Highlights
► Authors review the current and proposed nomenclature of filoviruses. ► Filovirus genome structure and content are reviewed. ► New filoviruses finds are discussed with regard to the current view of phylogeny. ► Swine as a newly identified host is discussed with regard to phylogenetic diversity. ► Integrations of filovirus-like sequences in host genomes are reviewed.
Keywords: Filoviridae, Ebola, Marburg, Phylogeny, Virus, Reston
Abstract
Sporadic fatal outbreaks of disease in humans and non-human primates caused by Ebola or Marburg viruses have driven research into the characterization of these viruses with the hopes of identifying host tropisms and potential reservoirs. Such an understanding of the relatedness of newly discovered filoviruses may help to predict risk factors for outbreaks of hemorrhagic disease in humans and/or non-human primates. Recent discoveries such as three distinct genotypes of Reston ebolavirus, unexpectedly discovered in domestic swine in the Philippines; as well as a new species, Bundibugyo ebolavirus; the recent discovery of Lloviu virus as a potential new genus, Cuevavirus, within Filoviridae; and germline integrations of filovirus-like sequences in some animal species bring new insights into the relatedness of filoviruses, their prevalence and potential for transmission to humans. These new findings reveal that filoviruses are more diverse and may have had a greater influence on the evolution of animals than previously thought. Herein we review these findings with regard to the implications for understanding the host range, prevalence and transmission of Filoviridae.
1. Introduction
1.1. Types of filoviruses
Filoviruses are associated with acute fatal hemorrhagic diseases of humans and/or non-human primates. The family consists of the two classic genera: Marburgvirus, discovered in 1967 (Siegert et al., 1967), comprised of various strains of the Lake Victoria marburgvirus including Marburg virus and Ravn virus, and the antigenically distinct genus Ebolavirus discovered in 1976 (Emond et al., 1977), comprised of five species partially including Sudan ebolavirus, Zaire ebolavirus, Ivory Coast ebolavirus also known as Cote d’Ivoire Ebolavirus or the Taï Forest ebolavirus, and the Reston ebolavirus (Towner et al., 2008). The fifth and most recent member of the Ebolavirus genus is the Bundibugyo ebolavirus identified in an outbreak of human disease in Uganda, Africa. In addition, a tentative, third genus of Filoviridae has been recently identified in Spain, in bats, with formal publication of the discovery and full genome characterization anticipated (Kuhn et al., 2010).
1.2. Classification of filoviruses
Filoviruses are large filamentous viruses, 80 nm in diameter and may vary in length from several hundred nm to as long as or longer than 1 μm. Their linear forms are pleomorphic and may be unbranched, branched, curved or straight. Indeed, genomic similarities between filoviruses and other negative-sense RNA viruses are pronounced, and are the basis for the current classification of filoviruses within the order Mononegavirales, proximal to Paramyxoviridae, Rhabdoviridae and Bornaviridae families.
1.3. Genome structure and content
Filovirus genomes are comprised of linear, non-segmented, negative-sense, single-stranded RNA approximately 19 kb in length (Regnery et al., 1980) (Fig. 1 ). The gene organization consists of a conserved 3′ non-coding region followed by seven genes that encode structural proteins and concluding in a conserved 5′ non-coding region (Fig. 1). In addition, the viral genes are flanked by extragenic regions containing promoters for transcription and replication (Feldmann et al., 1992) (Fig. 1). The nucleoprotein (NP), viral proteins VP35 and VP30 and the RNA-dependent RNA polymerase or L protein are bound to the viral genome, mediate transcription and replication and together form the nucleocapsid of the virus particle (Muhlberger, 2007). Another viral protein, VP40, serves as a matrix protein and plays a role in virus budding and release from the host cell (Urata et al., 2007). Virus protein VP24 represents yet another matrix protein and is involved in nucleocapsid formation and assembly (Noda et al., 2007). The seventh protein encoded by the virus is the envelope surface glycoprotein or spike glycoprotein (GP), which mediates attachment and entry into target cells and represents a major determinant of virus pathogenicity (Licata et al., 2004). A notable difference between Ebola and Marburg viruses is the use of mRNA editing in the expression of full length GP by Ebola viruses, but not Marburg viruses. As a result of RNA editing in Ebola viruses, two forms of envelope protein are expressed, the more abundant being a smaller soluble protein (sGP) and the less abundant being the full length GP (Fig. 1). The sGP has been shown to have antagonistic effects on TNF-α induced endothelial barrier functions (Falzarano et al., 2006, Sanchez et al., 1996, Volchkov et al., 1995, Wahl-Jensen et al., 2005). Regarding the full length GP, there is a strong correlation between the level of GP expression and severe vascular cell cytotoxicity that is characteristic of filovirus infections. In vitro, the mucin domain of GP itself has been found to elicit cytotoxicity of endothelial cells (Yang et al., 2000). In addition, high level expression of full-length GP on host cell surfaces has been shown to not only mask particular immune-reactive epitopes of the GP but also cause the down regulation of host cell surface immune surveillance markers including major histocompatibility complex class I molecules resulting in immune evasion (Reynard et al., 2009). Among the viral proteins, VP35 and VP24 are known to be involved in interference of host cellular antiviral interferon mechanisms by prevention of nuclear accumulation of the phosphorylated signal transduction and transcription activation element, STAT1(Basler et al., 2003, Reid et al., 2006, Valmas et al., 2010). In addition the VP40 of Marburg virus has been shown to inhibit interferon mediated antiviral mechanisms in a manner that is distinct from Ebola viruses through interference with the phosphorylation event of STAT1 and STAT2 and also has effects on Janus kinase dependent interferon pathways (Valmas et al., 2010).
Fig. 1.
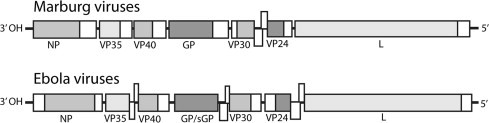
Genome representations of Marburg viruses and Ebola viruses. Open reading frames and intergenic regions are indicated as shaded and unshaded boxes, respectively. Overlapping intergenic regions are shown as split boxes.
2. Phylogeny of filoviruses
2.1. Newly proposed phylogenetic naming convention
Due to new discoveries, and a better understanding of the phylogeny, revisions have been recently proposed to the standard naming conventions used to describe members of Filoviridae (Kuhn et al., 2010). Using this newly proposed naming convention, Marburg viruses, which comprise multiple isolates of a single genus and species, would be written as Marburg marburgvirus to describe the genus and species, and Marburg virus (MARV) or Ravn virus (RAVV) to describe distinct viruses within the species (Table 1 ). Likewise, the genus Ebolavirus, comprised currently of multiple isolates of five representative species, would be similarly written and abbreviated to discriminate the genus, species, and individual virus isolate names (Table 1) (Kuhn et al., 2010). As a result of the recent genome characterization of a novel filovirus, Lloviu virus (LLOV), equidistantly related to Marburg and Ebola viruses and discovered in Schreiber’s long-fingered bats (Miniopterus schreibersii) in the Cueva del Lloviu Principality of Asturias in Spain, a new genus within Filoviridae, Cuevavirus, has been proposed (Kuhn et al., 2010).
Table 1.
Recent naming convention based on recent additions, and expanded phylogeny of Filoviridae as suggested by (Kuhn et al., 2010).
| Genus/species | Virus | Abbreviation |
|---|---|---|
| Marburg marburgvirus | Marburg virus | MARV |
| Marburg marburgvirus | Ravn virus | RAVV |
| Taï Forest ebolavirus | Taï Forest virus | TAFV |
| Reston ebolavirus | Reston virus | RESTV |
| Sudan ebolavirus | Sudan virus | SUDV |
| Zaire ebolavirus | Zaire virus | EBOV |
| Bundibugyo ebolavirus | Bundibugyo virus | BDBV |
| Lloviu cuevavirus | Lloviu virus | LLOV |
2.2. Host tropisms and transmission
Since the discovery of filoviruses, ostensibly random, sporadic and fatal outbreaks of disease in humans and non-human primates have evoked interest in delineation of host tropisms, potential reservoirs for disease transmission, and persistence in nature (Strong et al., 2008). While pathogenesis seems largely limited to humans and non-human primates, a few other animal species including swine have been found to be susceptible to infection by Ebola viruses (Barrette et al., 2009). Trace RT-PCR evidence and/or serological evidence suggest the potential susceptibility of duiker antelope to infection (Gonzalez et al., 2007, Rouquet et al., 2005). Candidate reservoir hosts for filoviruses are thought to be mammals of small body size, asymptomatic, and not likely to be companion animals (Peterson et al., 2004). Investigations into potential host species have also recently identified African fruit bats as possible reservoirs for Zaire ebolavirus (Leroy et al., 2005, Swanepoel et al., 1996) and Marburg virus (Swanepoel et al., 2007, Towner et al., 2007).
Transmission between susceptible hosts appears to be primarily through contact with infected individuals and can also occur by direct contact with infected carcasses (Leroy et al., 2009). For instance, it appears that the initial transition of the Ebola virus from primates to humans occurred primarily through handling of infected gorilla and chimpanzee carcasses (Walsh et al., 2005). Subsequent human to human transmission of Ebola virus has similarly been linked to close contact between immediate family members (Leroy et al., 2009, Walsh et al., 2005). Likewise Marburg virus outbreaks have been occasionally attributed to direct contact with infected mammalian tissues (Swanepoel et al., 2007). There have also been examples of human acquired infections of Marburg virus, upon entering caves where bats harboring the virus are known to reside. The route of infection for these cases is presumed to be through exposure to aerosol or bat excreta within the cave. (Anon, 2009, Swanepoel et al., 2007, Timen et al., 2009, Towner et al., 2009).
2.3. Swine as a new host tropism for Ebola virus
Reston ebolavirus, the only current member of Filoviridae to apparently not cause disease in humans, was identified in 1989 in the USA from a shipment of cynomolgus monkeys (Macaca fascicularis) from the Philippines (Jahrling et al., 1990, Morikawa et al., 2007). Subsequent outbreaks of disease occurred in the US in 1990 and 1996 and Italy in 1992 which were also traced back to a single facility in the Philippines (Miranda et al., 1999, Peters and LeDuc, 1999). After a gap of nearly 12 years, the Reston virus was identified in domestic swine in the Philippines during a naturally acquired co-infection with porcine reproductive and respiratory syndrome virus (PRRSV) (Barrette et al., 2009).
Viral genomes for Reston viruses identified from three samples at two geographically distinct locations, designated RESTV Philippines (swine) (Fig. 2 ) were significantly more divergent from each other (3.93% mean difference in nucleotide identity) than from the prototypical reference isolate from 1989 (2.5% mean difference in nucleotide identity), indicating polyphyletic origins of the Reston virus infections in swine at both locations. The lack of a phylogenetic clade, distinct from viruses in macaques, for the 2008 Reston virus infections in swine further suggested that Reston virus had been circulating since, and possibly before, its initial discovery in monkeys exported from the Philippines in 1989 (Barrette et al., 2009).
Fig. 2.

Consensus phylogenetic tree of filovirus complete genomes. Phylogenetic analysis was conducted on 33 aligned filovirus full genome nucleotide sequences using the Maximum Composite Likelihood method (Tamura et al., 2004). Evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987) in MEGA 4 (Tamura et al., 2007) and using software default settings with the complete deletion option enabled. The tree shown is the consensus of a 1000 randomly assorted bootstrap replicates (Felsenstein, 1985).
The prevalence of the circulating Reston virus in swine and intimate interconnection between man and domestic livestock were further predictive of transmission of Reston virus between swine and humans, and six individuals that worked with pigs on the affected farms were identified during the outbreak in swine to have developed serum IgG titers to Ebola virus confirming asymptomatic infection in humans (Barrette et al., 2009). The discovery of Reston virus in domestic swine had broad implications not only for our understanding of the potential host range of filoviruses and the newly recognized risk of their spread to food producing animals, but also to the greater prevalence and circulation of Ebola virus as directly inferred from the genetic diversity of the co-circulating viruses from swine as compared to the historical Reston virus isolates from monkeys (Barrette et al., 2009).
A recent publication by Kobinger et al. reported mucosal infection of swine with Zaire virus, resulting in severe lung pathology, as well as viral shedding from the oronasal mucosae. Virus was detectable for 14 days, and infected pigs were able to transmit infection to naïve animals. The report described severe hemorrhages within the heart and lungs of infected animals. In addition, it was identified that the virus had effects on cytokine levels, with a substantial downregulation of interferon-α (IFN-α). Although the pathology of the virus in swine appears to be distinct from that seen in humans and non-human primates, it does support a potential role for swine as a host in the virus transmission cycle (Kobinger et al., 2011).
2.4. Bundibugyo ebolavirus as a distinct new species
Newly discovered and characterized filoviruses have contributed to a greater resolution of diversity within the family. For instance, when comparing the full genomes of filoviruses, the Taï Forest ebolavirus (TAFV) and the recently identified Bundibugyo ebolavirus (BDBV) appear to have a common distant ancestor that in turn may have branched off from a common ancestor of Zaire ebolavirus (Fig. 2) (Towner et al., 2008). Similar to the complete genome, the phylogenetic relationship of VP35 displays an ancestral node for TAFV and BDBV that has an ancestral relationship that is closest to the Zaire ebolavirus (Fig. 3 ). While TAFV and BDBV group closely together in both analyses (Fig. 2, Fig. 3), their respective branch lengths as determined by the DNA Distance algorithm on the full length genome analysis clearly demonstrates the divergence of these two viruses leading to the classification of BDBV as a new distinct species (Fig. 2) (Towner et al., 2008). This distinction is less clearly defined in the more limited analysis of the VP35 gene (Fig. 3), highlighting the importance of full genome or multi-gene analyses in inferring phylogenetic relationships.
Fig. 3.
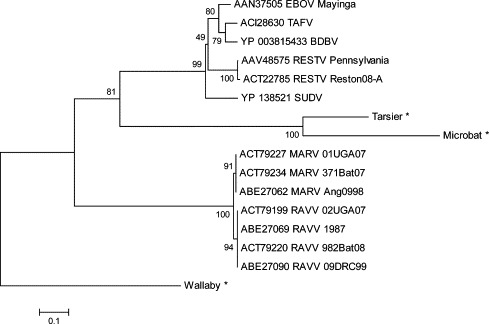
Consensus phylogenetic tree of endogenous and exogenous filovirus VP35 nucleotide sequences. VP35 sequences were analyzed using the Neighbor-Joining method (Saitou and Nei, 1987) in MEGA 4 (Tamura et al., 2007). The analysis was performed under the software default settings, except that evolutionary distances were computed using the Dayhoff matrix (Schwartz and Dayhoff, 1979) with the MEGA complete deletion option enabled. The rooted consensus tree was drawn based on 1000 randomly assorted bootstrap replicates (Felsenstein, 1985), with the Wallaby sequence integration being artificially designated as an outgroup based on its sequence containing multiple alignment gaps and stop codons. (∗) indicates sequence data derived from (Belyi et al., 2010).
2.5. Germline integrations of filovirus genomes in mammals
Another rather unexpected recent discovery is the apparent integration of RNA virus sequences into the germline genomes of a variety of mammalian species including the integration of bornavirus and filovirus-like gene sequences (Belyi et al., 2010, Horie et al., 1990, Taylor et al., 2010). These findings are particularly remarkable considering that most RNA viruses do not replicate their genomes in the host cell nucleus. In this regard, it is believed that co-expression of an endogenous or exogenous retrovirus might have led to the generation of cDNA from a co-infecting RNA virus in turn resulting in the integration of viral cDNA sequences into the genome of a host germ cell. For instance a genome integration of VP35 in Tarsier and microbat genomes appears to have originated from a common ancestor with the Ebolavirus genus (Fig. 3) (Belyi et al. 2010). In addition to integrations of viral VP35 sequences in the genomes of the tarsier, wallaby, and microbat, the NP gene has integrated into the guinea pig, microbat, shrew, opossum, and wallaby genomes, and an L gene remnant has been found in the opossum genome (Belyi et al. 2010). A large number of stop codons and gaps observed in the wallaby gene insertion may indicate an earlier integration event than observed in the tarsier and microbat. Similar integrations of genes from other positive or negative sense RNA viruses, likely facilitated by reverse transcription of endogenous or co-infecting retroviruses, have also been recently identified (Horie et al., 1990, Taylor et al., 2010). Based on the analysis of these paleoviral elements, found to be integrated, it is estimated that the association event with filovirus and their mammalian hosts occurred tens of millions of years prior to previous estimates (Taylor et al., 2010) In this regard a better understanding of why some species harbor integrated filovirus derived genes others do not, may one day have predictive value for identifying reservoir host species (Taylor et al. 2010). Transcription has also been identified from some of the integrated genes increasing the potential for evolutionary influence on the host.
3. Host reservoirs
3.1. Asymptomatic reservoir hosts and filovirus diversity
The phylogenetic origins of filoviruses have been of particular interest in light of the search for a reservoir host population. Initially, it was believed that the emergence of variants of Ebola virus was largely monophyletic, originating from a single event and diverging in a wave front distributed over time. However with the evidence of viral genome stability within an individual outbreak (Rouquet et al., 2005) coupled with the observation of multiple strains of Zaire ebolavirus occurring in a single outbreak, as seen in the Gabon/Republic of Congo outbreak in 2001 with up to 2–3% variation (Wittmann et al., 2007), it seems likely that some outbreaks may have more diverse origins than originally thought. Similarly, a pattern of multiple, parallel introductions of Reston virus in swine in the Philippines in 2008 (Barrette et al., 2009) lends credence to the observation that multiple individual introductions of unique viruses originating from a viral reservoir may be an integral part of the viral transmission cycle (Wittmann et al., 2007). This may occur as low pathogenic viruses may circulate undetected through the host population. This may be the outcome of a pool of divergent strains, which may indicate where the host and virus have co-evolved as to allow for viral propagation, with minimal pathogenicity. What is less clear is the mechanism by which the virus is able to emerge from a reservoir pool and cause disease in novel host species.
3.2. Co-infections with filoviruses
Identification of the definitive reservoir host has been particularly difficult, and may be due to a complex host relationship between a reservoir host, and possibly an amplification host. Additionally, there may be some incidents of coinfection which may be able to facilitate virus entry or persistence in different hosts. Fruit bat species Rousettus aegyptiacus, (Towner et al., 2009) and Hypsignathus monstrosus (Laminger and Prinz, 2010) in Africa, considered as likely reservoir hosts for Marburg and Ebola viruses respectively, have been shown to be seropositive for antibodies against the SARS coronavirus (SARS-CoV) (Muller et al., 2007). Likewise, within the insectivorous bats a single species, Miniopterus inflatus has been identified as a potential Ebola virus reservoir (Leroy et al., 2009) and has similarly been found to be seropositive for antibodies against the SARS-CoV (Muller et al., 2007). Coincidentally, both Ebola virus and SARS-CoV share a similarity upon virus entry as both are dependent upon cathepsin-L cleavage in the endosomal compartment of the host cell (Schornberg et al., 2006, Simmons et al., 2005). It is intriguing to speculate about how similar mechanisms of virus infectivity between the two viruses may affect host susceptibility to infection, disease, and/or synergistic or competing co-infection between the reservoir and new host.
Yet another speculative hypothesis for potential synergy between two viruses has been eluded to in findings of co-infections between the Reston virus and the Arteriviruses, simian hemorrhagic fever virus (SHFV) in Cynomolgus macaques (Dalgard et al., 1992, Jahrling et al., 1990) and PRRSV in pigs (Barrette et al., 2009). Despite the historical evidence of a co-infection, in Cynomolgus macaques, it has been demonstrated that the Reston virus alone is sufficient to cause fatal illness (Jahrling et al., 1996). Pathogenicity of Reston virus is less clear in pigs, where Reston virus was found in diseased lung tissue of pigs, likely contributing to disease, but was also only found in co-infection with the PRRSV suggesting that Reston virus alone may not be sufficient to manifest serious clinical signs in pigs (Barrette et al., 2009).
4. Conclusions
From this review of the recent literature, it is evident that filoviruses are more prevalent and more diverse than was once thought. Findings such as the identification of diverging Ebola viruses in domestic swine further highlight this point along with the potential risks for the evolution of emerging infectious diseases at the close interface between humans, domestic animals, and wildlife. Along with the recent discoveries highlighted in this review, an increased interest in surveillance for emerging diseases and an increased use of high throughput next generation sequencing will continue to uncover new filovirus members and potential host reservoirs at an ever increasing rate. Such discoveries that enhance our understanding of the phylogeny of filoviruses coupled to interdisciplinary approaches to the study of emerging infectious diseases at the human, animal, plant and environmental interfaces will no doubt be essential to elucidating new reservoir hosts and potential risk factors for transmission of filovirus infections to humans and livestock.
Acknowledgements
The authors are grateful to Drs. Ian Lipkin and Gustavo Palacios at the Center for Infection and Immunity, Mailman School of Public Health, Columbia University for advice in preparation of this review. The authors are also grateful to Drs. Paul Hauer and William White of the USDA, APHIS, National Veterinary Services Laboratories for critical reading of the manuscript.
References
- Anon, 2009. Imported case of Marburg hemorrhagic fever – Colorado, 2008. MMWR Morb Mortal Wkly Rep 58, 1377–1381. [PubMed]
- Barrette R.W., Metwally S.A., Rowland J.M., Xu L., Zaki S.R., Nichol S.T., Rollin P.E., Towner J.S., Shieh W.J., Batten B., Sealy T.K., Carrillo C., Moran K.E., Bracht A.J., Mayr G.A., Sirios-Cruz M., Catbagan D.P., Lautner E.A., Ksiazek T.G., White W.R., McIntosh M.T. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- Basler C.F., Mikulasova A., Martinez-Sobrido L., Paragas J., Muhlberger E., Bray M., Klenk H.D., Palese P., Garcia-Sastre A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyi V.A., Levine A.J., Skalka A.M. Unexpected inheritance. Multiple integrations of ancient bornavirus and ebolavirus/marburgvirus sequences in vertebrate genomes. PLoS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgard D.W., Hardy R.J., Pearson S.L., Pucak G.J., Quander R.V., Zack P.M., Peters C.J., Jahrling P.B. Combined simian hemorrhagic fever and Ebola virus infection in cynomolgus monkeys. Lab. Anim. Sci. 1992;42:152–157. [PubMed] [Google Scholar]
- Emond R.T., Evans B., Bowen E.T., Lloyd G. A case of Ebola virus infection. Br Med J. 1977;2:541–544. doi: 10.1136/bmj.2.6086.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., Krokhin O., Wahl-Jensen V., Seebach J., Wolf K., Schnittler H.-J., Feldmann H. Structure-Function Analysis of the Soluble Glycoprotein, sGP, of Ebola Virus. ChemBioChem. 2006;7:1605–1611. doi: 10.1002/cbic.200600223. [DOI] [PubMed] [Google Scholar]
- Feldmann H., Muhlberger E., Randolf A., Will C., Kiley M.P., Sanchez A., Klenk H.D. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez J.P., Pourrut X., Leroy E. Ebolavirus and other filoviruses. Curr. Top. Microbiol. Immunol. 2007;315:363–387. doi: 10.1007/978-3-540-70962-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M., Honda T., Suzuki Y., Kobayashi Y., Daito T., Oshida T., Ikuta K., Jern P., Gojobori T., Coffin, Jahrling P.B., Geisbert T.W., Dalgard D.W., Johnson E.D., Ksiazek T.G., Hall W.C., Peters C.J. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet. 1990;335:502–505. doi: 10.1016/0140-6736(90)90737-p. [DOI] [PubMed] [Google Scholar]
- Jahrling P.B., Geisbert T.W., Jaax N.K., Hanes M.A., Ksiazek T.G., Peters C.J. Experimental infection of cynomolgus macaques with Ebola-Reston filoviruses from the 1989–1990 U.S. Epizootic. Arch Virol Suppl. 1996;11:115–134. doi: 10.1007/978-3-7091-7482-1_11. [DOI] [PubMed] [Google Scholar]
- Jahrling P.B., Geisbert T.W., Dalgard D.W., Johnson E.D., Ksiazek T.G., Hall W.C., Peters C.J. Preliminary report: isolation of Ebola virus from monkeys imported to USA. Lancet. 1990;335:502–505. doi: 10.1016/0140-6736(90)90737-p. [DOI] [PubMed] [Google Scholar]
- Kobinger G.P., Leung A., Neufeld J., Richardson J.S., Falzarano D., Smith G., Tierney K., Patel A., Weingartl H.M. Replication, Pathogenicity, Shedding, and Transmission of Zaire ebolavirus in Pigs. J. Infect. Dis. 2011;204:200–208. doi: 10.1093/infdis/jir077. [DOI] [PubMed] [Google Scholar]
- Kuhn J.H., Becker S., Ebihara H., Geisbert T.W., Johnson K.M., Kawaoka Y., Lipkin W.I., Negredo A.I., Netesov S.V., Nichol S.T., Palacios G., Peters C.J., Tenorio A., Volchkov V.E., Jahrling P.B. Proposal for a revised taxonomy of the family Filoviridae: classification, names of taxa and viruses, and virus abbreviations. Arch. Virol. 2010;155:2083–2103. doi: 10.1007/s00705-010-0814-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laminger F., Prinz A. Fledertiere und andere Reservoirwirte der <i>Filoviridae</i>. Epidemiegefahr am afrikanischen Kontinent?–Eine deduktive Literaturanalyse. Wien. Klin. Wochenschr. 2010;122:19–30. doi: 10.1007/s00508-010-1434-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Epelboin A., Mondonge V., Pourrut X., Gonzalez J.P., Muyembe-Tamfum J.J., Formenty P. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis. 2009;9:723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Licata J.M., Johnson R.F., Han Z., Harty R.N. Contribution of ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 2004;78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda, M.E., Ksiazek, T.G., Retuya, T.J., Khan, A.S., Sanchez, A., Fulhorst, C.F., Rollin, P.E., Calaor, A.B., Manalo, D.L., Roces, M.C., Dayrit, M.M., Peters, C.J., 1999. Epidemiology of Ebola (subtype Reston) virus in the Philippines, 1996. J. Infect. Dis. 179(Suppl 1), S115–S119. [DOI] [PubMed]
- Morikawa S., Saijo M., Kurane I. Current knowledge on lower virulence of Reston Ebola virus (in French: Connaissances actuelles sur la moindre virulence du virus Ebola Reston) Comp. Immunol. Microbiol. Infect. Dis. 2007;30:391–398. doi: 10.1016/j.cimid.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Muhlberger E. Filovirus replication and transcription. Future Virology. 2007;2:205–215. doi: 10.2217/17460794.2.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M.A., Paweska J.T., Leman P.A., Drosten C., Grywna K., Kemp A., Braack L., Sonnenberg K., Niedrig M., Swanepoel R. Coronavirus antibodies in African bat species. Emerg. Infect. Dis. 2007;13:1367–1370. doi: 10.3201/eid1309.070342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Halfmann, P., Sagara, H., Kawaoka, Y., 2007. Regions in Ebola virus VP24 that are important for nucleocapsid formation. J. Infect. Dis. 196(Suppl 2), S247–S250. [DOI] [PubMed]
- Peters, C. J., LeDuc, J. W., 1999. An introduction to ebola: The virus and the disease. J. Infect. Dis.179, Six–Sxvi. [DOI] [PubMed]
- Peterson A.T., Carroll D.S., Mills J.N., Johnson K.M. Potential mammalian filovirus reservoirs. Emerg. Infect. Dis. 2004;10:2073–2081. doi: 10.3201/eid1012.040346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnery R.L., Johnson K.M., Kiley M.P. Virion nucleic acid of Ebola virus. J. Virol. 1980;36:465–469. doi: 10.1128/jvi.36.2.465-469.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S.P., Leung L.W., Hartman A.L., Martinez O., Shaw M.L., Carbonnelle C., Volchkov V.E., Nichol S.T., Basler C.F. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 2006;80:5156–5167. doi: 10.1128/JVI.02349-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard O., Borowiak M., Volchkova V.A., Delpeut S., Mateo M., Volchkov V.E. Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. J. Virol. 2009;83:9596–9601. doi: 10.1128/JVI.00784-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquet P., Froment J.M., Bermejo M., Kilbourn A., Karesh W., Reed P., Kumulungui B., Yaba P., Delicat A., Rollin P.E., Leroy E.M. Wild animal mortality monitoring and human Ebola outbreaks, Gabon and Republic of Congo, 2001–2003. Emerg. Infect. Dis. 2005;11:283–290. doi: 10.3201/eid1102.040533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The Neighbor-Joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanchez A., Trappier S.G., Mahy B.W., Peters C.J., Nichol S.T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci U S A. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of Endosomal Cathepsins in Entry Mediated by the Ebola Virus Glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, R., Dayhoff, M., 1979. Matrices for detecting distant relationships. National Biomedical Research Foundation.
- Siegert R., Shu H.L., Slenczka W., Peters D., Muller G. On the etiology of an unknown human infection originating from monkeys. Dtsch. Med. Wochenschr. 1967;92:2341–2343. doi: 10.1055/s-0028-1106144. [DOI] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong J.E., Wong G., Jones S.E., Grolla A., Theriault S., Kobinger G.P., Feldmann H. Stimulation of Ebola virus production from persistent infection through activation of the Ras/MAPK pathway. Proc Natl Acad Sci U S A. 2008;105:17982–17987. doi: 10.1073/pnas.0809698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R., Leman P.A., Burt F.J. Experimental inoculation of plants and animals with Ebola virus. Emerg. Infect. Dis. 1996;2:321–325. doi: 10.3201/eid0204.960407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel R., Smit S.B., Rollin P.E., Formenty P., Leman P.A., Kemp A., Burt F.J., Grobbelaar A.A., Croft J., Bausch D.G. Studies of reservoir hosts for Marburg virus. Emerg. Infect. Dis. 2007;13:1847–1851. doi: 10.3201/eid1312.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the Neighbor-Joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.J., Leach R.W., Bruenn J. Filoviruses are ancient and integrated into mammalian genomes. BMC Evol. Biol. 2010;10:193. doi: 10.1186/1471-2148-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timen A., Koopmans M.P., Vossen A.C., van Doornum G.J., Gunther S., van den Berkmortel F., Verduin K.M., Dittrich S., Emmerich P., Osterhaus A.D., van Dissel J.T., Coutinho R.A. Response to imported case of Marburg hemorrhagic fever, the Netherland. Emerg. Infect. Dis. 2009;15:1171–1175. doi: 10.3201/eid1508.090051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Amman B.R., Sealy T.K., Carroll S.A.R., Comer J.A., Kemp A., Swanepoel R., Paddock C.D., Balinandi S., Khristova M.L., Formenty P.B.H., Albarino C.G., Miller D.M., Reed Z.D., Kayiwa J.T., Mills J.N., Cannon D.L., Greer P.W., Byaruhanga E., Farnon E.C., Atimnedi P., Okware S., Katongole-Mbidde E., Downing R., Tappero J.W., Zaki S.R., Ksiazek T.G., Nichol S.T., Rollin P.E. Isolation of Genetically Diverse Marburg Viruses from Egyptian Fruit Bats. PLoS Pathog. 2009;5:e1000536. doi: 10.1371/journal.ppat.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Pourrut X., Albarino C.G., Nkogue C.N., Bird B.H., Grard G., Ksiazek T.G., Gonzalez J.P., Nichol S.T., Leroy E.M. Marburg virus infection detected in a common African bat. PLoS ONE. 2007;2:e764. doi: 10.1371/journal.pone.0000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner J.S., Sealy T.K., Khristova M.L., Albarino C.G., Conlan S., Reeder S.A., Quan P.L., Lipkin W.I., Downing R., Tappero J.W. Newly discovered ebola virus associated with hemorrhagic Fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata S., Noda T., Kawaoka Y., Morikawa S., Yokosawa H., Yasuda J. Interaction of Tsg101 with Marburg virus VP40 depends on the PPPY motif, but not the PT/SAP motif as in the case of Ebola virus, and Tsg101 plays a critical role in the budding of Marburg virus-like particles induced by VP40, NP, and GP. J. Virol. 2007;81:4895–4899. doi: 10.1128/JVI.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valmas C., Grosch M.N., Schumann M., Olejnik J., Martinez O., Best S.M., Krahling V., Basler C.F., Muhlberger E. Marburg virus evades interferon responses by a mechanism distinct from ebola virus. PLoS Pathog. 2010;6:e1000721. doi: 10.1371/journal.ppat.1000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkov V.E., Becker S., Volchkova V.A., Ternovoj V.A., Kotov A.N., Netesov S.V., Klenk H.D. GP mRNA of Ebola virus is edited by the Ebola virus polymerase and by T7 and vaccinia virus polymerases. Virology. 1995;214:421–430. doi: 10.1006/viro.1995.0052. [DOI] [PubMed] [Google Scholar]
- Wahl-Jensen V.M., Afanasieva T.A., Seebach J., Stroher U., Feldmann H., Schnittler H.-J. Effects of Ebola Virus Glycoproteins on Endothelial Cell Activation and Barrier Function. J. Virol. 2005;79:10442–10450. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P.D., Biek R., Real L.A. Wave-Like Spread of Ebola Zaire. PLoS Biol. 2005;3:e371. doi: 10.1371/journal.pbio.0030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T.J., Biek R., Hassanin A., Rouquet P., Reed P., Yaba P., Pourrut X., Real L.A., Gonzalez J.P., Leroy E.M. Isolates of Zaire ebolavirus from wild apes reveal genetic lineage and recombinants. Proc. Natl Acad. Sci. USA. 2007;104:17123–17127. doi: 10.1073/pnas.0704076104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.Y., Duckers H.J., Sullivan N.J., Sanchez A., Nabel E.G., Nabel G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]


