Abstract
Vectors based on different serotypes of adeno-associated virus hold great promise for human gene therapy, based on their unique tissue tropisms and distinct immunological profiles. A particularly interesting candidate is AAV8, which can efficiently and rapidly transduce a wide range of tissues in vivo. To further unravel the mechanisms behind AAV8 transduction, we used yeast two-hybrid analyses to screen a mouse liver complementary DNA library for cellular proteins capable of interacting with the viral capsid proteins. In total, we recovered approximately 700 clones, comprising over 300 independent genes. Sequence analyses revealed multiple hits for over 100 genes, including two encoding the endosomal cysteine proteases cathepsins B and L. Notably, these two proteases also physically interacted with the corresponding portion of the AAV2 capsid in yeast, but not with AAV5. We demonstrate that cathepsins B and L are essential for efficient AAV2- and AAV8-mediated transduction of mammalian cells, and document the ability of purified cathepsin B and L proteins to bind and cleave intact AAV2 and AAV8 particles in vitro. These data suggest that cathepsin-mediated cleavage could prime AAV capsids for subsequent nuclear uncoating, and indicate that analysis of additional genes recovered in our screen may help to further elucidate the mechanisms behind transduction by AAV8 and related serotypes.
Introduction
Adeno-associated virus is a non-pathogenic virus belonging to the parvovirus family, which also includes canine parvovirus, minute virus of mice, and the human B19 virus.1 However, AAV is distinguished from other family members, in that it is a dependovirus, able to replicate and produce viral particles only in the presence of a helper virus such as adenovirus. AAV particles are composed of a 4.7 kb single-stranded DNA genome enclosed within a non-enveloped icosahedral capsid of about 20 nm in diameter. Sixty subunits of the structural capsid proteins, VP1, VP2, and VP3, assemble at a 1:1:10 ratio to form the intact viral capsid.2
Vectors derived from AAV are particularly powerful and promising tools for human gene transfer, owing to their high safety (the vectors are devoid of wild-type genes, and integrate at low frequency, if at all) and their ability to mediate sustained gene expression in various cell types. Recently, the usefulness of AAV vectors has been further enhanced with the isolation of over 100 novel AAV serotypes.3, 4 Encapsidation of recombinant AAV genomes with these viral variants typically results in vector particles characterized by distinct tissue tropism and immunological profiles. An outstanding example is provided by AAV8, a serotype isolated from rhesus monkey. As shown by a recent plethora of reports, vectors pseudotyped with AAV8 can mediate unusually robust and sustained transgene expression in numerous tissues, including the liver, heart, and skeletal muscle.5, 6, 7 In particular, AAV8 can yield up to 20-fold higher transduction levels in the liver than the AAV prototype-2, despite an 83% amino-acid homology between the two capsids.6 It is not surprising that AAV8 is currently gaining increasing popularity as a vector for the in vivo transduction of multiple tissues and species, and is being considered for clinical evaluation in humans.
However, despite the growing interest in this particular AAV serotype, little is known about its mechanism of transduction. In fact, most of the insight into the various steps of cellular AAV infection stems from studies conducted with the AAV family prototype, AAV2. The initial step typically involves attachment to the cell surface via binding to one or more primary or secondary receptor molecules. For AAV2, these are heparan sulfate proteoglycans, αVβ5 integrin, and the fibroblast or hepatocyte growth factor receptors.8, 9, 10, 11 In contrast, AAV5 preferentially attaches to cells by binding to sialic acid and the platelet-derived growth factor receptor,12, 13 explaining the distinct tissue tropisms displayed by this serotype. Binding of AAV2 to the cell surface also appears to activate intracellular signaling pathways, which subsequently trigger internalization of the virus by receptor-mediated endocytosis. Rac1, a small GTP-binding protein, is a key player in this process, as its activity is essential to virus uptake into the cell.14
Subsequent to cell entry, the AAV2 particle proceeds to the endosomal compartment, where it can traffic to either the late or recycling endosomes. Two factors influencing the localization of the particles within the different endosomal subcompartments are the viral dose administered and the cell type.15, 16 In contrast to AAV2, AAV5 has been reported to accumulate in the Golgi compartment following its endocytosis from the cell surface,17 suggesting that AAV serotypes can utilize distinct transduction pathways. AAV2 trafficking to the endosome is a critical step, as particles injected directly into the cytoplasm do not express their viral genomes.18 In addition, treatments that block acidification of the endosomal compartment cause a significant decrease in AAV2 transduction.16 Collectively, these studies suggest that proper AAV endosomal processing may serve to “prime” the viral capsid for downstream steps, such as nuclear transport and/or uncoating of the virions.
Following endosomal escape, AAV2 particles travel to the perinuclear compartment, a step probably facilitated by an ATP-dependent molecular motor.19 The ubiquitin–proteasome pathway appears to be involved in the uncoating and/or nuclear translocation of AAV, as ubiquitination of the capsid proteins of both AAV2 and AAV5 was reported, as was the fact that proteasome inhibitors enhance nuclear AAV uptake.20, 21, 22 Inside the nucleus, AAV eventually uncoats to release its genome and allow its subsequent transcription and replication. In the case of AAV2, this process is slow and inefficient, and is probably one of the major limiting factors for transduction by recombinant AAV2. In contrast, AAV8 virions uncoat much more rapidly, likely explaining their very robust transduction efficiency.23
To further our understanding of the mechanisms behind AAV8 transduction, we conducted a yeast two-hybrid screen for host cellular proteins that are able to physically interact with AAV8 capsid proteins. Here, we report the identification of over 100 respective candidates, including two particularly interesting clones encoding the endosomal cysteine proteases cathepsins B and L. Using cathepsin-specific inhibitors, we were able to show that their activities are important for AAV8 transduction of mammalian cells, and that they play a similar role in AAV2. Moreover, in in vitro assays, we found that both cathepsins bound to and cleaved the capsids of either serotype, creating a virus-specific restriction pattern. These data suggest that cathepsins B and L are involved in the differential endosomal processing of AAV2 and AAV8, and that they may prime the virions for subsequent nuclear disassembly as a prerequisite for transgene expression.
Results
A two-hybrid screen to identify interaction partners of AAV8 capsid proteins
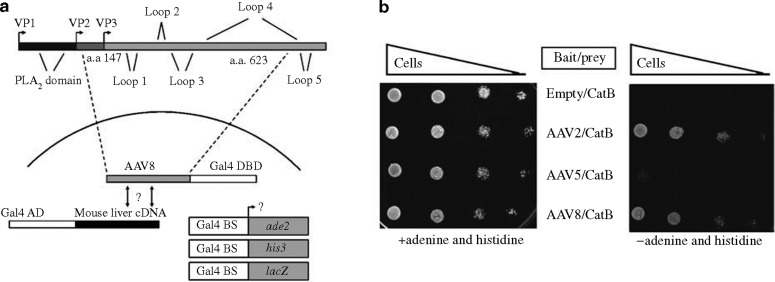
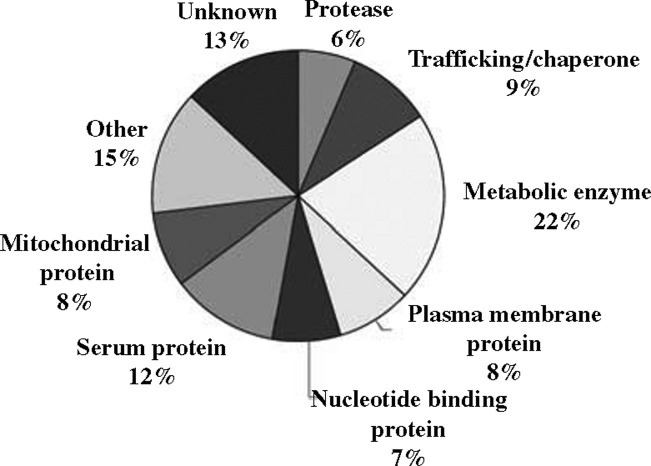
To gain insight into the mechanism of AAV8 transduction, we conducted a yeast two-hybrid screen to identify host cellular proteins capable of interacting with portions of the AAV8 capsid. We fused the Gal4p DNA-binding domain to portions of the AAV8 capsid homologous to regions of the AAV2 capsid that contain most of the exposed residues. The resulting bait (encoding residues 147–623 of the AAV8 VP1 capsid protein) was used to screen a mouse liver cDNA-Gal4p activation domain library under highly stringent conditions, requiring the activation of two different Gal4p-binding-dependent reporter genes (ade2 and his3). Sufficient yeast cells were transformed with both the bait and library (containing an estimated 3.5 million clones) plasmids to provide a 1.5-fold coverage of the library. We recovered 700 clones capable of activating both reporter genes, allowing the cells to grow on the selection media (Figure 1 ). Sequence analyses of the plasmid DNA from these clones revealed the identity of 313 different genes. Among these, 108 were repeatedly recovered from multiple independent colonies (listed in Table 1 ), suggesting that their interaction with the AAV8 bait was strong and genuine. The proteins encoded by these 108 genes belonged to a wide range of protein classes (Figure 2 ), including plasma membrane proteins (potentially acting as viral receptors on the cell surface) and proteases, which could theoretically contribute to viral uncoating during AAV cellular transduction. In fact, two particularly interesting candidates recovered in our screen were cathepsins B and L, endosomal proteins previously implicated in the uncoating of numerous other viruses, such as Ebola and reovirus.24, 25 As AAV transduction depends on virus trafficking through the endosome, we decided to further investigate the potential role of these two cathepsins in this step of the AAV life cycle.
Figure 1.
A yeast two-hybrid screen for proteins capable of interacting with the AAV8 capsid protein. (a) Portions of the AAV8 capsid were fused to the Gal4 DNA-binding domain and used as bait to screen prey encoding a mouse liver cDNA library fused to the Gal4 activation domain. The three reporter genes ade2, his3, and lacZ preceded by Gal4 binding sites (BS) were used to measure an interaction between bait and prey plasmids. (b) Yeast co-transformed with plasmids encoding the cathepsin B protease prey and the individual bait proteins were grown, serially diluted, and spotted onto media either containing or lacking adenine and histidine. The activation of both the ade2 and his3 reporter genes is necessary for growth on media lacking adenine and histidine.
Table 1.
Genes recovered from the screen of a mouse liver cDNA library with an AAV8 bait
| Gene name | Genbank No. | No. clones |
|---|---|---|
| Protease | ||
| Preprotrypsin, protease serine 2 | NM_009430 | 23 |
| cathepsin B | NM_007798 | 12 |
| proteasome subunit, beta type 4 | BC008241 | 9 |
| lysosomal pepstatin insensitive protease | AF111172 | 8 |
| cathepsin C | U74683 | 5 |
| trypsin 4 | BC061135 | 3 |
| cathepsin L | BC068163 | 2 |
| Trafficking/chaperone | ||
| chaperonin containing tcp-1, eta subunit | AB022160 | 6 |
| Selenium binding protein 1 | NM_009150 | 5 |
| KDEL ER protein retention recep. 1 | NM_133950 | 4 |
| trafficking protein particle complex 6A | BC037154 | 3 |
| dnaj(Hsp40), subfamily A, mem. 1 | NM_008298 | 2 |
| dnaj(Hsp40), subfamily C, mem. 14 | NM_028873 | 2 |
| heat-shock protein hsp84 | M36829 | 2 |
| rab1b | NM_029576 | 2 |
| importin 13 | NM_146152 | 2 |
| kinesin superfamily protein 1c | AK046574 | 2 |
| Metabolic enzyme | ||
| S-adenosylhomocysteine hydrolase | BC015304 | 26 |
| lecithin cholesterol acyltransferase | BC028861 | 19 |
| glutathione peroxidase 1 | BC086649 | 8 |
| glutathione S-transferase | BC046758 | 7 |
| pancreatic lipase | NM_026925 | 5 |
| glycine decarboxylase | NM_138595 | 4 |
| amylase 2 | BC100579 | 4 |
| phosphatidylehtanolamine n-methyltrans. | NM_008819 | 3 |
| branched chain ketoacid dehydrog. E1 | NM_199195 | 3 |
| UDP glucuronosyltransferase 2, B36 | NM_001029867 | 3 |
| aldehyde dehydrogenase 16, A1 | NM_145954 | 3 |
| betaine-homocysteine methyltransferase | NM_016668 | 3 |
| hydroxysteroid (17-beta) dehydrogenase 9 | NM_013786 | 3 |
| catalase | BC047126 | 3 |
| hydroxyacyl-coenzyme A dehydrogenase | NM_016763 | 3 |
| glucosidase, alpha, acid | NM_008064 | 3 |
| pyruvate dehydrogenase lipoamide beta | NM 024221 | 2 |
| spermidine synthase | BC005566 | 2 |
| sterol o-acyltransferase 2 | NM 146064 | 2 |
| cystathionine beta-synthase | BC026595 | 2 |
| phosphoenolpyruvate carboxykinase 1 | NM_011044 | 2 |
| phosphogluconate dehydrogenase | BC011329 | 2 |
| arylsulfatase A | BC011284 | 2 |
| Plasma membrane protein | ||
| asialoglycoprotein receptor 1 | BC022106 | 13 |
| laminin receptor | BC081461 | 12 |
| EGFR transcript variant 2 | NM_007912 | 8 |
| t-cell Ig and mucin domain containing 2 | NM_134249 | 4 |
| fibulin 5 | NM_011812 | 3 |
| procollagen type XVIII alpha 1 | NM_009929 | 3 |
| fibronectin 1 | BC025521 | 2 |
| Interferon induced transmemb. protein 3 | NM_025378 | 2 |
| thrombospondin, type I domain cont. 6 | BC034843 | 2 |
| Nucleotide binding protein | ||
| RNA binding motif protein 35b | NM_176838 | 7 |
| RNA binding protein, transcript variant 1 | NM_019733 | 4 |
| Activating transcription factor 5 | NM_030693 | 3 |
| RNA polymerase II, polypeptide G | BC005580 | 3 |
| polymerase (DNA-directed), delta 4 | NM_027196 | 3 |
| guanine nucleotide binding protein | NM_008143 | 3 |
| b-cell leukemia/lymphoma 3 | NM_033601 | 2 |
| cAMP responsive element bp 3 like 3 | NM_145365 | 2 |
| Serum protein | ||
| histidine-rich glycoprotein | AY137504 | 20 |
| alpha2-hs-glycoprotein | NM_013465 | 15 |
| complement component factor I | NM_007686 | 6 |
| albumin | NM_009654 | 5 |
| complement comp. 1, q subcomp. Gamma | NM_007574 | 4 |
| serum amyloid p-component | NM_011318 | 3 |
| selenoprotein p, plasma, 1 | NM_009155 | 3 |
| apolipoprotein A-II | BC031786 | 3 |
| coagulation factor XII | BC037085 | 3 |
| prealbumin or transthyretin | X03351 | 2 |
| α2- Macroglobulin | M93264 | 2 |
| ceruloplasmin | BC062957 | 2 |
| acid labile subunit insulin-like growth factor | U66900 | 2 |
| Mitochondrial protein | ||
| atp synthase, F1 complex, delta | BC008273 | 6 |
| succinate dehydrogenase complex, sub D | NM_025848 | 6 |
| thiosulfate sulfurtransferase | NM_009437 | 5 |
| atp synthase, F1 complex, beta | BC013253 | 4 |
| cytochrome c oxidase, sub. VIIIa | NM_007750 | 3 |
| ferredoxin 1 | NM_007996 | 2 |
| metaxin 2 | NM_016804 | 2 |
| ATPase, H+ transporting, V0 subunit C | NM_009729 | 2 |
| solute carrier family 25 (mitoch. carrier) | NM_013770 | 2 |
| Other | ||
| DAZ associated protein 2 | NM_011873 | 12 |
| TNF receptor-associated factor 2 | BC003801 | 8 |
| selenoprotein X 1 | NM_013759 | 7 |
| presenilin stabililzation factor b human | AY113699 | 7 |
| serine or cysteine peptidase inhibitor 1a | NM_009243 | 5 |
| serine or cysteine peptidase inhibitor 1c | BC010984 | 5 |
| fk506 binding protein 8 | AY187231 | 4 |
| prosaposin | AK151034 | 4 |
| interferon alpha responsive gene | BC039927 | 3 |
| solute carrier family 27, member 5 | NM_009512 | 2 |
| eukaryotic translation elongation factor 2 | BC013263 | 2 |
| prion interacting protein 1 | AK144107 | 2 |
| GDP dissociation inhibitor 2 | NM_008112 | 2 |
| ubiquitin associated protein 2-like | NM_028475 | 2 |
| nasal embryonic LHRH factor | AF266508 | 2 |
The Genbank accession numbers of fourteen genes of unknown function recovered in the screen are listed here, along with the numbers of clones recovered for each in parentheses: BC051545 (5), NM_025791 (5), NM_026690 (4), AK146231 (4), AK002249 (3), NM_026542 (3), NM_025708 (3), NM_026938 (2), AK004787 (2), AC139943 (2), NM_198411 (2), XM_510195 (2), NM_172541 (2), and AK220316 (2).
Figure 2.
Functional classifications of the 108 AAV8-binding partners identified by yeast two-hybrid analysis.
AAV2 and AAV8 capsid proteins interact specifically with the preys
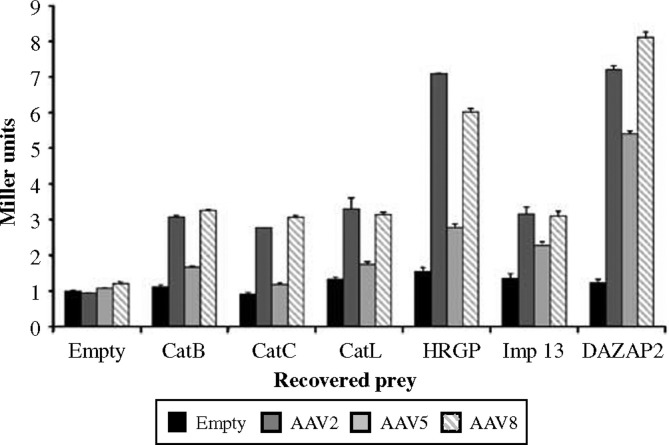
We first aimed to verify the specificity of the screen, and to determine the relative strength of the interactions. Therefore, we measured the activation of a Gal4p-dependent lacZ reporter gene in yeast transformed with the AAV8 bait and several clones of interest recovered from our screen. We included those encoding the cathepsin B and L proteases, as well as clones encoding histidine-rich glycoprotein (a serum adaptor protein thought to mediate attachment to cell surface heparan sulfate proteoglycans26), DAZ-associated protein 2 (a cytoplasmic factor thought to be involved in cell signaling27), and importin 13. The results were compared to bait plasmids containing the homologous regions of the AAV2 or AAV5 capsid (which are 83 and 61% similar to AAV8, respectively), or with an empty bait plasmid. As shown in Figure 3 , the activity of the recovered clones with the AAV8 bait was significantly higher (three- to seven-fold) as compared to the empty control, confirming that the interaction between these independent protein domains was indeed specific. Interestingly, the AAV2 bait was also able to interact with each of the candidate preys, a result that was not surprising in the light of the large sequence similarity between the AAV2 and AAV8 capsids. In contrast to both AAV2 and AAV8, reporter gene activation with the AAV5-based capsid bait was not significantly different from the empty plasmid control for most of the tested clones, including the cathepsin-encoding preys. From these examples, we conclude that the majority of the physical interactions identified in our screen were indeed specific.
Figure 3.
The recovered clones interact specifically with the AAV8 and AAV2 baits. Yeast transformed with the indicated combinations of bait and prey plasmids were tested for activation of the lacZ reporter gene using β-galactosidase assays (n=3).
Cathepsins B and L are involved in functional AAV2 and AAV8 transduction
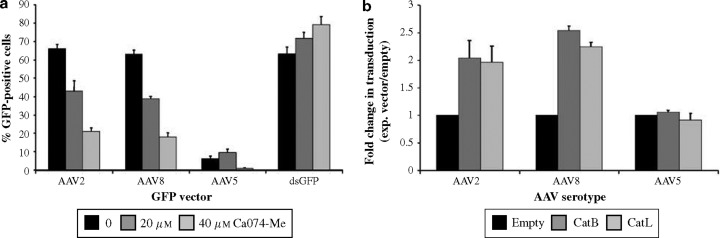
Having confirmed the interaction of AAV8 capsid proteins with cathepsin B and L in yeast, we sought to determine whether these two proteases played a role in AAV8-mediated transduction of mammalian cells. To this end, we incubated murine fibroblast NIH-3T3 cells with Ca074-Me, a cell-permeable cathepsin inhibitor that preferentially blocks cathepsin B activity, and also inhibits cathepsin L at high doses. Four hours later, gfp-encoding AAV2 or AAV8 vectors were added to the cells. The number of green fluorescent protein GFP-expressing cells was determined 1 day later and compared to samples infected in the absence of inhibitor. Interestingly, both AAV2- and AAV8-mediated transduction was significantly reduced up to three times in a dose-dependent manner by Ca074-Me treatment (Figure 4a ), suggesting that cathepsin B and L activity is important for transduction with these two serotypes. This result was further validated using a series of control experiments. Firstly, when the assay was conducted with AAV5(gfp), we only observed a decrease in vector transduction at a high dose of inhibitor (40 μM). This apparent inhibitory effect was most likely non-specific as the use of a lower inhibitor dose (20 μM) actually resulted in a slight increase in AAV5(gfp) transduction. Secondly, when we transfected cells with a plasmid expressing a destabilized GFP protein (with a half-life of 2 h) before Ca074-Me treatment, we likewise observed a marginal increase in the number of GFP-positive cells, ruling out inhibitory effects on gene expression from the drug per se. Thirdly, when we tested the non-cell-permeable version of the drug, Ca074, as well as the cathepsin L inhibitor Z-Phe-Tyr(t-Bu)-diazomethyl ketone, we found no significant change in transduction efficiency by the AAV2 and AAV8 vectors (data not shown). To further study the role of cathepsins B and L in AAV2- and AAV8-mediated transduction, NIH-3T3 cells were transfected with cathepsin B or L expression plasmids before being infected with the same AAV(gfp) vectors used above. Western blot analysis confirmed that cells transfected with the expression vectors had increased levels of cathepsin B or L as compared to control cells transfected with an empty plasmid (data not shown). Transduction by AAV2 and AAV8 vectors was increased at least twofold in cells overexpressing cathepsin B or L, whereas no significant change was seen with the AAV5 vector (Figure 4b), confirming that cathepsin B and L activity is important for AAV2 and AAV8 infection.
Figure 4.
Cathepsins B and L are involved in AAV2- and AAV8-mediated transduction of mammalian cells. (a) NIH-3T3 cells were transiently transfected with an eGFP-expressing plasmid, pd2EGFP-N1, or transduced with AAV2, AAV5, or AAV8 GFP-expressing vectors, in the presence of Ca074-Me (a cathepsin B and L inhibitor), and then analyzed by fluorescent-activated cell sorting to determine the number of GFP-positive cells (n=3). (b) NIH-3T3 cells were either transiently transfected with a cathepsin B, cathepsin L, or an empty expression vector, before being incubated with AAV2, AAV5, or AAV8 GFP-expressing vectors. The number of GFP-positive cells was determined by FACS analysis (n=3).
In vitro cleavage of AAV particles by cathepsins B and L
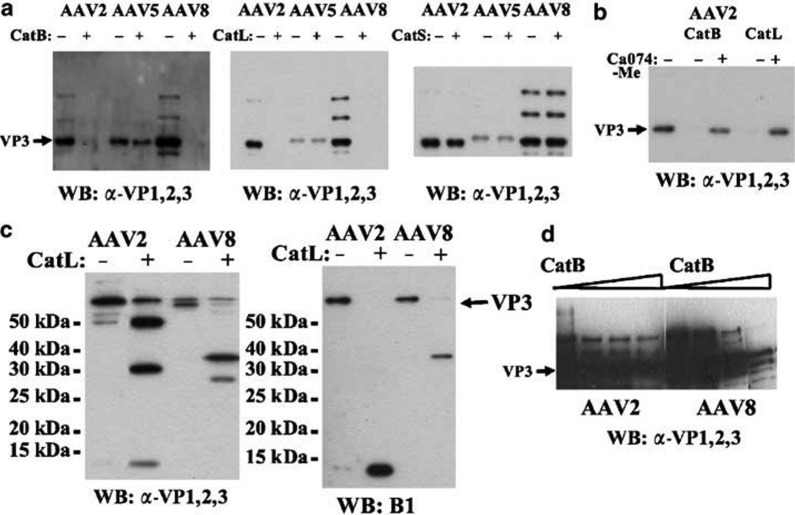
Having established a functional role of cathepsins B and L in cellular transduction by AAV2 and AAV8, we next measured the ability of purified cathepsins to bind and cleave the assembled AAV particles. To this end, we treated 1 × 109 recombinant particles of AAV serotypes 2, 5 or 8 with purified cathepsin B, cathepsin L, or a control cysteine protease not recovered in our screen, cathepsin S. Viral proteins were then analyzed by Western blotting, using a polyclonal antibody that recognizes the capsid proteins of all three serotypes. As expected, we found a strong reduction in the levels of AAV2 and AAV8 full-length VP3 protein, after incubation with cathepsin B or L, a result consistent with the predicted cleavage of the viral capsid proteins by these two proteases (Figure 5a ). Importantly, the specificity of this cleavage was confirmed with the cathepsin S negative control, where VP3 levels remained stable under otherwise identical experimental conditions. Moreover, addition of the Ca074-Me cathepsin inhibitor to the reaction abolished AAV capsid cleavage by cathepsin B or L, confirming that their activity was responsible for the observed digestion (Figure 5b). Finally, none of the three tested cathepsins were able to cleave the AAV5 VP3 protein. This result is consistent with our two-hybrid data, which indicated that the AAV2- and AAV8-based capsid baits, but not the AAV5-based counterpart, could physically interact with the cathepsin preys.
Figure 5.
Purified cathepsins B and L cleave intact AAV2 and AAV8 capsids, but not AAV5. (a) AAV2, AAV5, and AAV8 capsids were treated with either cathepsin B (left panel), L (middle panel), or S (right panel), before being resolved on a polyacrylamide gel and visualized by Western blot analysis with the α-VP1, 2, and 3 polyclonal antibody (this antibody was raised against AAV2 capsid proteins, and as AAV5 has little homology to AAV2, AAV5 capsid proteins were recognized inefficiently) (n=4). (b) The cathepsin inhibitor Ca074-Me was added to the AAV2 capsids during treatment with the purified cathepins B and L. The levels of VP3 were determined by Western blot analysis with α-VP1, 2, 3 after resolving the proteins on a polyacrylamide gel (n=2). (c) The molecular mass of the fragments generated by cathepsin L digestion of AAV2 and AAV8 capsids was determined by Western blot analysis using the α-VP1, 2, and 3 (left panel) or B1 (right panel) antibodies, after resolving the proteins on a high-percentage (12.5%) polyacrylamide gel. The monoclonal B1 antibody specifically recognizes a conserved epitope in the VP3 C-terminus (n=3). (d) AAV2 and AAV8 capsids were incubated with increasing amounts of cathepsin B (from left to right, no enzyme, 0.3, 3, and 20 μM). The levels of VP3 were then determined by Western blot analysis with α-VP1, 2, and 3 after resolving the proteins on a polyacrylamide gel (n=2).
Cathepsin cleavage patterns are AAV serotype specific
Although cathepsins B and L generated similar cleavage products within a given AAV serotype (2 or 8), we observed that the exact peptide pattern differed between the two viral isolates. To analyze this phenomenon in more detail, we resolved the digested capsid proteins on a high-percentage (12.5%) polyacrylamide gel. This was followed by immunoblotting with two distinct antibodies, allowing us to differentiate between C-terminal peptide fragments and the more upstream cleavage products. We found that the AAV8 capsid was digested into two fragments of approximately 35 and 27 kDa in size, with the upper band being the C-terminal fragment, as evidenced by its recognition by the B1 antibody (which binds an epitope found in the VP3 C-terminus) (Figure 5c). In contrast, AAV2 particle digestion resulted in three predominant bands with sizes of approximately 50, 30, and 13 kDa, as detected by the polyclonal α-VP1,2,3 serum. Additionally, we noted a fourth band of around 20 kDa upon overexposure of the blot (as it was fainter than the other bands, we hypothesize that this polypeptide contained fewer epitopes bound by the polyclonal antibody). Interestingly, upon overexposure of the blots, the undigested AAV2 and AAV8 samples also displayed trace amounts of the same cleavage products. One possible explanation for this observation is that the purified AAV vectors contain a small sub-population of viral capsids that had been processed by cellular cathepsins before purification. As the sum of the AAV2 cleavage product bands exceeded the size of the full-length AAV2 VP3 protein (63 kDa), we reasoned that one or more of them probably represented a partial cleavage product. Indeed, subsequent immunoblotting with the B1 antibody revealed that only the smallest 13 kDa peptide contained the C-terminal B1 epitope, suggesting that together with the N-terminal 50 kDa product it formed the full-length VP3 protein. We, moreover, hypothesized that the two intermediate bands of 30 and 20 kDa detected with the polyclonal antibody resulted from secondary cleavage of the large 50 kDa fragment, especially as none of these three bands was recognized by the B1 antibody. Accordingly, the presence of a substantial amount of the 50 kDa precursor peptide would indicate that this particular reaction occurs inefficiently, or that this cleavage site is only accessible on a subset of all VP3 subunits within the assembled AAV2 capsid. In contrast to this apparent two-step cleavage process for AAV2, our finding of only two bands for AAV8, whose sizes add up to full-length VP3, suggests that this particular serotype is cleaved only once and more efficiently than AAV2. This idea was indeed confirmed when the cleavage rates of the two vectors were compared, by partially digesting them with increasing amounts of cathepsin B. Intriguingly, AAV8 VP3 levels were reduced more readily than those of AAV2 when both were incubated with the same amount of enzyme, further indicating that AAV8 capsid cleavage may be more efficient than that of AAV2 (Figure 5d). As will be discussed below, the differential cleavage rates and patterns could explain some of the differences in the transduction rate and kinetics seen with AAV2 and AAV8.
AAV particles remain intact upon cathepsin cleavage
Previous studies indicated that despite AAV capsid processing in the endosome, particles escape from this compartment as assembled virions. We thus sought to investigate how cathepsin cleavage affected the stability of the AAV capsid, by assessing the ability of different monoclonal antibodies to bind either digested or undigested virions. To this end, AAV particles of serotypes 2 or 8 were incubated with cathepsin B as indicated above, and then transferred onto a nitrocellulose membrane under non-denaturing conditions. As a control, we included undigested viral samples that were incubated at 75°C, based on previous reports that this treatment triggers dissociation of the AAV2 capsid.28 The particles were then detected by immunoblotting with either the B1 or A20 antibodies. The former binds an epitope in the C-terminus of VP3 that is inaccessible in assembled capsids, but becomes available for binding once the capsid proteins dissociate. In contrast, the A20 antibody solely binds to intact AAV2 capsids, as its three-dimensional epitope is disrupted once the subunit proteins disassemble. Interestingly, we did not observe any differences between the digested and undigested samples with the B1 antibody, as a very minimal amount of protein was recognized in both cases (Figure 6 ). Consistent with this result, we obtained a strong signal for both AAV2 samples with the A20 antibody, whose specificity for intact particles was confirmed by its lack of reaction with the heat-denatured particles. There was only a slight decrease in the A20 signal intensity for the digested sample, perhaps owing to a change in the virion confirmation upon cathepsin cleavage of the individual capsid proteins. Moreover, we observed that upon addition to cultured cells, digested AAV2 and AAV8(gfp) vectors displayed similar or even slightly higher transduction efficiencies as compared to undigested particles (data not shown). Collectively, these data strongly suggest that AAV particles remain intact after cathepsin cleavage. This supports the idea that this process does not lead to capsid dissociation, but is rather a prerequisite for efficient downstream processing of the virions.
Figure 6.
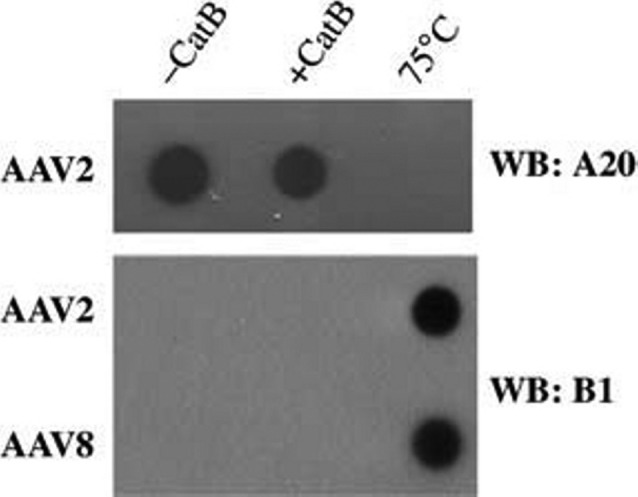
AAV particles do not disassemble upon cathepsin cleavage. AAV particles incubated with or without cathepsin B were transferred to a nitrocellulose membrane under non-reducing conditions and detected by immunoblotting with the B1 or A20 (only recognizes intact AAV2 particles) monoclonal antibodies. Capsids denatured at 75°C for 30 min were also included as a control (n=2).
Discussion
This study was performed to better understand the transduction mechanism of AAV8, an AAV serotype currently in preclinical development for use in human gene therapy. A key component of AAV transduction is the ability of the AAV capsid to interact with specific host proteins that enable the proper uptake, trafficking, and uncoating of the virus. To date, only a small number of these host proteins were identified, and only for individual serotypes (excluding AAV8), leaving us with a limited understanding of the mechanisms behind AAV transduction. The yeast two-hybrid screen described in this report identified a number of proteins capable of directly interacting with portions of the AAV8 capsid, which should help to shed light on the complex processes leading to AAV gene expression.
Several previous studies indicated that endosomal processing is necessary for transduction by the AAV family prototype, AAV2, whose capsid protein sequence is highly homologous to AAV8. Interestingly, our screen recovered two endosomal proteases, cathepsins B and L, which either alone or in tandem act as uncoating factors for a number of other viruses, such as reovirus, SARS coronavirus, hepatitis B, Ebola, Nipah, and Hendra.24, 25, 29, 30, 31, 32 In this report, we show that these cathepsins appear to play a similar role in AAV8 and AAV2, by demonstrating that both proteases can bind to and cleave their capsids, and that their activity is important for efficient cellular transduction by both serotypes. Our results also indicate a possible redundancy in function by these two proteases, as both cleaved the capsid proteins of a given serotype similarly. Moreover, cellular transduction was reduced by a cathepsin inhibitor that is able to inhibit both cathepsins B and L efficiently. Furthermore, our assays showed that AAV5-mediated cellular transduction is not only independent of these proteins, but the AAV5 capsid is neither bound nor cleaved by the cathepsins. A higher dose of the Ca074-Me inhibitor diminished transduction by AAV5, but its inhibition profile was clearly distinct from that of AAV2 and AAV8. Ca074-Me is a cathepsin B inhibitor at low doses and was reported to inhibit cathepsin L upon dosage escalation.25 This suggests that another cellular protease important for AAV5 transduction was also inhibited at the tested higher dose. Clearly, AAV5's mechanism of transduction differs from that of AAV2 and AAV8, which are more closely related to each other and both seem to depend on cathepsins B and L for efficient gene expression.
The demonstration of the functional significance of the cathepsins for AAV8 transduction has validated the usefulness of our screen. This is further substantiated by the fact that in another study,33 we show that an additional frequently recovered clone, the 37/67 kDa laminin receptor, is also involved in AAV8 infection. We demonstrate that this receptor is important for cellular binding and transduction by AAV8 in vitro and in vivo, providing the first report of a cellular receptor for AAV8. To our knowledge, no previous studies have identified proteins capable of directly binding to the AAV8 capsid. As summarized in Figure 7 , our two-hybrid screen has helped uncover proteins involved in both extra- and intracellular steps of AAV8 infection, i.e., receptor binding and endosomal processing.
Figure 7.
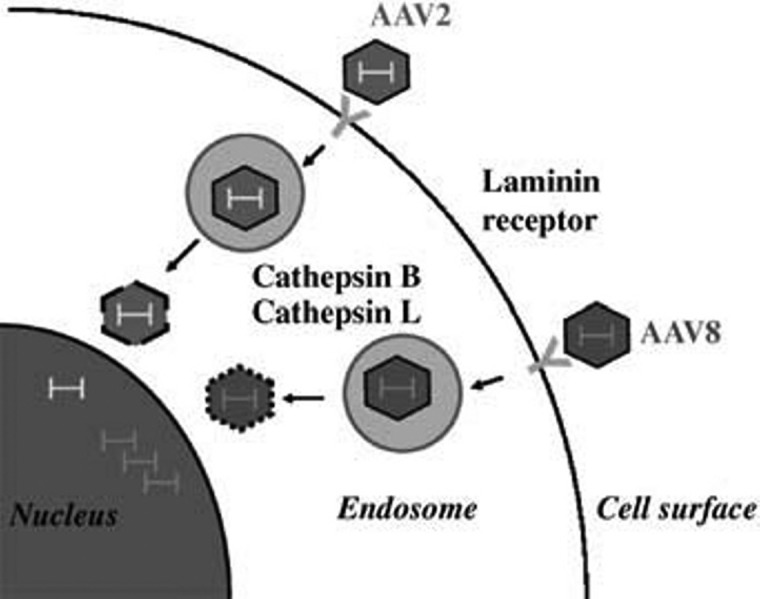
A model of cellular transduction by AAV2 and AAV8. Shown are the proteins recovered in our screen for which we subsequently confirmed the ability to bind to AAV2 and AAV8 particles. The laminin receptor acts as a cellular receptor for both serotypes, allowing them to bind to and subsequently enter the cell. Once the particle enters the endosome, cathepsins B and L bind to and cleave both capsids, but the AAV8 and AAV2 capsids appear to be cleaved differently. Following particle processing, they are released from the endosome and traffic to the nucleus, where AAV8 has previously been shown to uncoat more readily.23
Despite the many similarities between AAV2 and AAV8 reported in this and other studies, previous work showed that the in vivo transduction rate of AAV8 is much more robust than that of AAV2, possibly owing to a faster uncoating rate. AAV8 reaches maximal expression levels more rapidly than AAV2 and can yield up to 20-fold higher transduction levels in the liver.23 Interestingly, our study demonstrated that AAV2 and AAV8 capsids are processed differently by certain endosomal proteases. The AAV8 capsid protein appears to be cleaved efficiently at a single site close to the center of the VP3 protein, whereas the AAV2 VP3 has two distinct cleavage sites. The C-terminal site is cleaved more efficiently in our assay than the more N-terminal site, resulting in a partially digested capsid fragment. Even when 10-fold less AAV2 virions were incubated with the same amount of enzyme, the 50 kDa precursor fragment was still readily visible upon Western blot analyses (data not shown). In addition, the AAV8 capsid appears to be cleaved by the cathepsins at a quicker rate than AAV2. This indicates that distinct cathepsin cleavage of AAV2 and AAV8 may account for some of the differences in their uncoating and transduction efficiencies.
Previous studies showed that AAV2 can traffic through different subcompartments within the endosome, in a dose- and cell line-dependent manner.15, 16 As cathepsins B and L were reported to localize mostly in the late endosome, and to a lesser degree in the early endosome,34, 35 the manner and efficiency of AAV particle processing may depend on which compartment it shuttles to. It is possible that only a subset of the internalized particles gets cleaved by cathepsins B and L, whereas the remainder are processed by a different set of proteins, or not at all. In a previous study, it was reported that although AAV2 particles traffic through late endosomes in both HEK-293 and NIH-3T3 cells, they do so to a much lesser degree in the latter (partly explaining why they are less permissive for AAV2 infection).16 Perhaps in more permissive cell lines where the AAV particles traffic more frequently to the late endosome, the capsids are cleaved even more efficiently by the cathepsins because of their increased colocalization in the endosomal compartment. In addition, Ding et al. 15 reported that particles shuttled to the recycling endosome are more competent for transgene expression on a per genome basis than those in the late endosome. The authors speculated that this is due to more efficient processing in the recycling endosome. As cathepsins B and L have not been reported to be present in this compartment so far, AAV particles may be processed by other proteases or ubiquitin-associated proteins present in this subcompartment. Other proteases recovered in our screen are potential candidates for this activity (Table 1), perhaps acting synergistically with the cathepsins, or in an independent and compartment-specific fashion.
Our two-hybrid screen implicated a number of proteins from many different classes as factors able to interact with the AAV8 capsid. As our screen was conducted in a yeast system, it is unlikely that all of the recovered genes would bind the intact virus in a mammalian cellular setting. However, using a number of independent assays, we showed that this screen accurately predicted the ability of the AAV8 capsid to bind some of these proteins. This convinces us that further analysis of the other recovered clones is clearly warranted. Although this study focused only on proteins involved in AAV transduction, it is easy to envision how various other proteins could interact with the AAV capsid and thereby influence many additional processes relevant to AAV biology, such as the regulation of capsid synthesis, viral replication, and/or viral transmission. In addition, as other members of the parvovirus family share many similarities to AAV, some of the proteins identified in our screen may also be important for their normal function. Further analysis of these proteins might ultimately provide valuable insights into the treatment of viruses such as the pathogenic B19 virus, whose infection can lead to fetal loss in pregnant women, and can cause severe anemia in immuno-compromised individuals.36
In summary, we have furthered our understanding of AAV8 transduction by identifying a number of candidate host cell proteins that can interact with its capsid. We have revealed the importance of several of these proteins in AAV8- and AAV2-mediated transduction. Further study into the role of these and the other recovered clones in the AAV8 life cycle may have important implications for the use of this powerful gene transfer vector.
Materials and Methods
Plasmids. The AAV2, AAV5, and AAV8 bait plasmids were constructed by first PCR amplifying homologous regions (nucleic acid residues 439–1,860 (AAV2), 436–1,827 (AAV5), 442–1,869 (AAV8) of each AAV cap gene) from the AAV helper plasmids pDG,37 pDP5,38 and pE5/V18,5 respectively, using the following primers: AAV2 (CGGAATTCGAGCACTCTCCTGTGGAGCCA and CGGGATCCGCTTTGCCCAGATGGGCCCCTG); AAV5 (CGGAATTCGACCACTTTCCAAAAAGAAAG and GAAGATCTACTTGGCCCAGATGGGTCCTTG); and AAV8 (CGGAATTCCCATCACCCCAGCGTTCTCCA and GAAGATCTGCTTGGCCCAGATGGGACCCTG). The PCR products were digested with BamHI and EcoRI for AAV2, or BglII and EcoRI for AAV5 and AAV8, before ligation into the BamHI–/EcoRI-linearized plasmid pGBK-T7 (BD Clontech, Palo Alto, CA). In the resulting constructs, the cap genes were fused in-frame with a Gal4 DNA-binding domain. The mouse cathepsin B and L cDNAs were amplified from a mouse liver cDNA library (BD Clontech) using the following primers: cathepsin B (CGGAATTCACCATGTGGTGGTCCTTGATCCT and CCGCTCGAGTTAGAATCTTCCCCAGTACT), and cathepsin L (ACGCCAATTGACCATGAATCTTTTACTCTTTT and CCGCTCGAGTCAATTCACGACAGGATAGC). The cathepsin B and L PCR products were digested with EcoRI and XhoI, or MfeI and XhoI, respectively, and then cloned into the pcDNA3 (Invitrogen, Carlsbad, CA) vector digested with EcoRI and XhoI.
Yeast two-hybrid screen. The initial two-hybrid screen was conducted using the AAV8 bait plasmid, the mouse liver Matchmaker cDNA library (BD Clontech), and the yeast strain AH109 (MATa, trp1-901, leu2-3, leu2-112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1 UAS -GAL1 TATA -HIS3, GAL2 UAS -GAL2 TATA -ADE2, URA3::MEL1 UAS -MEL1 TATA -lacZ) (BD Clontech). Clones were selected for growth on SD media (0.67% yeast nitrogen base without amino acids, 2% glucose, and supplemented with the appropriate amino acids at a concentration of 0.004%), lacking adenine, histidine, leucine, and tryptophan. To isolate the plasmid DNA from yeast, cells were resuspended in 200 μl lysis buffer (2% Triton X-100, 1% SDS, 100 mM NaCl, 10 mM Tris (pH 8), 1 mM EDTA (pH 8)), before the addition of glass beads and 200 μl phenol. After vortexing for 2 min, the cells were centrifuged for 5 min at 15,000 r.p.m. The DNA was ethanol precipitated from the aqueous phase, resuspended in water, and transformed into Escherichia coli. Library plasmid DNA was isolated, sequenced, and then analyzed using the National Center for Biotechnology Information BLAST program.
β-Galactosidase assays. Activity of the lacZ reporter gene was determined using β-galactosidase assays with chlorophenolred-D-galactopyranoside as a substrate according to BD Clontech. Briefly, the cells were grown overnight in selective SD media lacking leucine and tryptophan, and then diluted in YPD media (1% bacto-yeast extract, 2% bacto-tryptone, 2% glucose). After 4 h, the OD600 of the yeast culture was measured (later used to calculate the Miller units, see below) before resuspending the cells in buffer 1 (2.38% HEPES, 0.9% NaCl, 0.065% L-Aspartate, 1% BSA, and 0.05% Tween 20), and subsequently lysing them via three freeze–thaw cycles in liquid nitrogen. The CPRG substrate was dissolved to a concentration of 0.1335% in buffer 1 and added to the lysed cells. The reaction was stopped using 3 mM zinc chloride, and the OD578 of the sample was taken to calculate the Miller units=(1,000 × OD578)/(time (min) × OD600).
AAV transduction in mammalian cells. 3.75 × 104 NIH-3T3 cells per well were grown overnight in 24-well plates in Dulbecco's modified Eagle's medium with 10% fetal calf serum, 2 mM L-glutamine, and 1% antibiotic/antimycotic. Cells were transfected with the cathepsin B or L expression vectors, or pd2EGFP-N1 (BD Clontech) using Polyfect (Qiagen, Valencia, CA) according to the manufacturer's instructions. The cells were incubated for 4 h with the cathepsin inhibitor Ca074-Me (Calbiochem, San Diego, CA) before addition of the AAV2, AAV5 or AAV8(gfp) vectors. The AAV(gfp) vector construct, consisting of a double-stranded genome expressing the gfp gene from an Rous sarcoma virus promoter, will be described in detail elsewhere (Grimm et al., manuscript in preparation). Briefly, it was engineered by assembling a full-length AAV2 ITR together with a subfragment of an AAV4 inverted terminal repeat , in a pBlueScriptII (Stratagene, La Jolla, CA) backbone. The AAV4 fragment contained the entire ITR sequence except for the terminal resolution site, to prevent DNA nicking during vector DNA replication. The dsAAV(gfp) vector plasmid was pseudotyped with the three different capsids using a standard triple plasmid transfection approach.39 The virus was purified using cesium chloride density gradient centrifugation and quantitated by dot blot assay. The resulting virus particles were then added to the cells at the following ratio (particles/cell): 5 × 106 for AAV5 or AAV8(gfp), or 1 × 105 for AAV2(gfp). The cells were trypsinized 24 h later and analyzed by fluorescent-activated cell sorting.
Treatment of AAV with purified cathepsin. An 8.5 μg portion of purified bovine spleen cathepsin B, 7.8 μg of purified bovine kidney cathepsin L in reaction buffer (50 mM sodium acetate pH 5.0, 3 mM dithiothreitol), or 6 μg of human recombinant cathepsin S in reaction buffer (50 mM sodium acetate pH 7.4, 3 mM DTT) (all from Calbiochem, San Diego, CA, USA) was incubated with AAV2, AAV5, or AAV8 particles for 8 h at 37°C in a final volume of 30 μl. Ca074-Me 10 nmol was added to individual reactions to inhibit cathepsin B or L activity. Proteins were denatured by boiling for 5 min after the addition of Laemmli buffer. The samples were resolved on a 7.5 or 12.5% polyacrylamide gel and analyzed by immunoblotting with α-VP1,2 and ,3 antibody or the monoclonal B1 antibody (both from American Research Products, Belmont, MA). For experiments shown in Figure 5d, 2 × 1010 recombinant particles of AAV2 or AAV8 were incubated with increasing amounts of cathepsin B in a 20 μl reaction volume for 16 h at 37°C.
Determination of AAV capsid stability after cathepsin digestion. 1 × 109 particles of AAV2 or AAV8 in PBS were incubated at 75°C for 30 min, or digested with cathepsin B as indicated above, and then vacuum-transferred onto a nitrocellulose membrane under non-denaturing conditions. The AAV proteins were then detected by immunoblotting with the B1 or A20 monoclonal antibodies (both from American Research Products).
Acknowledgments
BA was supported by a fellowship from the Canadian Institute of Health Research. This work was supported by NIH Grant HL66948 (MAK).
References
- 1.Fisher RE, Mayor HD. The evolution of defective and autonomous parvoviruses. J Theor Biol. 1991;149:429–439. doi: 10.1016/s0022-5193(05)80091-8. [DOI] [PubMed] [Google Scholar]
- 2.Muzyczka N. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr Top Microbiol Immunol. 1992;158:97–129. doi: 10.1007/978-3-642-75608-5_5. [DOI] [PubMed] [Google Scholar]
- 3.Gao G. Clades of adeno-associated viruses are widely disseminated in human tissues. J Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimm D, Kay MA. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr Gene Ther. 2003;3:281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- 5.Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- 8.Kashiwakura Y. Hepatocyte growth factor receptor is a coreceptor for adeno-associated virus type 2 infection. J Virol. 2005;79:609–614. doi: 10.1128/JVI.79.1.609-614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qing K, Mah C, Hansen J, Zhou S, Dwarki V, Srivastava A. Human fibroblast growth factor receptor 1 is a co-receptor for infection by adeno-associated virus 2. Nat Med. 1999;5:71–77. doi: 10.1038/4758. [DOI] [PubMed] [Google Scholar]
- 10.Summerford C, Bartlett JS, Samulski RJ. Vβ5 integrin: a co-receptor for adeno-associated virus type 2 infection. Nat Med. 1999;5:78–82. doi: 10.1038/4768. [DOI] [PubMed] [Google Scholar]
- 11.Summerford C, Samulski RJ. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J Virol. 1998;72:1438–1445. doi: 10.1128/jvi.72.2.1438-1445.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Pasquale G. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med. 2003;9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 13.Kaludov N, Brown KE, Walters RW, Zabner J, Chiorini JA. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J Virol. 2001;75:6884–6893. doi: 10.1128/JVI.75.15.6884-6893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanlioglu S, Benson PK, Yang J, Atkinson EM, Reynolds T, Engelhardt JF. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J Virol. 2000;74:9184–9196. doi: 10.1128/jvi.74.19.9184-9196.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding W, Zhang LN, Yeaman C, Engelhardt JF. rAAV2 traffics through both the late and the recycling endosomes in a dose-dependent fashion. Mol Ther. 2006;13:671–682. doi: 10.1016/j.ymthe.2005.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen J, Qing K, Srivastava A. Adeno-associated virus type 2-mediated gene transfer: altered endocytic processing enhances transduction efficiency in murine fibroblasts. J Virol. 2001;75:4080–4090. doi: 10.1128/JVI.75.9.4080-4090.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bantel-Schaal U, Hub B, Kartenbeck J. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J Virol. 2002;76:2340–2349. doi: 10.1128/jvi.76.5.2340-2349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding W, Zhang L, Yan Z, Engelhardt JF. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 2005;12:873–880. doi: 10.1038/sj.gt.3302527. [DOI] [PubMed] [Google Scholar]
- 19.Seisenberger G, Ried MU, Endress T, Buning H, Hallek M, Brauchle C. Real-time single-molecule imaging of the infection pathway of an adeno-associated virus. Science. 2001;294:1929–1932. doi: 10.1126/science.1064103. [DOI] [PubMed] [Google Scholar]
- 20.Duan D, Yue Y, Yan Z, Yang J, Engelhardt JF. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J Clin Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Z, Zak R, Luxton GW, Ritchie TC, Bantel-Schaal U, Engelhardt JF. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Z. Distinct classes of proteasome-modulating agents cooperatively augment recombinant adeno-associated virus type 2 and type 5-mediated transduction from the apical surfaces of human airway epithelia. J Virol. 2004;78:2863–2874. doi: 10.1128/JVI.78.6.2863-2874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas CE, Storm TA, Huang Z, Kay MA. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert DH, Deussing J, Peters C, Dermody TS. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J Biol Chem. 2002;277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- 26.Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein: a novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol Cell Biol. 2005;83:106–118. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 27.Shi Y, Luo S, Peng J, Huang C, Tan D, Hu W. The structure, expression and function prediction of DAZAP2, a down-regulated gene in multiple myeloma. Genomics Proteomics Bioinformatics. 2004;2:47–54. doi: 10.1016/S1672-0229(04)02007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bleker S, Sonntag F, Kleinschmidt JA. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J Virol. 2005;79:2528–2540. doi: 10.1128/JVI.79.4.2528-2540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cooper A, Shaul Y. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsid-like core particles. J Biol Chem. 2006;281:16563–16569. doi: 10.1074/jbc.M601418200. [DOI] [PubMed] [Google Scholar]
- 30.Huang IC. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pager CT, Jr, Craft WW, Patch J, Dutch RE. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pager CT, Dutch RE. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J Virol. 2005;79:12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akache B, Grimm D, Pandey K, Yant SR, Xu H, Kay MA. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J Virol. 2006;80:9831–9836. doi: 10.1128/JVI.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing R, Mason RW. Design of a transferrin-proteinase inhibitor conjugate to probe for active cysteine proteinases in endosomes. Biochem J. 1998;336(Part 3):667–673. doi: 10.1042/bj3360667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corcoran A, Doyle S. Advances in the biology, diagnosis and host-pathogen interactions of parvovirus B19. J Med Microbiol. 2004;53:459–475. doi: 10.1099/jmm.0.05485-0. [DOI] [PubMed] [Google Scholar]
- 37.Grimm D, Kern A, Rittner K, Kleinschmidt JA. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 38.Grimm D, Kay MA, Kleinschmidt JA. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 39.Grimm D. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods. 2002;28:146–157. doi: 10.1016/s1046-2023(02)00219-0. [DOI] [PubMed] [Google Scholar]