of a calicivirus antigenically related to feline caliciviruses from feces of a dog with diarrhea. J. Vet. Med. Sci. 1996;58:1041–1043. doi: 10.1292/jvms.58.10_1041. [DOI] [PubMed] [Google Scholar]
- Gu W., Zhou T., Ma J., Sun X., Lu Z. Analysis of synonymous codon usage in SARS coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S.K., Ghosh T.C. Gene expressivity is the main factor in dictating the codon usage variation among the genes in Pseudomonas aeruginosa. Gene. 2001;273:63–70. doi: 10.1016/s0378-1119(01)00576-5. [DOI] [PubMed] [Google Scholar]
- Jenkins G.M., Pagel M., Gould E.A., Pm D.A.Z., Holmes E.C. Evolution of base composition and codon usage bias in the genus Flavivirus. J. Mol. Evol. 2001;52:383. doi: 10.1007/s002390010168. [DOI] [PubMed] [Google Scholar]
- Jiang P., Sun X., Lu Z. Analysis of synonymous codon usage in Aeropyrum pernix K1 and other Crenarchaeota microorganisms. J. Genet. Genomics. 2007;34:275–284. doi: 10.1016/S1673-8527(07)60029-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles J.O., Gaskell R.M., Gaskell C.J., Harvey C.E., Lutz H. Prevalence of feline calicivirus, feline leukaemia virus and antibodies to FIV in cats with chronic stomatitis. Vet. Rec. 1989;124:336. doi: 10.1136/vr.124.13.336. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liu Y.S., Zhou J.H., Chen H.T., Ma L.N., Pejsak Z., Ding Y.Z., Zhang J. The characteristics of the synonymous codon usage in enterovirus 71 virus and the effects of host on the virus in codon usage pattern. Infect. Genet. Evol. Journal of Molecular Epidemiology & Evolutionary Genetics in Infectious Diseases. 2011;11:1168–1173. doi: 10.1016/j.meegid.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Guo W., Qi T., Ma J., Zhao S., Tian Z., Pan J., Zhu C., Wang X., Xiang W. Genetic analysis of the PB1-F2 gene of equine influenza virus. Virus Genes. 2013;47:250–258. doi: 10.1007/s11262-013-0935-x. [DOI] [PubMed] [Google Scholar]
- Marín A., Bertranpetit J., Oliver J.L., Medina J.R. Variation in G + C-content and codon choice: differences among synonymous codon groups in vertebrate genes. Nucleic Acids Res. 1989;17:6181–6189. doi: 10.1093/nar/17.15.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moratorio G., Iriarte A., Moreno P., Musto H., Cristina J. A detailed comparative analysis on the overall codon usage patterns in West Nile virus. Infect. Genet. Evol. 2013;14:396–400. doi: 10.1016/j.meegid.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Morla S., Makhija A., Kumar S. Synonymous codon usage pattern in glycoprotein gene of rabies virus. Gene. 2016;584:1–6. doi: 10.1016/j.gene.2016.02.047. [DOI] [PubMed] [Google Scholar]
- Nasrullah I., Butt A.M., Tahir S., Idrees M., Tong Y. Genomic analysis of codon usage shows influence of mutation pressure, natural selection, and host features on Marburg virus evolution. BMC Evol. Biol. 2015;15:1–15. doi: 10.1186/s12862-015-0456-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden J. 1999. Analysis of Codon Usage [PhD Dissertation] [Google Scholar]
- Pesavento P.A., Maclachlan N.J., Dillardtelm L., Grant C.K., Hurley K.F. Pathologic, immunohistochemical, and electron microscopic findings in naturally occurring virulent systemic feline calicivirus infection in cats. Vet. Pathol. 2004;41:257. doi: 10.1354/vp.41-3-257. [DOI] [PubMed] [Google Scholar]
- Prikhodko V.G., Sandoval-Jaime C., Abente E.J., Bok K., Parra G.I., Rogozin I.B., Ostlund E.N., Green K.Y., Sosnovtsev S.V. Genetic characterization of feline calicivirus strains associated with varying disease manifestations during an outbreak season in Missouri (1995–1996) Virus Genes. 2014;48:96–110. doi: 10.1007/s11262-013-1005-0. [DOI] [PubMed] [Google Scholar]
- Radford A.D., Coyne K.P., Dawson S., Porter C.J., Gaskell R.M. Feline calicivirus. Vet. Res. 2007;38:319–335. doi: 10.1051/vetres:2006056. [DOI] [PubMed] [Google Scholar]
- Romero H., Zavala A., Musto H., Bernardi G. The influence of translational selection on codon usage in fishes from the family Cyprinidae. Gene. 2003;317:141–147. doi: 10.1016/s0378-1119(03)00701-7. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Li W.H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 1986;24:28. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Singh N.K., Tyagi A., Kaur R., Verma R., Gupta P.K. Characterization of codon usage pattern and influencing factors in Japanese encephalitis virus. Virus Res. 2016;221:58–65. doi: 10.1016/j.virusres.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. U. S. A. 1988;85:2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Bi Y., Wong G., Gray G.C., Gao G.F., Li S. Epidemiology, Evolution, and Recent Outbreaks of Avian Influenza Virus in China. J. VIROL. 2015;89:8671–8676. doi: 10.1128/JVI.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. TRENDS MICROBIOL. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. The ‘effective number of codons’ used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- Zhao S., Zhang Q., Liu X., Wang X., Zhang H., Wu Y., Jiang F. Analysis of synonymous codon usage in 11 Human Bocavirus isolates. Biosystems. 2008;92:207–214. doi: 10.1016/j.biosystems.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zheng H., Xu A., Yan D., Jiang Z., Qi Q., Sun J. Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genomics. 2016;17:677. doi: 10.1186/s12864-016-3021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.H., Gao Z.L., Zhang J., Ding Y.Z., Stipkovits L., Szathmary S., Pejsak Z., Liu Y.S. The analysis of codon bias of foot-and-mouth disease virus and the adaptation of this virus to the hosts. Infect. Genet. Evol. 2013;14:105–110. doi: 10.1016/j.meegid.2012.09.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables