Abstract
Caliciviruses are important human and animal pathogens. Novel caliciviruses have been identified recently in dogs, raising questions about their pathogenic role and concerns regarding their zoonotic potential. By screening stool samples of young or juvenile dogs using RT-PCR assays, sapoviruses (SaVs) were found in 7/320 (2.2%) samples of animals with acute gastroenteritis while they were not detected in healthy animals (0/119). The sequence of a nearly 3 kb portion at the 3′ end of the genome, encompassing the RNA-dependent RNA polymerase (RdRp), the capsid region (ORF1) and the ORF2 were determined for three strains. A distinctive genetic feature in canine SaVs was a 4-nucleotide (nt) interval between the ORF1 and ORF2. Two strains (Bari/4076/07/ITA and Bari/253/07/ITA) were very closely related in the RdRp and capsid regions to the strain AN210D/09/USA (90.4–93.9% nt), while strain Bari/5020/07/ITA displayed only 71.0–72.0% nt identity to this group of canine SaVs and 76.0% to strain AN196/09/USA. Overall, these findings indicate that the canine SaVs detected in Italy may represent distinct capsid types, although all currently known SaVs segregate into the novel proposed genogroup, tentatively named as GXIII.
Keywords: Sapovirus, Dogs, Enteritis, Genomic analysis
Highlights
-
•
Sapoviruses (SaVs), Caliciviridae family, have been recently discovered in dogs.
-
•
Canine SaVs were detected in 2.2% of diarrheic dogs but not in asymptomatic dogs.
-
•
The viruses were genetically related to canine SaV prototypes detected in USA.
Sapoviruses (SaVs), Caliciviridae family, are an important cause of human acute gastroenteritis worldwide (Glass, 2013). The SaV genome is a poly-adenylated, single-stranded, positive-sense RNA of 7.3 to 7.5 kb in length and contains two main open reading frames (ORFs). ORF1 encodes a polyprotein that undergoes protease processing to produce several nonstructural proteins, including an RNA-dependent RNA polymerase (RdRp) and a capsid protein (VP1). ORF2 encodes a small basic protein with an unknown function. A third ORF (ORF3), overlapping the 5′ end of the capsid gene, is present only in GI, GIV, and GV SaVs (Hansman et al., 2007) and in bat SaVs (Tse et al., 2012). Based on the full-length VP1 sequence, SaVs have been classified into five genogroups (GI–GV). More recently, nine additional genogroups (GVI–GXIV) have been proposed (Scheuer et al., 2013) but only GI, GII, GIV and GV are known to infect humans. Animal SaVs genetically closely related to human SaVs (GI, GV and GVIII) have been identified in chimpanzee (GI) (Mombo et al., 2014), pigs (GV and GVIII) (Martella, V., et al., 2008b, L'Homme, Y., et al., 2010, Scheuer, K.A., et al., 2013) and rodents (GII) (Firth et al., 2014) raising public health concerns of potential cross-species transmission.
Metagenomic investigation of canine fecal virome in the USA has revealed the presence of SaVs in diarrheic dogs (Li et al., 2011). SaVs have been also identified in a study in Japan in 2/97 (2.06%) dogs with enteritis (Soma et al., 2014). Information on the epidemiology and genetic heterogeneity of these newly described canine caliciviruses is limited at the moment and it is not clear whether these viruses may play a role as enteric pathogens of dogs and to which extent they impact on canine health. In this study, we investigated the prevalence of canine SaVs in the stools of dogs with or without diarrhea.
A total of 320 stool samples (collection A) were collected between January and December 2007 from young household dogs (aged 1 to 6 months) hospitalized with signs of mild to severe gastroenteritis. In addition, 119 fecal specimens from asymptomatic household dogs (1–8 months of age) receiving routine care at 3 veterinary clinics (collection B), were included in the study. SaV RNA was detected by RT-PCR using the broadly reactive consensus primers for caliciviruses, p289–p290 (Jiang et al., 1999), targeting the highly conserved motifs “DYSKWDST” and “YGDD” of the RdRp region. All fecal samples collected from dogs with enteritis signs were also tested for the presence of canine parvovirus type 2 (CPV-2), canine coronavirus (CCoV) and norovirus (NoV) by either gel-based PCR or quantitative PCR and reverse transcription (RT)-PCR (Pratelli, A., et al., 1999, Vennema, H., et al., 2002, Decaro, N., et al., 2005).
SaV RNA was detected in 7 diarrheic fecal samples, with an overall prevalence of 2.2% (7/320). In four samples, SaV RNA was found in mixed infection with CPV-2 and CCoV (1 = 0.3%) or in conjunction with CPV-2 and NoVs (3 = 0.9%). Three (0.9%) SaV positive specimens also contained CCoV, but not other viral pathogens. SaV RNA was not detected in asymptomatic dogs (0/119) (collection B). Upon statistical analysis using the χ2 test, with Yates' correction (for continuity) for dichotomous variables and the Rule of Three (Tuyl et al., 2009), SaV prevalence did not significantly differ between diarrheic and healthy dogs.
These findings demonstrate firmly that these novel caliciviruses circulate in dogs in Italy. In our analysis SaVs were detected only in diarrheic animals, but all the positive cases were mixed infections with other enteric viruses such as CPV-2 or CCoV or NoVs. Also, the prevalence of SaV in diarrheic dogs was low (2.2%). As we used calicivirus broadly reactive consensus primers for our screening, in the attempt to increase the chances of identifying genetically divergent SaV strains, it is possible that this approach affected the sensitivity of the study, preventing the identification of SaV RNA in samples with a low viral load. Whether SaVs can act as a primary causative agent of gastrointestinal disease or whether they can trigger mechanisms of synergism in co-infections should be demonstrated in experimental infections.
Partial RdRp sequences were determined from all the SaV positive samples. By FASTA and BLAST analyses, five strains (201-07/IT, 253-07/IT, 4076-07/IT, 4078-07/IT and 324-07/IT) shared 78.6–100% nt and 90.2–100% aa identities to each other and displayed the closest relatedness (73.2–90.4% nt and 88.5–100% aa identities) to the SaVs AN210D and AN196 detected in the US (Li et al., 2011) and to other canine SaV-like sequences currently available in the databases (GenBank accession KM001673 and KM001674; unpublished data). Two strains, 5020-07/IT and 53-07/IT, showed 86.6% nt and 92.3% aa identities to each other, but were more distantly related to the other canine SaVs (72.6% nt and 75.0–78.0% aa identities) (Fig. 1 ).
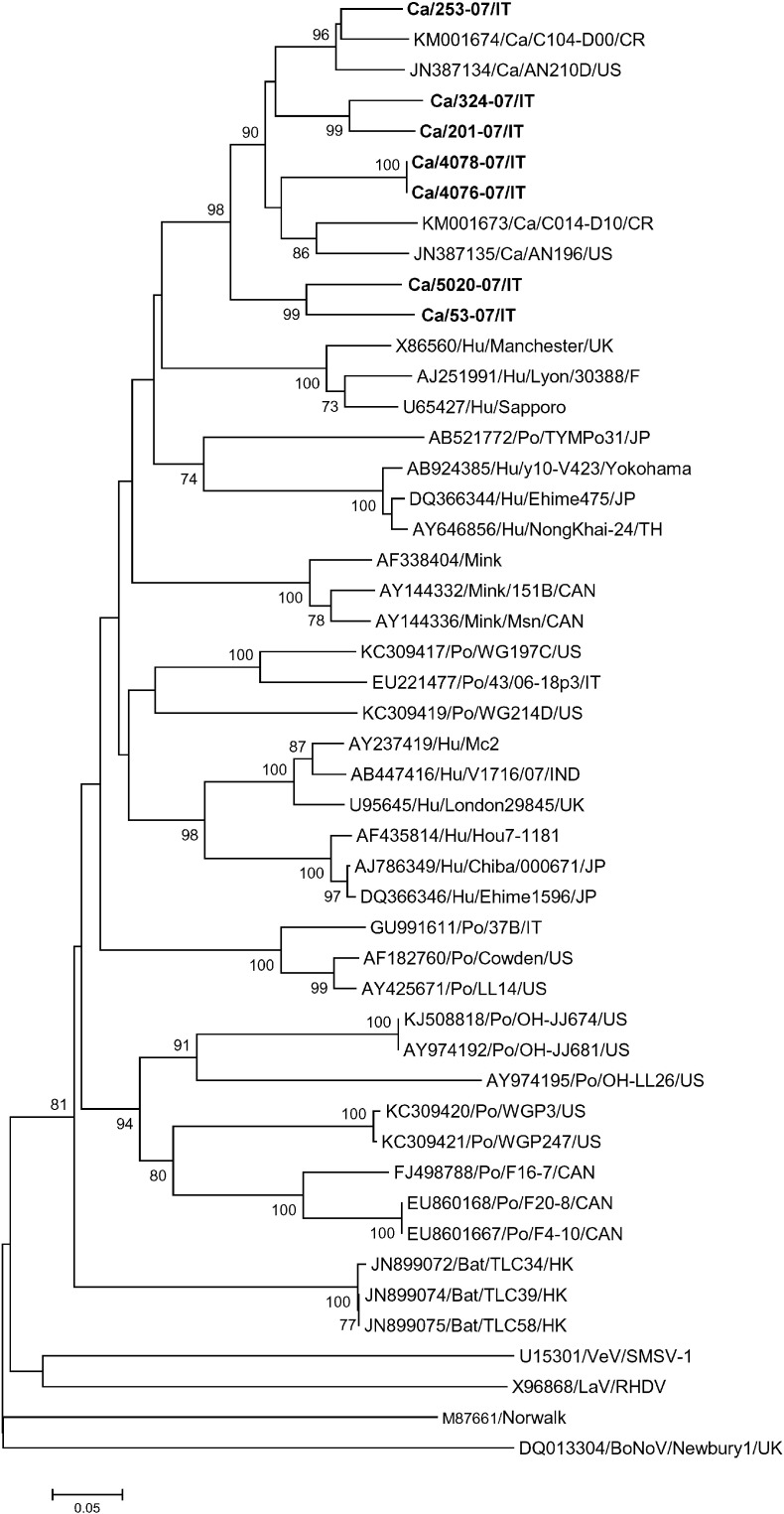
Fig. 1.
Phylogenetic tree based on the RdRp gene of SaVs. The tree was generated on a 270 nt-long portion of the RdRp region, using the neighbor joining method and Kimura-2 parameter correction, supplying statistical support with bootstrapping over 1000 replicates. The scale bar indicates the number of nucleotide substitutions per site. The sequences detected in this study are in boldface.
For three strains, 253-07/IT, 4076-07/IT and 5020-07/IT (accession numbers KT207827, KT207828 and KT207829, respectively), the 3′ ends of their genome, including the partial RdRp gene, the capsid gene, the ORF2 and the 3′ untranslated region (3′ UTR), were determined by 3′ RACE protocol and primer-walking strategy, as previously described (Wang et al., 2005).
In the 0.8 kb portion of the analyzed RdRp region, strains Bari/253/07/ITA and Bari/4076/07/ITA displayed 91.3% nt and 99.5% aa identities to each other, 91.3–93.9% nt and 99.0–99.5% aa identities to the SaV strain AN210D/09/USA, and only 71.3–72.6% nt and 74.6–77.6% aa to strain Bari/5020/07/ITA, which, in turn, was more related (72.6% nt and 77.6% aa identities) to the SaV strain AN210D/09/USA.
Strains 253-07/IT, 4076-07/IT and 5020-07/IT had deduced capsid (555 aa) and ORF2 (166 aa) proteins identical in size, while the 3′ UTR was 142 nt, 144 nt and 141 nt in length, respectively. As observed in the prototype canine strain AN210D, a 4-nt interval was found between ORF1 and ORF2, while the other members of the Sapovirus genus overlap by 1–8-nt in the ORF1–ORF2 region (Scheuer et al., 2013). By sequence comparison in the full-length capsid protein, strains 253-07/IT and 4076-07/IT shared 89.4% nt and 96.2% aa identities to each other and were more related genetically to the SaV strain AN210D (90.0–92.8% nt and 94.0–95.0% aa identities), while identity to strain 5020-07/IT was 71.0–72.0% nt and 79.5–79.7% aa, respectively. Strain 5020-07/IT showed the highest identity to the SaV AN196 (76.0% nt and 88.0% aa identities). Neighbor-joining phylogenetic analysis was performed with a selection of capsid sequences representative of the Sapovirus genus, including canine, human, porcine, mink, bat and rodent SaVs. Based on the inspection of the tree (Fig. 2 ) and according to the pairwise distance analyses, all the Italian strains segregated with the canine SaVs AN210D and AN196 (bootstrap value, 100%) into the novel proposed genogroup, tentatively named as GXIII (≥ 60% pairwise aa inter-genogroup identity) (Scheuer et al., 2013). This genogroup of SaVs appeared genetically closer to GII human and rodent strains (mean aa identity of 48.0–56.0%) than to other SaV genogroups (Table 1 ). Interestingly, genetic distance calculations using the short polymerase region did not support the inclusion of the strain 5020-07/IT into the novel proposed genogroup, as there was < 80% aa identity with the other canine SaVs (Wang, Q.H., et al., 2005, L'Homme, Y., et al., 2010). The clustering inconsistencies observed between the RdRp and capsid regions reinforce the notion that a definitive characterization/classification of SaV genogroups necessarily relies on sequence analysis of the full-length capsid gene. Analysis of 59 representative capsid sequences of human SaV has allowed calculating precisely the cutoff values for genotypes (≤ 83.1% nt identity) (Oka et al., 2015). Accordingly, based on our analysis, the canine SaVs can be classified into at least 3 genotypes, with strains, 25307/IT, 407607/IT and AN210D being classified within the same genotype, and with strains 5020-07/IT and AN196 representing additional two genotypes. Overall, these findings indicate that canine SaVs are genetically highly heterogeneous, thus posing a challenge for the selection of reliable molecular diagnostic tools. Gathering more sequence information from canine SaVs will be helpful to optimize the diagnostic instruments.
Fig. 2.
Phylogenetic tree based on the capsid protein of SaVs. The tree was using the aa sequence of the complete capsid region of SaVs, using the neighbor joining method and Poisson correction, and supplying a statistical support with bootstrapping over 1000 replicates. The scale bar indicates the number of aa substitutions per site. Black triangles indicate the canine SaV strains detected in this study.
Table 1.
Amino acid sequence identity in the complete capsid protein between the canine SaV strains detected in this study (5020/07/ITA, 253/07/ITA and 4076/07/ITA) and SaV genogroups.
| SaV genogroups |
% aa identity |
|---|---|
| GI | 41.7–48.4 |
| GII | 48.0–56.0 |
| GIII | 40.4–43.7 |
| GIV | 48.6–52.5 |
| GV | 48.6–51.9 |
| GVI | 37.6–39.6 |
| GVII | 36.8–38.5 |
| GVIII | 41.7–48.4 |
| GIX | 38.7–51.1 |
| GX | 38.2–39.0 |
| GXI | 37.1–38.2 |
| GXII | 41.5–42.3 |
| GXIII | 79.5–96.2 |
| GXIV | 41.5–43.1 |
In conclusion, genetically diverse canine SAVs with a distinct and conserved genome organization were found only in diarrheic dogs although with a low prevalence.
Human SaVs have been associated with sporadic cases of gastroenteritis in pediatric population and with small to large outbreaks of gastroenteritis in human communities, although the prevalence rates by these enteric pathogens in children are usually low (Medici et al., 2006). On the contrary, SaV prevalence in piglets with weaning and post-weaning enteritis is high, although co-infections by other enteric viruses are quite common in piglets (Martella, V., et al., 2008a, Dufkova, L., et al., 2013). While comparing SaV epidemiological patterns in different species may be of difficult interpretation, conclusive information on the pathogenic role of SaVs in dogs could only be obtained by experimental infections.
Conflict of interest statement
All authors declare that there are no financial or other relationships that might lead to a conflict of interest. All authors have seen and approved the manuscript and have contributed significantly to the work.
Acknowledgments
This work was supported by the grants “Studio sui vesivirus nei cani — Fondi Ateneo 2010”; “Calicivirus nei carnivori e nell'uomo: caratterizzazione molecolare, epidemiologia, implicazioni zoonosiche — PRIN 2008”. KB was supported by the Momentum program (awarded by the Hungarian Academy of Sciences).
References
- Decaro N., Elia G., Martella V., Desario C., Campolo M., Trani L.D., Tarsitano E., Tempesta M., Buonavoglia C. A real-time PCR assay for rapid detection and quantitation of canine parvovirus type 2 in the feces of dogs. Vet. Microbiol. 2005;105:19–28. doi: 10.1016/j.vetmic.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Dufkova L., Scigalkova I., Moutelikova R., Malenovska H., Prodelalova J. Genetic diversity of porcine sapoviruses, kobuviruses, and astroviruses in asymptomatic pigs: an emerging new sapovirus GIII genotype. Arch. Virol. 2013;158:549–558. doi: 10.1007/s00705-012-1528-z. [DOI] [PubMed] [Google Scholar]
- Firth C., Bhat M., Firth M.A., Williams S.H., Frye M.J., Simmonds P., Conte J.M., Ng J., Garcia J., Bhuva N.P., Lee B., Che X., Quan P.L., Lipkin W.I. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York city. Mbio. 2014;5 doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass R.I. Beyond discovering the viral agents of acute gastroenteritis. Emerg. Infect. Dis. 2013;19:1190–1191. doi: 10.3201/eid1908.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansman G.S., Oka T., Katayama K., Takeda N. Human sapoviruses: genetic diversity, recombination, and classification. Rev. Med. Virol. 2007;17:133–141. doi: 10.1002/rmv.533. [DOI] [PubMed] [Google Scholar]
- Jiang X., Huang P.W., Zhong W.M., Farkas T., Cubitt D.W., Matson D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods. 1999;83:145–154. doi: 10.1016/s0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- L'Homme Y., Brassard J., Ouardani M., Gagné M.J. Characterization of novel porcine sapoviruses. Arch. Virol. 2010;155:839–846. doi: 10.1007/s00705-010-0651-y. [DOI] [PubMed] [Google Scholar]
- Li L., Pesavento P.A., Shan T., Leutenegger C.M., Wang C., Delwart E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 2011;92:2534–2541. doi: 10.1099/vir.0.034611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martella V., Bànyai K., Lorusso E., Bellacicco A.L., Decaro N., Mari V., Saif L., Costantini V., De Grazia S., Pezzotti G., Lavazza A., Buonavoglia C. Genetic heterogeneity of porcine enteric caliciviruses identified from diarrhoeic piglets. Virus Genes. 2008;36:365–373. doi: 10.1007/s11262-008-0198-0. [DOI] [PubMed] [Google Scholar]
- Martella V., Lorusso E., Banyai K., Decaro N., Corrente M., Elia G., Cavalli A., Radogna A., Costantini V., Saif L.J., Lavazza A., Di Trani L., Buonavoglia C. Identification of a porcine calicivirus related genetically to human sapoviruses. J. Clin. Microbiol. 2008;46:1907–1913. doi: 10.1128/JCM.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici M.C., Martinelli M., Abelli L.A., Ruggeri F.M., Di Bartolo I., Arcangeletti M.C., Pinardi F., De Conto F., Izzi G., Bernasconi S., Chezzi C., Dettori G. Molecular epidemiology of norovirus infections in sporadic cases of viral gastroenteritis among children in northern Italy. J. Med. Virol. 2006;78:1486–1492. doi: 10.1002/jmv.20723. [DOI] [PubMed] [Google Scholar]
- Mombo I.M., Berthet N., Bouchier C., Fair J.N., Schneider B.S., Renaud F., Leroy E.M., Rougeron V. Characterization of a genogroup I sapovirus isolated from chimpanzees in the Republic of Congo. Genome Announc. 2014;17:2. doi: 10.1128/genomeA.00680-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T., Wang Q., Katayama K., Saif L.J. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 2015;28:32–53. doi: 10.1128/CMR.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Tempesta M., Greco G., Martella V., Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J. Virol. Methods. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuer K.A., Oka T., Hoet A.E., Gebreyes W.A., Molla B.Z., Saif L.J., Wang Q. Prevalence of porcine noroviruses, molecular characterization of emerging porcine sapoviruses from finisher swine in the United States, and unified classification scheme for sapoviruses. J. Clin. Microbiol. 2013;51:2344–2353. doi: 10.1128/JCM.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma T., Nakagomi O., Nakagomi T., Mochizuki M. Detection of norovirus and sapovirus from diarrheic dogs and cats in Japan. Microbiol. Immunol. 2014;59:123–128. doi: 10.1111/1348-0421.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H., Chan W.M., Li K.S., Lau S.K., Woo P.C., Yuen K.Y. Discovery and genomic characterization of a novel bat sapovirus with unusual genomic features and phylogenetic position. PLoS One. 2012;7 doi: 10.1371/journal.pone.0034987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuyl F., Gerlach R., Mergersen K. The Rule of Three, its variants and extensions. Int. Stat. Rev. 2009;77:266–275. [Google Scholar]
- Vennema H., de Bruin E., Koopmans M. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 2002;25:233–235. doi: 10.1016/s1386-6532(02)00126-9. [DOI] [PubMed] [Google Scholar]
- Wang Q.H., Han M.G., Cheetham S., Souza M., Funk J.A., Saif L.J. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 2005;11:1874–1881. doi: 10.3201/eid1112.050485. [DOI] [PMC free article] [PubMed] [Google Scholar]