Abstract
Bats are notorious reservoirs of genetically-diverse and high-profile pathogens, and are playing crucial roles in the emergence and re-emergence of viruses, both in human and in animals. In this report, we identified and characterized previously unknown and diverse genetic clusters of bat coronaviruses in the Atlantic Forest Biome, Brazil. These results highlight the virus richness of bats and their possible roles in the public health.
Highlights
-
•
We describe the circulation of CoV lineages, 13 α-CoV and two β-CoV in AFB bats.
-
•
Coronaviruses were detected in 15 bat samples from eight bat species (50% positivity).
-
•
A cluster of α-CoVs were detected in Sturnira bat that grouped with α-CoVs lineage-1.
-
•
We identified two distinct β-CoV that clustered within MERS-CoV containing lineage C.
-
•
Results highlight the virus richness of bats and their possible roles in the public health.
1. Introduction
Brazil harbours 15% of the world's bat diversity and carries 178 distinct bats species (Nogueira et al., 2014). Out of these, a total of 113 species exist only in the Atlantic Forest Biome (AFB), which is the second largest rain forest of the South America, and is one of the unique regions with highest biodiversity in the world (Paglia et al., 2012).
Bats are historically unique and widespread mammals playing essential roles in the emergence and re-emergence of viruses of both veterinary and public health importance. Although viruses of diverse genetic backgrounds can co-exist asymptomatically in bats, majority of these viruses are single stranded RNA viruses (Calisher et al., 2006). Coronaviruses (CoVs) are enveloped, positive-sense, single-stranded RNA viruses in the family Coronaviridae, and are usually associated with respiratory, enteric, hepatic and neurological pathologies of varying severity (Woo et al., 2012). Coronaviruses are classified into four genera; Alphacoronavirus (α-CoV) and Betacoronavirus (β-CoV) have been exclusively identified in mammals, whereas Gammacoronavirus and Deltacoronavirus are mainly detected in avian species (Woo et al., 2012, ICTV, 2015). Based on the genetic relatedness, the β-CoVs can further be subdivided into four clades: A to D (Drexler et al., 2014).
All CoVs that can potentially infect human were originated from animal reservoirs, and four of such CoVs are believe to be transmitted to human through bats including α-CoVs (229E and NL63) and highly pathogenic β-CoV (Severe Acute Respiratory Syndrome and Middle East Respiratory Syndrome) (Bolles et al., 2011, Chan et al., 2015, Corman et al., 2013, Huynh et al., 2012). Recently, a number of novel bats CoVs have been identified, primarily from African, Asian and European bats (Calisher et al., 2006, Chu et al., 2006, Drexler et al., 2014), as well as from South American countries including Costa Rica, Panama, Ecuador, Mexico and Brazil (Corman et al., 2013, Goes et al., 2013). Collectively, these studies indicate the co-existence of bats and viruses at the interface of viral evolution and bats ecology.
A limited number of studies have been conducted in Brazil to map the nature and breath of bats in harbouring viral populations. In previous studies, a total of five distinct CoV lineages have been detected in just 10% of local bats (15 species), and most of these are belonging to α-CoV (Brandao et al., 2008, Corman et al., 2013, Goes et al., 2013). These attributes, and the existence of a large number of human beings (120 million) in the Atlantic Forest Biome, Brazil, clearly highlight the potential of bats in not only carrying zoonotic viruses but also in possible transmission of viruses to human beings.
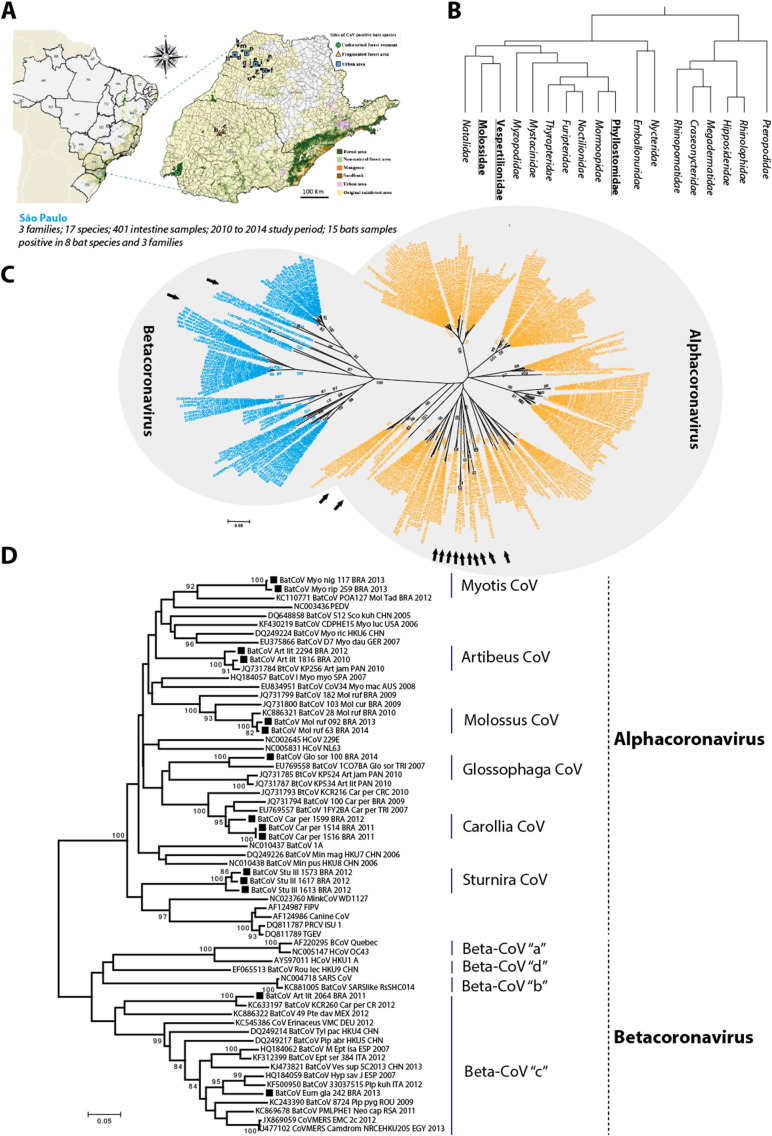
To ascertain the diversity of bats CoVs circulating in the Brazilian bats, a total of 401 intestine tissues from 17 bat species were collected from 2010 to 2014 encompassing bats with distinct diet habits (Table 1 , Fig. 1A, B). Bats from urban (N = 192) and rural (N = 10) areas were received from 14 municipalities of the Northwestern State of São Paulo by the Rabies Laboratoryof Universidade Estadual Paulista (UNESP), Araçatuba-SP. Additionally, 99 and 100 samples were collected from Iguaçu National Park, and from two distinct disturbed landscape sites, respectively, and provided by our collaborators of the Zoology Department at UNESP, Rio Claro-SP. All activities were authorized and approved by the Ethic Committee of Institute of Biomedical Research from University of São Paulo (56-18-03/2014) and bats species were identified based on the morphological characteristics including head-boby, forearm size, and dental arch as described by Vizotto and Taddei, 1973, Gregorin and Taddei, 2002 and Miranda et al. (2011).
Table 1.
Bat species tested for CoV RNA in different ecological regions of Brazil.
| Species | No of samples tested/no positive (%) | Sex - no of samples tested/no positive (%) |
Location - no of samples tested/no CoV positive (%) |
Bats family | Bats diet types | |||
|---|---|---|---|---|---|---|---|---|
| Male | Female | Undist. Forest | Fragm. Forest | Urban area | ||||
| Artibeus fimbriatus | 3/0 | 3/0 | 0/0 | 3/0 | 0/0 | 0/0 | Phyllostomidae | Frugivorous |
| Artibeus lituratus | 126/3 (2.4%) | 79/0 | 44/3 (6.8%) | 60/2 (3.3%) | 60/1 (1.6%) | 6/0 | Phyllostomidae | Frugivorous |
| Artibeus obscurus | 1/0 | 1/0 | 0/0 | 0/0 | 0/0 | 1/0 | Phyllostomidae | Frugivorous |
| Artibeus planirostris | 4/0 | 4/0 | 0/0 | 2/0 | 1/0 | 1/0 | Phyllostomidae | Frugivorous |
| Carollia perspicillata | 44/3 (6.8%) | 30/2 (6.7%) | 14/1 (7.1%) | 12/0 | 32/3 (9.4%) | 0/0 | Phyllostomidae | Frugivorous |
| Desmodus rotundus | 10/0 | 8/0 | 2/0 | 0/0 | 10a/0 | 0/0 | Phyllostomidae | Hematophagous |
| Eptesicus furinalis | 5/0 | 3/0 | 2/0 | 0/0 | 0/0 | 5/0 | Vespertilionidae | Insectivorous |
| Eumops glaucinus | 27/1 (3.7%) | 11/0 | 16/1 (6.2%) | 0/0 | 0/0 | 27/1 (3.7%) | Molossidae | Insectivorous |
| Glossophaga soricina | 3/1 (33.3%) | 2/1 (50%) | 1/0 | 0/0 | 0/0 | 3/1 (33.3%) | Phyllostomidae | Nectivorous |
| Lasiurus cinereus | 1/0 | 1/0 | 0/0 | 0/0 | 0/0 | 1/0 | Vespertilionidae | Insectivorous |
| Molossus molossus | 80/0 | 42/0 | 38/0 | 0/0 | 0/0 | 80/0 | Molossidae | Insectivorous |
| Molossus rufus | 56/2 (3.6%) | 37/1 (2.7%) | 19/1 (5.2%) | 0/0 | 0/0 | 56/2 (3.6%) | Molossidae | Insectivorous |
| Myotis nigricans | 8/1 (12.5%) | 7/1 (14.3%) | 1/0 | 0/0 | 0/0 | 8/1 (12.5%) | Vespertilionidae | Insectivorous |
| Myotis riparius | 1/1 (100%) | 1/0 | 1/1 (100%) | 0/0 | 0/0 | 1/1 (100%) | Vespertilionidae | Insectivorous |
| Platyrrhinuslineatus | 3/0 | 1/0 | 2/0 | 0/0 | 0/0 | 3/0 | Phyllostomidae | Frugivorous |
| Sturnira lilium | 28/3 (10.7%) | 19/0 | 9/3 (33.3%) | 22/2 (9.1%) | 6/1 (16.6%) | 0/0 | Phyllostomidae | Frugivorous |
| Vampyressa pusilla | 1/0 | 1/0 | 0/0 | 0/0 | 1/1 (100%) | 0/0 | Phyllostomidae | Frugivorous |
| Total | 401/15 (3.7%) | 251/5 (2%) | 149/10 (6.7%) | 99/4 (4%) | 110/5 (4.5%) | 192/6 (3.1%) | ||
Undist. Forest (Undisturbed Forest), Fragm. Forest (Fragmented Forest); bold font indicates species positive for CoV RNA.
Sample obtained from rural area.
Fig. 1.
A: Brazil's map and sites for bats capture/collection. Bats were collected from 17 sites (a–q) in the Atlantic Forest Biome from two adjacent Paraná and São Paulo states, Brazil. B: Based on morphological characteristics, bats were classified into different families (positive families were bold and underlined). Phylogenetic tree was adapted from Simmons (2005). C: Phylogenetic analysis of bat CoVs detected in the AFB, and reported previously around the world. Brazilian sequences reported in this study are marked with arrows. D: A higher resolution tree of the alpha and betacoronaviruses. Sequences studied in this report are marked with black filled square. Original map available in http://mapas.sosma.org.br from Atlas of the Atlantic Forest Remains (Fundação SOS Mata Atlântica/Instituto Nacional de Pesquisas Espaciais-INPE).
Nucleic acids extracted from 30 mg of intestine tissue of individual bat using NucliSENS® easyMAG® were subjected to first strand cDNA synthesis with random hexamers and RT-PCR High-Capacity cDNA Archive Kit (Applied BioSystems). The cDNA preparations were screened by pancoronavirus Nested PCR assay targeting the RNA-dependent RNA polymerase (RdRp) gene (Chu et al., 2011). Positive samples (n = 15) were sequenced and final sequences were submitted to the GenBank (KT717380-KT717394) (Supplementary table). A dataset, consisting of sequence generated in this study and all publically available bats CoVs, was phylogenetically analysed by neighbour-joining method in MEGA6 software using Kimura's two-parameter correction and 10,000 bootstrap values (Tamura et al., 2013).
Coronaviruses were detected in 15 bat intestines samples from eight bat species with distinct diet habit, demonstrating a marked potential of CoVs distribution among bat species in AFB that harbours 9% of world's bat diversity. All CoVs-positive bat species were geographical distributed in the Neotropics and in anthropogenic area apparently affected by fragmented forests, and bats-abundance urbanized areas. The detection of CoVs varied by bat species; undisturbed forest remnant (4%), fragmented forests (4.5%) and urban areas (3.1%) (Table 1).
Phylodynamics analyses indicated the circulation of 13 α-CoV and 2 β-CoV in AFB bats (Fig. 1C). This is of primary interest as majority of bat CoVs surveillance studies conducted in the New World bats detected only α-CoVs (Corman et al., 2013, Drexler et al., 2014, Osborne et al., 2011). A higher resolution analysis indicated a distinct distribution and diversity of α-CoV and β-CoV lineages (Fig. 1D).
α-CoV sequences obtained from bats of same genus presented high nucleotide sequence similarity (e.g. Artibeus, Glossophaga, Carollia, Molossus, Myotis and Sturnira) (Fig. 1D and Supplementary table), even with sequences detected in other studies from bats of geographically distant regions. This relation can be exemplified by the high similarity of the CoV RNA partial sequences (91,9%) detected in Carollia perspicillata species from Fenix-PR, Brazil (KT717385), and Fyzabad, Trinidad and Tobago (EU769557) (Carrington et al., 2008), located at a distance of at least 3900 km. Similar results were previously reported for a variety of bat CoVs and are taken as evidence of co-evolution of CoV genotypes and specific host genera (Drexler et al., 2014, Corman et al., 2013, Anthony et al., 2013).
Moreover, two previously uncharacterized α-CoV from Myotis nigricans and Myotis riparius and one β-CoV cluster were identified in studied bats (Fig. 1C and D). CoV sequences from Myotis bats genera presented high nt sequence similarity (98.9%) and were most closely related to bat CoVs (KC110771) collected from Brazilian Molossus molossus and Tadarida brasiliensis. A distinct cluster of α-CoVs were also detected in Sturnira bat that grouped with α-CoVs lineage 1, a group with an evolutionary history of recombination and cross-species transmission between domestic and livestock animals, such as feline, canine and swine α-CoVs (Lorusso et al., 2008). This clade presents nt similarity between 73.9% and 74.4% with transmissible gastroenteritis virus (DQ811789) and feline coronavirus (AF124987) depending on the Sturnira lineage (Supplementary table). This branching pattern may possibly explain the common-ancestral origin of α-CoVs lineage 1 species from bats and other animal species. However, extensive evolutionary studies on complete genome sequences of these isolates are required to provide information on the virus origin and divergence.
Out of two β-CoVs, the virus from Eumops glaucinus clustered within MERS-CoV containing lineage C. The second β-CoV detected in Artibeus lituratus shown highest similarity (96.4%) with CoV reported from Costa Rica (Corman et al., 2013) and lower similarity (69.3%) with MERS-CoV (Supplementary table). These sequences showed sequence similarities with β-CoV detected in Pipistrellus bats that poses high homology with human MERS-CoV (JX869059). Notably to observer that the Eumops bat positive for CoV was found on an urban area and was predated by a domesticated cat. It was not possible to rule out any event of virus transmission to human, however, this highlight the potential of domesticated animals in virus transmission and disease dynamics at the virus-animal-human interface. This is of special importance due to the established role of animals in the transmission of viruses to human being (Johnson et al., 2015). Taken together, these results represent the first detection of lineage C β-CoV in South American bats. Despite of close relationship between lineage C β-CoVs in Asia, Africa and Europe, cumulative data indicate that this lineage showed a more diversified host family distribution in Americas in Mormoopidae, Phyllostomidae and Molossidae bat species (Anthony et al., 2013, Corman et al., 2013, Goes et al., 2013).
2. Conclusions
We present a great diversity of CoV genotypes and clusters in Brazilian bats, highlighting a biogeographic distribution of bats CoVs in the region. It is indispensable in future to investigate the evolutionary events in genetically diverse bats CoVs using complete genome sequences, and their possible transmission potentials to human being. Although it is not possible to calculate the risk of “spill over” events of Brazilian bats CoVs to humans, our results reinforce the need for expanded and continuing surveillance of CoVs in bat fauna, including those in the AFB regions of Brazil.
The following is the supplementary data related to this article.
List and information's of positive samples bat samples detected in our study.
Acknowledgements
This study was supported by FAPESP (São Paulo Research Foundation) process number 2013/11006-0 and British Council grant number 172710323. Cruz-Neto A.P was supported by FAPESP grant process number 2008/57687-0. We thank Luiz Aurélio de Campos Crispin and Mariana Cristine Pereira de Souza for their contributions to this study.
Contributor Information
Luiz Gustavo Bentim Góes, Email: lgbgoes@usp.br.
Angélica Cristine de Almeida Campos, Email: camposac@usp.br.
Cristiano de Carvalho, Email: criscar@fmva.unesp.br.
Guilherme Ambar, Email: guilhermeambar@gmail.com.
Luzia Helena Queiroz, Email: lhqueiroz@fmva.unesp.br.
Ariovaldo Pereira Cruz-Neto, Email: ariovaldopcruz@gmail.com.
Muhammad Munir, Email: muhammad.munir@pirbright.ac.uk.
Edison Luiz Durigon, Email: eldurigo@usp.br.
References
- Anthony S.J., Ojeda-Flores R., Rico-Chavez O., Navarrete-Macias I., Zambrana-Torrelio C.M., Rostal M.K., Epstein J.H., Tipps T., Liang E., Sanchez-Leon M., Sotomayor-Bonilla J., Aguirre A.A., Avila-Flores R., Medellin R.A., Goldstein T., Suzan G., Daszak P., Lipkin W.I. Coronaviruses in bats from Mexico. J. Gen. Virol. 2013;94:1028–1038. doi: 10.1099/vir.0.049759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M., Donaldson E., Baric R. SARS-CoV and emergent coronaviruses: viral determinants of interspecies transmission. Curr. Opin. Virol. 2011;1:624–634. doi: 10.1016/j.coviro.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandao P.E., Scheffer K., Villarreal L.Y., Achkar S., Oliveira Rde N., Fahl Wde O., Castilho J.G., Kotait I., Richtzenhain L.J. A coronavirus detected in the vampire bat Desmodus rotundus. Braz. J. Infect. Dis. 2008;12:466–468. doi: 10.1590/s1413-86702008000600003. [DOI] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington C.V., Foster J.E., Zhu H.C., Zhang J.X., Smith G.J., Thompson N., Auguste A.J., Ramkissoon V., Adesiyun A.A., Guan Y. Detection and phylogenetic analysis of group 1 coronaviruses in South American bats. Emerg. Infect. Dis. 2008;14:1890–1893. doi: 10.3201/eid1412.080642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin. Microbiol. Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Poon L.L., Chan K.H., Chen H., Guan Y., Yuen K.Y., Peiris J.S. Coronaviruses in bent-winged bats (Miniopterus spp.) J. Gen. Virol. 2006;87:2461–2466. doi: 10.1099/vir.0.82203-0. [DOI] [PubMed] [Google Scholar]
- Chu D.K., Leung C.Y., Gilbert M., Joyner P.H., Ng E.M., Tse T.M., Guan Y., Peiris J.S., Poon L.L. Avian coronavirus in wild aquatic birds. J. Virol. 2011;85:12815–12820. doi: 10.1128/JVI.05838-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Rasche A., Diallo T.D., Cottontail V.M., Stocker A., Souza B.F., Correa J.I., Carneiro A.J., Franke C.R., Nagy M., Metz M., Knornschild M., Kalko E.K., Ghanem S.J., Morales K.D., Salsamendi E., Spinola M., Herrler G., Voigt C.C., Tschapka M., Drosten C., Drexler J.F. Highly diversified coronaviruses in neotropical bats. J. Gen. Virol. 2013;94:1984–1994. doi: 10.1099/vir.0.054841-0. [DOI] [PubMed] [Google Scholar]
- Drexler J.F., Corman V.M., Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014;101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goes L.G., Ruvalcaba S.G., Campos A.A., Queiroz L.H., de Carvalho C., Jerez J.A., Durigon E.L., Davalos L.I., Dominguez S.R. Novel bat coronaviruses, Brazil and Mexico. Emerg. Infect. Dis. 2013;19:1711–1713. doi: 10.3201/eid1910.130525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorin R., Taddei V.A. Mastozoologia-Neotropical. Vol. 9. 2002. Chave Artificial para a Identificação de Molossídeos Brasileiros (Mammalia, Chiroptera) pp. 13–32. [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., Frieman M.B., Baric R.S., Donaldson E.F. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICTV . 2015. Virus Taxonomy 2015 Release. ( http://www.ictvonline.org/virusTaxonomy.asp accessed 30 May 2016) [Google Scholar]
- Johnson C., Hitchens P.L., Smiley Evans T., Goldstein T., Thomas K., Clements A., Joly D.O., Wolfe N.D., Daszak P., Karesh W.B., Mazet J.K. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 2015;5:14830. doi: 10.1038/srep14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorusso A., Decaro N., Schellen P., Rottier P.J., Buonavoglia C., Haijema B.J., de Groot R.J. Gain, preservation, and loss of a group 1a coronavirus accessory glycoprotein. J. Virol. 2008;82:10312–10317. doi: 10.1128/JVI.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.M.D., Bernardi I.P., Passos F.C. 2011. Chave ilustrada para determinação dos morcegos da Região Sul do Brasil.Curitiba, João M.D. Miranda. (51p) [Google Scholar]
- Nogueira M.R., de Lima I.P., Moratelli R., Tavares V.D.C., Gregorin R., Peracchi A.L. Check List. Vol. 10. 2014. Checklist of Brazilian bats, with comments on original records; pp. 808–821. [Google Scholar]
- Osborne C., Cryan P.M., O'Shea T.J., Oko L.M., Ndaluka C., Calisher C.H., Berglund A.D., Klavetter M.L., Bowen R.A., Holmes K.V., Dominguez S.R. Alphacoronaviruses in New World bats: prevalence, persistence, phylogeny, and potential for interaction with humans. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paglia A.P., Fonseca G.A.B., Rylands A.B., Herrmann G., Aguiar L.M.S., Chiarello A.G. Conservation International; Arlington, VA: 2012. Lista Anotada dos Mamíferos do Brasil/Annotated Checklist of Brazilian Mammals. Occasional Papers in Conservation Biology, No. 6. [Google Scholar]
- Simmons N.B. Evolution. An Eocene big bang for bats. Science. 2005;307:527–528. doi: 10.1126/science.1108871. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizotto L.D., Taddei V.A. Chave para determinação de quirópteros brasileiros. Bol Cienc. 1973;1:1–72. [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel mammalian and avian coronaviruses in the genus Deltacoronavirus supports bat coronaviruses as the gene source of Alphacoronavirus and Betacoronavirus and avian coronaviruses as the gene source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List and information's of positive samples bat samples detected in our study.