Abstract
Environmental modifications are leading to biodiversity changes, loss and habitat disturbance. This in turn increases contacts between wildlife and hence the risk of transmission and emergence of zoonotic diseases. We analyzed the environment and land use using remote spatial data around the sampling locations of bats positive for coronavirus (21 sites) and astrovirus (11 sites) collected in 43 sites. A clear association between viruses and hosts was observed. Viruses associated to synanthropic bat genera, such as Myotis or Scotophilus were associated to highly transformed habitats with human presence while viruses associated to fruit bat genera were correlated with natural environments with dense forest, grassland areas and regions of high elevation. In particular, group C betacoronavirus were associated with mosaic habitats found in anthropized environments.
Keywords: Bat viruses, Cambodia, Lao PDR, South East Asia, Environmental analysis, Evolution of environment, Emerging diseases
Graphical abstract
Highlights
-
•
Deforestation and anthropization generate high diversity of bats.
-
•
Anthropized environment increases the risk of bat-borne virus transmission.
-
•
Highly pathogenic αCoV_2 & βCoV_C are linked to anthropogenic habitat.
-
•
Murine astroviruses are associated with human settlements.
-
•
Ungulates astroviruses are associated with wild areas (elevated grasslands).
1. Introduction
South-East Asia (SEA) is considered a hotspot for emerging infectious diseases (Jones et al., 2008). The region is undergoing major demographic and economic development with major impacts on environment and biodiversity (Sodhi et al., 2004). Outbreaks of Nipah virus infections and severe acute respiratory syndrome (SARS) pandemic in SEA both originated from bats (Chua et al., 2000, Wang et al., 2006, Field, 2009). SEA is hosting a high bat diversity and is habitat for 30% of the known global bat fauna (Kingston, 2010). In SEA, bats are often hunted for food (Mildenstein et al., 2016). In Cambodia, bat guano is collected on guano farms to be used as a plant fertilizer (Chhay, 2012).
Logging and conversion of forests into agricultural lands, monoculture plantations and urban areas have impacted the land cover configuration (Flint, 1994, Sodhi et al., 2004, DeFries et al., 2010). The Greater Mekong Subregion has lost 30% of its forest since 1970 (Costenbader et al., 2015). SEA is predicted to lose 75% of its original forest and 42% of its mammal species by 2100 (Sodhi et al., 2004). The deforestation rates in SEA are the highest in any tropical regions which could bring bats even closer to humans (Achard et al., 2002, Sodhi et al., 2004, Stibig et al., 2007).
Bats were discovered as a reservoir for the progenitor virus of the SARS-coronavirus (SARS-CoV) species, responsible for the SARS pandemic between 2002 and 2004 (Wang et al., 2006). Coronaviruses belonging to alpha-CoV and beta-CoV, including MERS-like viruses have been recently detected in Cambodia and Lao bat populations (Lacroix et al., 2017a). Several other viral families have also been detected, including astroviruses, flaviviruses, lyssaviruses, bunyaviruses or henipaviruses (Salaün et al., 1974, Olson et al., 2002, Osborne et al., 2003, Reynes et al., 2004, Reynes et al., 2005, Lacroix et al., 2017b).
Other works have shed light on how anthropogenic and environmental changes may impact the dynamic of virus transmission and public health (Calisher et al., 2006, Han et al., 2015). Bats are usually sensitive to environmental changes which can modify the dynamic of populations with a consequent impact on the risk of virus transmission. In order to investigate if bat viruses can be found in any kind of environment or are associated with specific landscapes, we conducted a multivariate analysis of isolated bat viruses, bats and environments previously described (Lacroix et al., 2017a, Lacroix et al., 2017b).
2. Materials and methods
2.1. Description of the study area
The study was carried out in Cambodia and Lao PDR which experience a rainy season from May to October, and a dry season from November to April. The thickly forested Lao PDR landscape consists of rugged mountains (up to 2800 m) surrounded by plains and plateaus. The Mekong River represents the “lifeline” of the country, and is the major economic corridor (Sisouphanthong and Taillard, 2000). In Cambodia, the landscape is characterized by a low-lying central plain including the Tonle Sap Lake and the upper areas of the Mekong River delta dominated by irrigated cultures. These areas are flooded during the rainy season (Wildlife, 2014). They are surrounded by uplands and low mountains, and thinly forested and transitional plains at around 200 m above sea level (Wildlife, 2014). Cambodia and Lao PDR experienced an important forest loss and fragmentation: 22% and 24% of their forest cover disappeared between 1973 and 2009, respectively (WWF, 2013). In 2010, the forest covered 57.2% and 77.2% of Cambodia and Lao PDR respectively (Food and Agriculture Organization, World Bank, 2016, World Development Indicators, 2016).
2.2. Bats collection
Bats were collected from 43 sites in Cambodia and Lao PDR between 2010 and 2013 as previously described (Lacroix et al., 2017a, Lacroix et al., 2017b) (Supplementary Table 1; Fig. 1). Sampling was conducted in two phases (Lacroix et al., 2017a, Lacroix et al., 2017b). Phase 1 was performed in 2010 by the Institut Pasteur in Cambodia (IPC). Bats were captured using harp traps, stored in Mist Net bags and humanely euthanized under supervision of the National Veterinary Research Institute in full compliance with local ethical and legal guidelines at the sampling sites in the Cambodian provinces of Ratanakiri, Stung Treng and Preah Vihear. Phase 2 of sampling was performed by the Wildlife Conservation Society (WCS), as part of USAID PREDICT project, from 2011 to 2013. Samples were taken at the human-bat interface and thus mostly from bats dedicated to human consumption, in markets, restaurants or directly from hunters. Fresh guano samples were also taken from guano farms in Cambodia. Geographical coordinates of each collection site were recorded using a Global Positioning System (GPS).
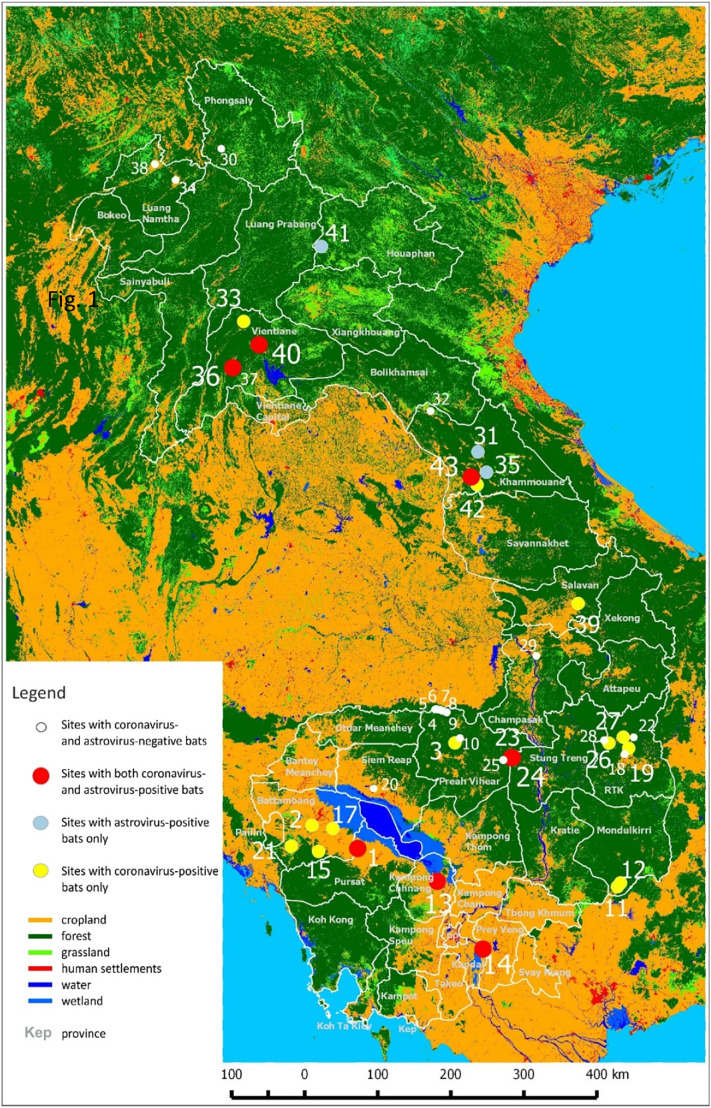
Fig. 1.
Location of the sampling points in Cambodia and Lao PDR.
White circle: Sites with coronavirus- and astrovirus-negative bats.
Red circle: Sites with both coronavirus- and astrovirus-positive bats.
Blue circle: Sites with astrovirus-positive bats only.
Yellow circle: Sites with coronavirus-positive bats only.
Dark green: Forest; light green: grassland; orange: cropland; red: human settlements; blue: wetland; dark blue: water. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. Detection of coronavirus and astrovirus
Viral RNA was extracted from rectal swabs, oral swabs and lung samples as previously described (Lacroix et al., 2017a, Lacroix et al., 2017b) (Table 1 ). The detection was based on broadly reactive reverse transcription-polymerase chain reaction (RT-PCR) assays, following previously described procedures (Chu et al., 2008, Watanabe et al., 2010).
Table 1.
Summary of the detection of astrovirus and coronavirus in bats collected from 43 sites in Lao PDR and Cambodia between 2010 and 2013.
| Country | Site | No of bat collected | No of bat tested (no of positive) | Bat genus positive | Coronavirus clustersa | No of bat tested (no of positive) | Bat genus positive | Astrovirus strainsb |
|---|---|---|---|---|---|---|---|---|
| Cambodia | 1 | 72 | 5 (1) | Sco | αCoV-1 | 72 (15) | Sco | Bat astV, cluster 8 astV |
| 2 | 24 | 24 (5) | Cyn, Mgl | βCoV-D1, βCoV-D2, βCoV-D3 | 24 (0) | – | ||
| 3 | 43 | 43 (4) | Cyn | βCoV-D2 | 43 (0) | – | ||
| 4 | 12 | 12 (0) | – | 12 (0) | – | |||
| 5 | 7 | 6 (0) | – | 7 (0) | – | |||
| 6 | 11 | 11 (0) | – | 11 (0) | – | |||
| 7 | 17 | 17 (0) | – | 17 (0) | – | |||
| 8 | 39 | 36 (0) | – | 39 (0) | – | |||
| 9 | 1 | 1 (0) | – | 1 (0) | – | |||
| 10 | 40 | 39 (0) | – | 39 (0) | – | |||
| 11 | 9 | 9 (1) | Pip | αCoV-1 | 9 (0) | – | ||
| 12 | 20 | 20 (1) | Pip | βCoV-C | 20 (0) | – | ||
| 13 | 47 | 50 (2) | Myo | αCoV-2 | 47 (20) | Myo | Bat astV | |
| 14 | 392 | 392 (28) | Sco | αCoV-1 | 390 (17) | Sco | Bat astV | |
| 15 | 112 | 112 (3) | Cyn | βCoV-D3 | 112 (0) | – | ||
| 16 | 6 | 6 (0) | – | 6 (0) | – | |||
| 17 | 53 | 53 (2) | Sco | αCoV-1 | 53 (0) | – | ||
| 18 | 17 | 17 (0) | – | 19 (0) | – | |||
| 19 | 28 | 28 (2) | Eon, Cyn | βCoV-D1, βCoV-D2 | 28 (0) | – | ||
| 20 | 16 | 16 (0) | – | 16 (0) | – | |||
| 21 | 7 | 7 (1) | Mgp | βCoV-D2 | 7 (0) | – | ||
| 22 | 14 | 14 (0) | – | 14 (0) | – | |||
| 23 | 34 | 34 (1) | Cyn | βCoV-D2 | 34 (2) | Mgd | Bat astV | |
| 24 | 59 | 59 (2) | Rhin, Rs | αCoV-4, βCoV-D2 | 59 (7) | Tph, Hip, Rhin, Rs | Bat astV, cluster7 astV | |
| 25 | 9 | 9 (0) | – | 9 (0) | – | |||
| 26 | 12 | 12 (4) | Cyn, Eon | βCoV-D1, βCoV-D2 | 12 (0) | – | ||
| 27 | 25 | 25 (4) | Cyn, Eon | βCoV-D1, βCoV-D2 | 25 (0) | – | ||
| 28 | 2 | 2 (0) | – | 2 (0) | – | |||
| Lao PDR | 29 | 3 | 3 (0) | – | 3 (0) | – | ||
| 30 | 2 | 2 (0) | – | 2 (0) | – | |||
| 31 | 32 | 32 (0) | – | 32 (1) | Ia | Bat astV | ||
| 32 | 3 | 3 (0) | – | 3 (0) | – | |||
| 33 | 281 | 281 (6) | Eon, Rs | βCoV-D1, βCoV-D3, βCoV-D4 | 131 (0) | – | ||
| 34 | 14 | 14 (0) | – | 14 (0) | – | |||
| 35 | 9 | 9 (0) | – | 9 (1) | Rs | Bat astV | ||
| 36 | 100 | 100 (3) | Eon, Rs | βCoV-D3 | 84 (9) | Eon, Rs | Bat astV, cluster7 astV | |
| 37 | 10 | 10 (0) | – | 10 (0) | – | |||
| 38 | 23 | 22 (0) | – | 23 (0) | – | |||
| 39 | 74 | 74 (2) | Hip | αCoV-4 | 33 (0) | – | ||
| 40 | 176 | 176 (17) | Eon, Rs | βCoV-D3, βCoV-D4 | 143 (12) | Eon, Rs, Rhin, Hip | Bat astV, cluster7 astV | |
| 41 | 107 | 105 (0) | – | 107 (3) | Rhin | Bat astV, cluster7 astV | ||
| 42 | 6 | 6 (1) | Rs | βCoV-D3 | 6 (0) | – | ||
| 43 | 29 | 29 (3) | Rs | βCoV-D3 | 29 (6) | Rs | Bat astV, cluster7 astV |
Hip: Hipposideros; Eon: Eonycteris; Rs: Rousettus; Rhin: Rhinolophus; Cyn: Cynopterus; Mgp: Megaerops; Mgl: Macroglossus; Mgd: Megaderma; Pip: Pipistrellus; Myo: Myotis; Sco: Scotophilus.
From the data previously presented in Lacroix et al. (2017a).
From the data previously presented in Lacroix et al. (2017b).
2.4. Environmental indices
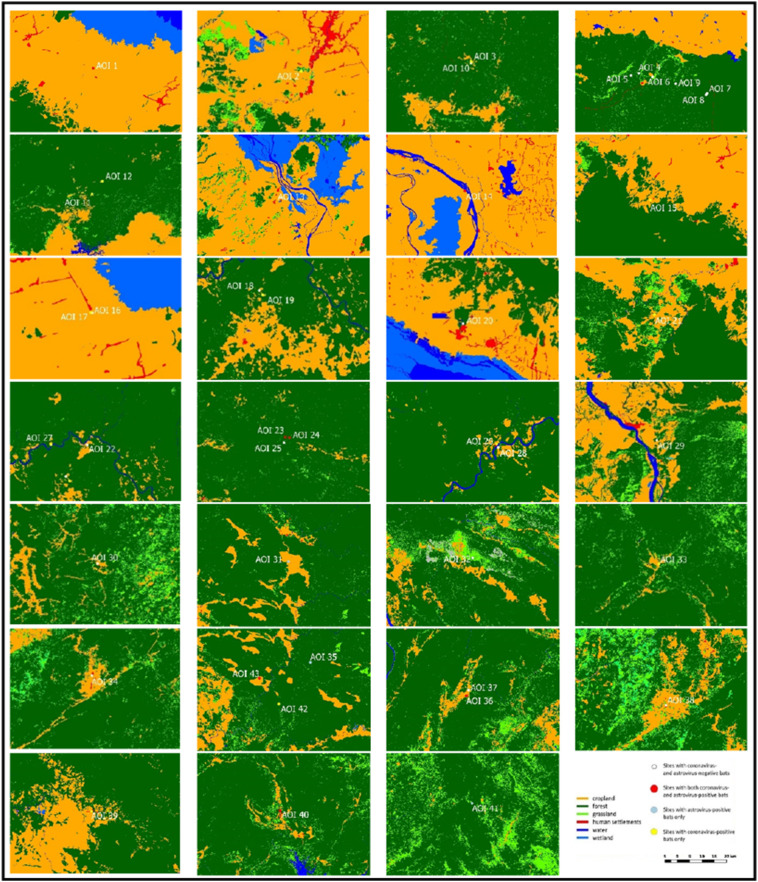
Land cover data were obtained from GlobeLand30 (GLC30) service operated by the National Geomatics Center of China (NGCC, 2014). Initial data were produced in 2010 and updated in 2014. Images were from Landsat8. Land cover map showed the main classes of land use as a synthesis of various material types, i.e. natural attributes and features on earth used for GlobeLand30 (GLC30) classification from 30 × 30 m multispectral satellite images. Schemes were merged into six land use categories: crop land, forest, grassland, wetland, water bodies and settlement areas. Elevation map was obtained from Shuttle Radar Topography Mission (SRTM) of the Consortium for Spatial Information (CGIAR-CSI). Most bats were obtained from markets or directly from hunters and have been hunted in surrounding areas (Lacroix et al., 2017a, Lacroix et al., 2017b). It was thus important to estimate the area of origin of these hunted bats. According to road connectivity and cost effectiveness, the distance between hunting and selling points was assumed to be less than 20 km. A 20-km radius was in agreement with the ecology and potential flying distances of the collected bats (Flemming and Eby, 2003, Kunz and Fenton, 2005, IUCN, 2017) and should then encompass the original environment of the hunted bats. In this study, this 1256 km2 (20 km radius) buffer zone was designated as “Area Of Interest (AOI)”. AOIs are shown in Fig. 2 . Data were extracted using the Patch Analyst extension in Geographical Information Systems (GIS) software ArcMap 10.3.1. Forest cover was examined more precisely in each AOI: 1) as a simple proportion of forest to each land cover type, 2) using the Shannon diversity index (SDI), 3) using the Fg fragmentation index defined as the ratio of the surface of the forest to the edge length of the forest (Fortin et al., 2005) and 4) using the ED edge density index which is the density of forest borders divided by the forest surface. Administrative data were obtained from the GADM database of Global Administrative Areas (version 2.8, November 2015). Main road networks in Cambodia and Lao PDR were obtained from the Digital Chart of the World (DCW) available on the Diva-gis online resource (www.diva-gis.org) and Google Earth. The network density in each AOI was assessed by measuring the total road length and the number of intersections. Connectivity index shows the density of roads connections for each AOI as the total length of the main roads (L) and the number of intersections (I). Connectivity = log (L ∗ l).
Fig. 2.
Closer landscape view of the areas of interest.
White circle: Sites with coronavirus- and astrovirus-negative bats.
Red circle: Sites with both coronavirus- and astrovirus-positive bats.
Blue circle: Sites with astrovirus-positive bats only.
Yellow circle: Sites with coronavirus-positive bats only.
Dark green: forest; light green: grassland; orange: cropland; red: human settlements; blue: wetland; dark blue: water. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.5. Statistical analysis
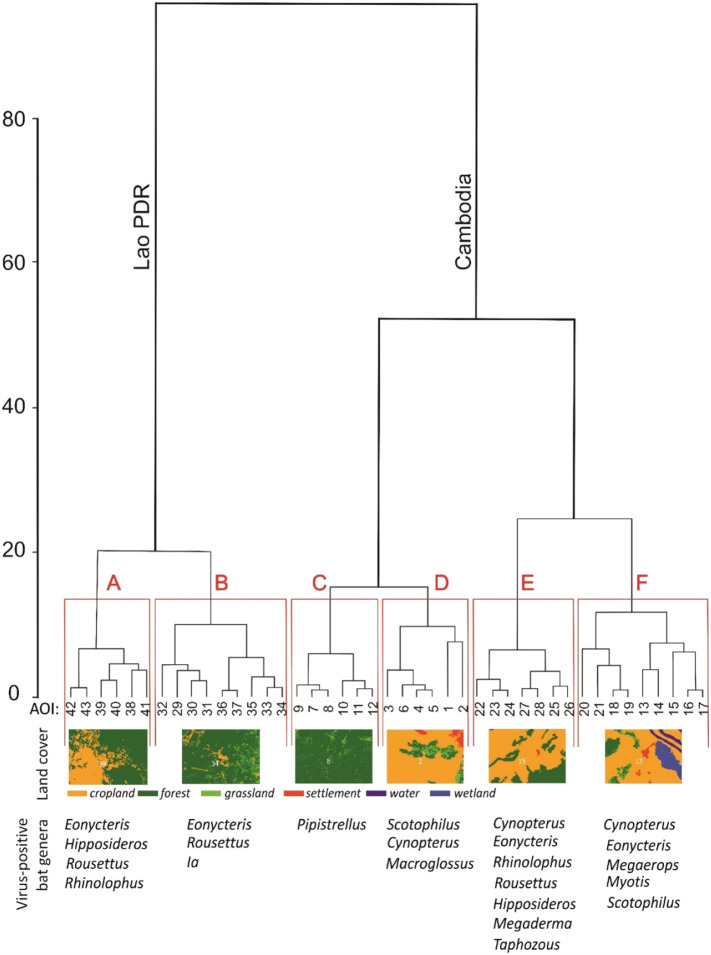
A hierarchical analysis, also known as cluster analysis, based on environmental parameters in each AOI was performed in order to describe the different type of habitats using the software R version 3.3.0 (www.r-project.org). The principle of hierarchical analysis is to build a binary tree of data that successively merges similar groups of points. From a statistical point of view each group is described by the index of similarity. Parameters used for hierarchical analysis were: Land cover [% forest cover, % settlement cover, % grassland, % surface water cover, % cropland cover and % wetland cover]; Mean elevation; and Connectivity. Similarity is described by the distance on the Y axis, in this work a Euclidean distance. As a first step, data are grouped into most similar pairs (expressed by a correlation index). As a second step, pairs are grouped into larger groups of similarity. In this work, Euclidean distances between 8 and 12 were yielded 6 independent clusters and each cluster comprised from 6 to 9 statistically similar AOIs. Principal Component Analyses (PCA) were performed using Statistica v.12. PCA analysis shows statistical, multidimension links between several variables and is used to emphasize variations. PCA requires statistically independent factors. The information is shown as a set of new orthogonal variables, the principal components (p). The pattern of similarity of the observed variables is displayed as points on a surface. In this case we used PCA to assess the correspondence of spatial environment and anthropogenic factors with respect to (a) astrovirus and coronavirus found on bats, and (b) to spatial environment and anthropogenic factors. As a first step all 11 factors corresponding to spatial analysis and considered in each AOI were assessed for correlation using a correlation matrix. Out of 11 factors considered, 3 were strongly correlated and dependent, i.e. forest, ED, SDI (r = 0.56–0.76). Out of these three factors, SDI (Shannon Index) provided a more complete information integrated both forest cover and density. SDI was therefore retained for PCA analysis while forest and ED were excluded from this analysis. PCA analysis was therefore performed with 9 environment-linked factors: SDI, Fragmentation, Connectivity, Settlements, Cropland, Wetland, Grassland, Water, and Elevation. PCA analysis was preceded by data normalization. Finally, three sets of supplementary variables were investigated, i.e. (i) bats sampled in each AOI, (ii) coronavirus isolated from sampled bats, and (iii) astrovirus isolated from sampled bats.
3. Results
3.1. Sampling sites
A total of 1997 bats were sampled, 1128 in Cambodia and 869 in Lao PDR at the interface between humans and bats (Table 1, Fig. 1, Fig. 2). For animals collected in the environment and in guano farms, the exact environmental description of the sampling location was available. For samples collected in restaurants and meat shops or directly from hunters, the exact original location of animals remained unknown but they were hunted for market trade near the selling place, i.e., mostly from areas at the border of deep forests, mixed agricultural zones with sparse forests or protected forest areas, close to water surfaces or in limestone karsts areas with mountain forests (Fig. 1) (Lacroix et al., 2017a, Lacroix et al., 2017b). Most investigated areas, i.e. 75% (31 sites) were located in lowland areas 500 m including 8 location under 150 m, in Cambodia. 7 AOI were located between 500 m and 1000 m, mostly in north Cambodia and Lao PDR (mostly rivers valleys) whereas 5 AOI were located above 1000 m in northern Lao PDR. AOI were described by nine statistically independent parameters: i.e. water cover, settlements cover, cropland cover, wetland cover, grassland cover, elevation, forest Shannon index, forest fragmentation index and connectivity index (Table 2 ). Hierarchical analysis based on these 9 parameters resulted into 6 clusters of landscape parametrization (Fig. 3 ). The main parameters for cluster characterization were: grass cover, cropland, Shannon index, fragmentation index and connectivity index. Cluster A included 6 AOIs displaying medium to high elevation, forest percentage of ca. 70–80%, low fragmentation, low water and wetland percentage and high connectivity index (2.5%–2.9%). Only one AOI (38) was negative for viruses. AOI 39, 40, 41, 42 and 43 were positive for coronavirus while only two sites (40 and 43) were positive for astrovirus. Cluster B comprised 9 AOIs (33, 35, 36), including four sites positive for at least one virus. In these areas, forest cover (80–95%) and Shannon index were higher than for cluster A and the human settlements cover was small or not detected in a 30 × 30 m resolution, indicating that villages were small and scattered. Both clusters A and B were located in Lao PDR. Euclidean distance indicated that clusters A and B segregated independently from the other 4 clusters (Fig. 3). These remaining clusters were characterized by low cropland cover, low human settlement cover and high Shannon index. Clusters C, D, E and F corresponded to AOI located in Cambodia. The cluster pairs C & D and E & F were the most similar. Cluster C included 6 AOIs with only two (11, 12) where only coronavirus were detected while no astrovirus was present. The landscape corresponded to 75 to 92% of forest cover, 1.5 to 2% of grass cover and a similar elevation (300 m above sea level). For both AOI 11 and 12, the connectivity index was high whereas the human settlement cover was less than 0.25% indicating that villages were small and scattered. Cluster D comprised 6 AOIs (1–6) displaying cultivated areas with high cropland cover and one of the highest settlement cover (7.2% for AOI 2). Three AOIs (1, 2, 3) were positive for astrovirus, including AOI 1 which displayed a high wetland cover and one of the highest cropland cover. Cluster E covered 7 AOIs (22–27) dominated by forest (85–98%) with the lowest connectivity index, one of the lowest forest fragmentation index and limited cultivated and grassland areas. The Shannon index was also among the lowest. Cluster E represented a wilderness parts of Cambodia. Cluster E also displayed the highest virus richness, i.e. 4 coronavirus sub-clusters and 5 astrovirus sub-clusters. The last cluster, F, comprised 9 AOIs (13–21) out of which 6 were positive for viruses (13, 14, 15, 17, 19, 21). All displayed mostly cultivated land with patches of forest, and the highest connectivity index and wetland cover, corresponding to highly transformed environments. The human settlement cover was rather low (0.1–3.5%) indicating that villages were concentrated in clusters. Cluster F also displayed the highest rate of coronavirus detection (37, including 28 αCoV_1 in AOI 14) and astrovirus (37 Bat_AstVs, located only in AOIs 13 and 14). Distances indicated that clusters E and F were the most divergent (Fig. 3). Another conclusion from this analysis is the higher bat biodiversity observed in Cambodia associated to more anthropized environment than in LAO PDR where dense forest was predominant (Fig. 3).
Table 2.
Variables used in the statistical analysis.
| Variable type | Variable | Name of variable | Description | Source |
|---|---|---|---|---|
| Active | Cultiv_land | Cropland | Percentage of crop lands in the AOI | GlobeLand30 (NGCC, 2014) |
| Wetland | Irrigated culture | Percentage of irrigated culture lands in the AOI | GlobeLand30 (NGCC, 2014) | |
| Grassland | Grassland | Percentage of grassland in the AOI | GlobeLand30 (NGCC, 2014) | |
| Water | Water bodies | Percentage of water in the AOI | GlobeLand30 (NGCC, 2014) | |
| Human_set | Human settlement | Percentage of human settlement in the AOI | GlobeLand30 (NGCC, 2014) | |
| SDI | Shannon index | Measure of forest patch diversity | GlobeLand30 (NGCC, 2014) | |
| Frag | Fragmentation | Ratio of perimeter/area of forest patches | GlobeLand30 (National Geomatics Center of China NGCC, 2014, Fortin et al., 2005) | |
| Elevation | Elevation | Average elevation of the AOI | Consortium for Spatial Information (CGIAR-CSI) | |
| Connectivity | Road connectivity | Density of roads connections for the AOI | Digital Chart of the World, www.diva-gis.org | |
| Supplementary (Set 1) | Eon | Eonycteris | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) |
| Cyn | Cynopterus | Bat genus collected in the AOI | ||
| (Lacroix et al., 2017a, Lacroix et al., 2017b) | ||||
| Hip | Hipposideros | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Ia | Ia | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Mgl | Macroglossus | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Mgd | Megaderma | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Mgp | Megaerops | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Myo | Myotis | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Pip | Pipistrellus | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Pt | Pteropus | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Rhn | Rhinolophus | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Rs | Rousettus | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Sco | Scotophilus | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Tph | Taphozous | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Tyl | Tylonycteris | Bat genus collected in the AOI | (Lacroix et al., 2017a, Lacroix et al., 2017b) | |
| Supplementary (Set 2) | bCoV_D1 | Beta-coronavirus from the subcluster D1 of the bCoV_D lineage | (Lacroix et al., 2017a) | |
| bCoV_D2 | Beta-coronavirus from the subcluster D2 of the bCoV_D lineage | (Lacroix et al., 2017a) | ||
| bCoV_D3 | Beta-coronavirus from the subcluster D3 of the bCoV_D lineage | (Lacroix et al., 2017a) | ||
| bCoV_D4 | Beta-coronavirus from the subcluster D4 of the bCoV_D lineage | (Lacroix et al., 2017a) | ||
| bCoV_C | Beta-coronavirus from the bCoV_C lineage | (Lacroix et al., 2017a) | ||
| aCoV_1 | Alpha-coronavirus from the subcluster 1 | (Lacroix et al., 2017a) | ||
| aCoV_2 | Alpha-coronavirus from the subcluster 2 | (Lacroix et al., 2017a) | ||
| aCoV_4 | Alpha-coronavirus from the subcluster 4 | (Lacroix et al., 2017a) | ||
| Supplementary (Set3) | Ung_AstV | Astroviruses from the cluster 7, including diverse mammal astV (ungulates, porcupine) | (Lacroix et al., 2017b) | |
| Mur_AstV | Astroviruses from the cluster 8, including murine astVs | (Lacroix et al., 2017b) | ||
| Bat_AstV | Astroviruses from the clusters 1, 2, 3, 4, 5, 6, including only bat astVs | (Lacroix et al., 2017b) | ||
Supplementary variables are shown in italic.
Fig. 3.
Hierarchical classification of landscapes and definition of areas of interest (AOI).
Orange: cropland; dark green: forest; light green: grassland; red: human settlements; blue: wetland; dark blue: water. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Landscape typology of virus-positive AOI
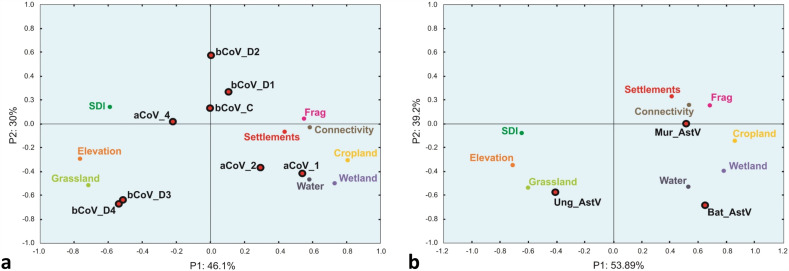
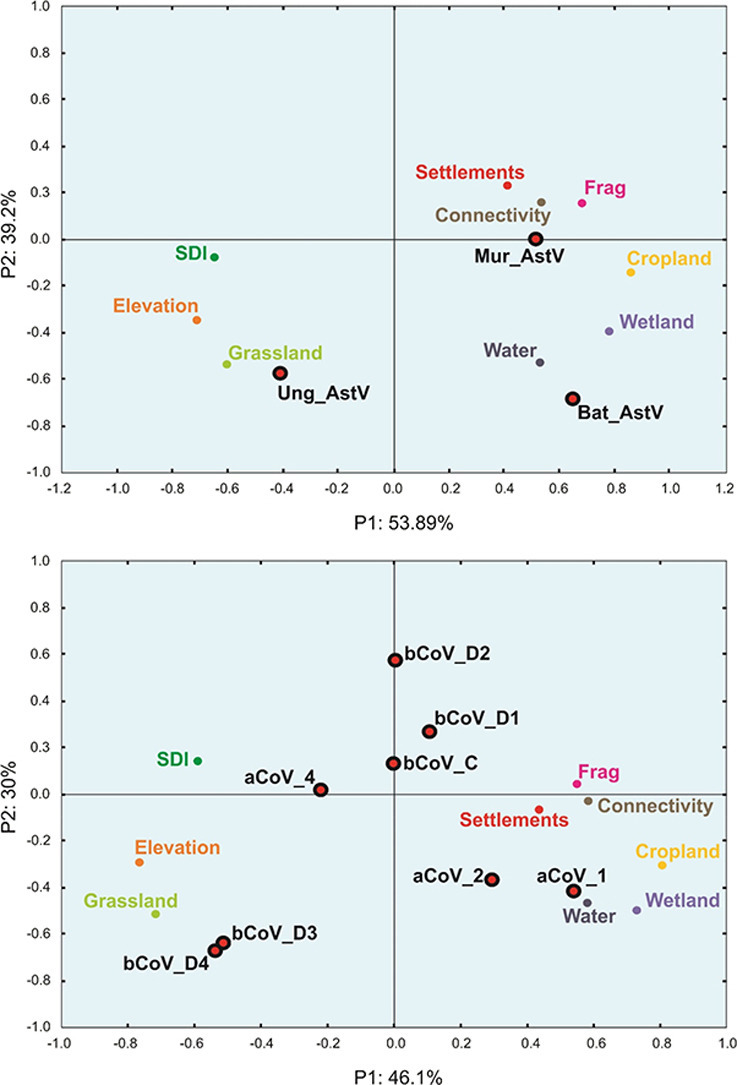
PCA were performed with the nine selected active independent variables (i.e. water cover, settlements cover, cropland cover, wetland cover, grassland cover, elevation, forest Shannon index (SDI), forest fragmentation index and connectivity index) for coronavirus subclusters (Fig. 4a) and for astrovirus subclusters (Fig. 4b). Owing to the fact that only 8 AOIs out 43 harbored bats positive for both viruses, the PCA relating to coronaviruses and astroviruses differed slightly. Although they were established with the same parameters, the positive sites being different, the values associated with these parameters were also slightly different. With respect to the coronavirus–positive bats (Fig. 4a), the first axis of the PCA (PC1) accounted for 46.1% of the total variability while the second axis (PC2) accounted for 30%, so a total variability of 76.1%. The contributions of the environmental variables to the construction of this axis were the highest for elevation, SDI and grassland which were correlated on the negative side and connectivity, forest fragmentation, cropland cover, water cover, wetland cover and settlement cover which were distributed on the positive side (Fig. 4a). The principal dimension of variability of the PCA opposed anthropogenic transformed lands on the positive side to more natural environments (i.e. forest, higher elevation areas and grassland areas, with low fragmentation) on the negative side (Fig. 4a). The second component of the PCA (PC2) accounted for 30% of the total variability. Fragmentation index, settlement cover and connectivity index were close to the axis and did not participate to the topology of the PCA. Grassland cover, water cover bodies, wetland cover, elevation and cropland cover participated the most to topology on the negative side. SDI was located on the positive side with a more limited weight (Fig. 4a). When considering the PCA representing the astrovirus-positive AOIs, contributions were slightly different (Fig. 4b). The first two axes contributed for a total of 93.09% with axis 1 representing 53.89% of the dispersion and axis 2 representing 39.2%. The distribution with respect to axis 2 was not affected and remained the same as for coronavirus-positive AOIs. The axis 1 displayed however a slightly different topology (Fig. 4b). SDI moved to the negative side but very close to axis 1 and therefore did not bear any weight in the analysis. Cropland cover remained in the negative side but closer to axis 1. Settlements cover, connectivity index and fragmentation index moved to the positive side to be moderately distant from the axis and gaining thus in representativeness (Fig. 4b).
Fig. 4.
Principal component analysis of the distribution of viruses with respect to environment types.
a. Distribution of coronaviruses.
b. Distribution of astroviruses.
3.3. Correlation of coronavirus and astrovirus with environmental parameters
The betacoronaviruses βCoV_D3 and βCoV_D4 were strongly associated with natural habitats and in particular with grassland and high elevation (Fig. 4a). The betacoronavirus clusters βCoV_D2 and, to a lower extent, βCoV_D1 and βCoV_C, seemed to be more correlated with the forest density gradient. βCoV-D2 was the most influenced by higher forest density (Fig. 4a). The alphacoronavirus αCoV_1 and αCoV_2 were strongly associated to anthropogenic environments with wetlands, whereas αCoV_4 was in contrary slightly more associated with more natural habitats (Fig. 4b). Astroviruses also displayed a clear differential distribution associated with environmental parameters. The Ung_AstV cluster was strongly associated to natural environments, in particular grassland and higher elevation, whereas the Mur_AstV cluster was strongly associated to an anthropogenic habitat (Fig. 4b). The Bat_AstV cluster was clearly associated to open areas close to water and wetlands (Fig. 4b).
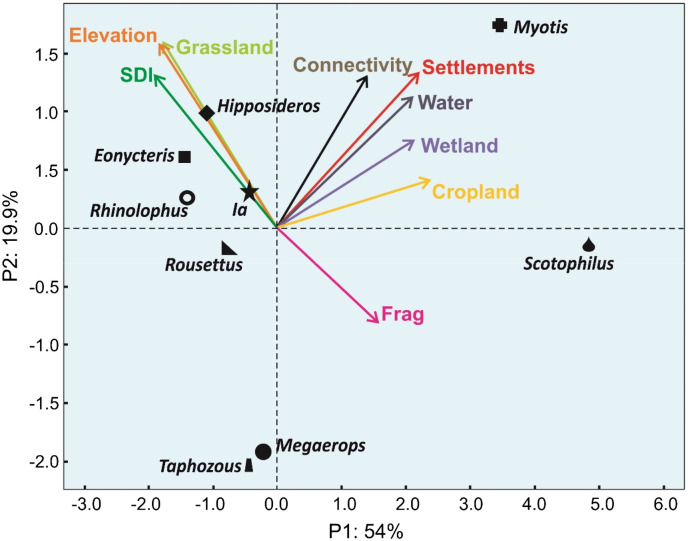
3.4. Correspondence between bat genera and environmental data
Bat genera displayed a strong correspondence with specific environmental parameters (Fig. 5 ). The two main axes of the PCA represented a total of 73.9% of inertia, with 54% and 19.9% for axis 1 and axis 2, respectively. The genus Myotis was strongly correlated with anthropogenic environments, water areas and wetlands. The distribution of Scotophilus was similarly influenced by forest fragmentation, water areas and anthropogenic environments. Conversely, the genera Hipposideros, Eonycteris, Ia and Rhinolophus were correlated with more natural habitats comprising dense forests and grassland area at higher elevation. This correlation was stronger for the former two. The genus Rousettus was close to the center, indicating the distribution of this genus was not strongly influenced by the parameters. Megaderma and Taphozous were strongly opposed to all environmental parameters analyzed indicating that their distribution was influenced by a distinct parameter not considered in this work.
Fig. 5.
Principal component analysis of the distribution of bats genera with respect to environment types.
4. Discussion
This work demonstrates both a clear distribution of bats and bat-borne viruses depending on environmental factors. For example, the Lesser Asian bat (Scotophilus) shows correlation with the anthropogenic, fragmented environments, close to water areas. The Mouse-eared bat (Myotis) also tends to be collected in anthropogenic areas around lakes or biggest regional river corridors regularly flooded. These observations are in accordance with their biology and ecology: Myotis and Scotophilus can adapt to many habitats but preferentially settle in anthropogenic habitats, in old buildings or crevasses. They rely on water bodies, which attract prey for these insectivorous bats (Bates et al., 2008b, Rosell-Ambal et al., 2008). Scotophilus bats are also reared in farms for their guano (Chhay, 2012). The presence of Hipposideros, Macroglossus and Rhinolophus, in environments with forest areas mixed with other categories of land cover (mosaic landscape) is also in accordance with their ecology (Bates et al., 2008a, Hutson et al., 2008, Walston et al., 2008). As a consequence, the two coronavirus sub-clusters αCoV_1 and αCoV_2, detected specifically in Scotophilus and Myotis respectively (Lacroix et al., 2017a) displayed the same trend, i.e. a high correlation with anthropogenic environment. Fruit and forest-dependent bat genera testing positive for viruses (Rousettus, Eonycteris) were correlated with an environment mostly consisting of densely forested areas at a higher elevation. The betacoronavirus clusters βCoV_D3 and βCoV_D4 which were found mostly in bats from the genera Rousettus and Eonycteris also correlated with the same type of environment. Finding coronavirus sub-clusters specific to a given bat genus in similar environment types indicates that the distribution of viruses is likely to follow the distribution of their hosts. The highest diversity of bats observed in anthropized environments emphasizes furthermore the risk associated with environmental changes. Deforestation and anthropization instead of leading to the elimination of bats generate conversely yield to a higher diversity. This might be explained by the complexity of the anthropized environments which offer opportunities to different groups of ubiquity bat species whereas natural environments might be more selective and suited for species with more strict ecological requirements. This higher biodiversity occurring by definition at the human interface, the risk of virus transmission is therefore increasing as well.
A particular attention has to be paid to the sub-cluster αCoV_2 and βCoV_C. These sub-clusters are composed by viruses which are highly pathogenic for pigs and humans respectively. The sub-cluster αCoV_2 comprises the coronaviruses responsible for the porcine epidemic diarrhea, which threatens pig farming activities around the world (Song et al., 2015, Lacroix et al., 2017a). βCoV_C comprises the zoonotic strain of coronavirus responsible for the Middle East Respiratory Syndrome (MERS) (Omrani et al., 2015). The sub-cluster αCoV_2 and lineage βCoV_C are linked to anthropogenic habitats in accordance with the presence of their hosts (Myotis and Pipistrellus, respectively) in the same environment (Csorba et al., 2008, Rosell-Ambal et al., 2008). The correspondence with host habitat is even more evident with astroviruses. Ungulate astrovirus found in ungulates and bats are associated with grasslands at higher elevation while murine astroviruses found in rodents and bats are associated with human settlements, indicating that the host bats are present in these same habitats. Bat astroviruses, found only in bats, are associated with water surface and irrigated cultures. This could be explained by the fact that irrigated cultures provide an important insect biomass and food resources for insectivorous bat populations which are harboring mainly arboviruses (Kunz and Fenton, 2005, Lacroix et al., 2017b).
Fragmentation of the habitat has been reported as a factor having a major importance in the spatial distribution of species. The distribution pattern of bat population has been found to also impact the viral richness of the host (Henle et al., 2004, Meyer et al., 2008, Gay et al., 2014). The fragmentation of bat populations was linked to a decrease in viral species richness (Gay et al., 2014). However, this work indicates that if fragmentation is an important parameter, it is not sufficient by itself to explain the distribution and must be considered jointly with the Shannon Diversity Index (SDI index) to yield a picture accurate enough. In this work, combining these parameters allowed us to clearly associate certain clusters of viruses to given environments and this is to our knowledge, the first time it is achieved in SEA. This correlation is a direct consequence of the association between bats hosting these viruses and specific environments. This work should thus be accompanied by the modeling of the modification of landscapes (Jung and Threlfall, 2016) over the next decades in order to assess and predict which part of the current biodiversity may increase or be at risk.
From a methodological standpoint, large sampling was not possible because of the status of protected species attached to the reservoir (IUCN, 2017). On another hand, avoiding the sampling of protected reservoirs for the sake of conservation and thus avoiding early detection and risk assessment is putting human populations at risk and contrary to public health policy (Morse, 2012, Lipkin and Anthony, 2015). In this work we considered both aspects at once. The limited sample size and limited quality of the bat material are a direct consequence of their status of protected species which was respected and enforced. Indeed, the bat sample collection was opportunistic, and mostly based on animals hunted by local populations (Lacroix et al., 2017a, Lacroix et al., 2017b). One of the consequences was the difficulty to reach the level of species identification but this could be easily achieved in the future by implementing procedures integrating molecular typing of bats (Clare et al., 2011, Korstian et al., 2016).
However, if the species level could not be reached with certainty in a significant part of the samples, except for Ia io, Pipistrellus coromandra and Megaerops niphanae, the ecology of the species present in South East Asia was similar within a given genus. The IUCN red list of threatened species provides the basis for a comparative analysis (IUCN, 2017). Scotophilus species found in South East Asia are reported to roost in crevices, cracks in walls, or roof of old buildings as well as leaves and crowns of palms, hollows of trees and among leaves of banana. This is in conformity with the analysis of the distribution of Scotophilus reported in this work as being influenced by forest fragmentation, water areas and anthropogenic environments. The genus Myotis was found in this work to strongly correlate with anthropogenic environments, water areas and wetlands. The ecology of Myotis species in South East Asia is matching this description (IUCN, 2017). M. annectans was found on a river valley, M. horsfieldii and M. ater are found near to water source and streams in lowland forest as well as disturbed forest and agricultural areas. M. rosseti is found in disturbed areas while M. siligorensis has been collected in lowland second growth forests over streams. M. pilosus is a fish-eating bat and therefore strictly dependent on water. We reported in this work that the genera Hipposideros, Eonycteris, Ia and Rhinolophus as correlating with more natural habitats comprising dense forests and grassland area at higher elevation. Hipposideros scutinares is known only from caves in limestone areas while H. cineraceus is roosting in hollows of trees in forests in South East Asia. H. galeritus is found in lowland forests as well as rubber plantations in Southeast Asia (IUCN, 2017). H. pomona ecology is not well known but in Southeast Asia it roosts essentially in caves. Similarly not much is known about H. rotalis which is considered to live in dry forests (IUCN, 2017). Eonycteris spelaea is found in caves in forested areas. However, it is a forest nectar eating bat which has adapted to agricultural and orchard crops (IUCN, 2017). With respect to Rhinolophus, similar features are observed. R. coelophyllus is found in forest, R. lepidus is associated with intact lowland tropical moist forest. R. malayanus is found in caves in secondary forest and degraded habitat. R. paradoxolophus was found in dry pine forest, while R. affinis is living in primary and secondary forest (IUCN, 2017). The two Rousettus species described in the IUCN database as present in Lao PDR and Cambodia, Rousettus amplexicaudatus and Rousettus leschenaultia, share the same ecology and roost in caves, old and ruined buildings and disused tunnels in forest, agricultural areas, disturbed habitats and at the forest edge. This matches the position described for Rousettus in this work. Finally, with respect to Taphozous theobaldi and Taphozous melanopogon, they both share the same forest habitat with roosting in caves or abandoned buildings and mines (IUCN, 2017). This short review of the ecology of the bats genera described in this work indicates that the species of a given genus share the same ecological traits and therefore that the lack of identification at the species is not impairing the conclusions on correlation between bat-borne viruses and environmental patterns.
Another potentially biasing aspect is that the estimated distance to the actual origin of the sampled bats was based on an assumption. This could have been underestimated or overestimated for some collection sites. Each bat genus is likely to present specificities in terms of migrations, flying distance and how they explore their environment (flights during days or nights, occurrences of flights for food search, etc.) (Smith et al., 2011). One must also take into account if species associated to similar environment shelter together. This will influence the presence and prevalence of bat-borne viruses which is not only a consequence of environmental change but also of ethology and ecology of bats. This can thus influence the occurrence of contacts with humans and should be further investigated in order to consider the specificity of each bat genus. However, this problem has been overcome with the definition of AOI considering both economic and ecological aspects. Another key methodological aspect is the resolution and quality of spatial data. This resolution must be adapted to the observed differing density and a 30 × 30 m resolution seems to be a good compromise between quality, definition, data access and calculation burden. Therefore, despite this limitation, the statistical approach implemented in this work allowed us to obtain the information needed on association with virus and environment. This can help designing prospective risk scenarios based on the expected evolution of landscape.
This brings us to the last aspect which is the possibility to model and develop scenarios of risk management. The prediction of landscape modification, climate change and anthropogenic behaviors (urbanization, roads, development of croplands, animal husbandry, etc.) can be achieved by economic planning and using GIS analysis. The association between environmental patterns and viral sub-clusters described in this work will therefore permit to develop a typology of situations at risk depending on (i) the virus involved, (ii) on the evolution of risk of contact, and (iii) the risk of emergence in the different situations which could arise. Preventive and protective actions could therefore be proposed to reduce this risk and hinder the evolution and development of hazardous contexts. This work is a preliminary step and deeper analyses and modeling must be further investigated. Nevertheless it provides opportunities for developing prospective approaches capable of managing protection of bats and wildlife while protecting human populations from potentially devastating emerging bat-borne viral diseases.
The following are the supplementary data related to this article.
Correspondence between names of the areas of interest (AOI), corresponding to the 43 collection sites described in the related articles (Lacroix et al., 2017a, Lacroix et al., 2017b).
Acknowledgments
Acknowledgements
A. Lacroix was supported by the USAID Emerging Pandemic Threats PREDICT project (cooperative agreement number GHN-A-OO-09-00010-00). We are very grateful to Philippe Dussart, Institut Pasteur in Cambodia, for his support.
Conflict of interest
Philippe Buchy is currently an employee of GSK vaccines.
References
- Achard F., Eva H.D., Stibig H.J., Mayaux P., Gallego J., Richards T., Malingreau J.P. Determination of deforestation rates of the world's humid tropical forests. Science. 2002;297:999–1002. doi: 10.1126/science.1070656. [DOI] [PubMed] [Google Scholar]
- Bates P., Bumrungsri S., Francis C., Csorba G. Hipposideros armiger. IUCN Red List Threat. Species 2008 ET10110A3162617. 2008. Available: (WWW Document) [DOI]
- Bates P., Kingston T., Francis C., Rosell-Ambal G., Heaney L., Gonzales J.C., Molur S., Srinivasulu C. Scotophilus kuhlii. IUCN Red List Threat. Species 2008 ET20068A9142479. 2008. Available: (WWW Document) [DOI]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhay S. Cambodian bats: a review of farming practices and economic value of lesser Asiatic yellow house bat Scotophilus kuhlii (Leach, 1821), in Kandal and Takeo Provinces, Cambodia. Camb. J. Nat. Hist. 2012;2012:164. [Google Scholar]
- Chu D.K.W., Poon L.L.M., Guan Y., Peiris J.S.M. Novel astroviruses in insectivorous bats. J. Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W.J., Goldsmith C.S., Gubler D.J., Roehrig J.T., Eaton B., Gould A.R., Olson J., Field H., Daniels P., Ling A.E., Peters C.J., Anderson L.J., Mahy B.W.J. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- Clare E.L., Lim B.K., Fenton M.B., Hebert P.D.N. Neotropical bats: estimating species diversity with DNA barcodes. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costenbader J., Broadhead J., Yasmi Y., Durst P.B. FAO, USAID and LEAF; Bangkok: 2015. Drivers Affecting Forest Change in the Greater Mekong Subregion (GMS): An Overview. [Google Scholar]
- Csorba G., Bates P., Furey N., Bumrungsri S., Molur S., Srinivasulu C. Pipistrellus coromandra (Coromandel Pipistrelle, Indian Pipistrelle, Little Indian Bat). IUCN Red List Threat. Species 2008 ET17335A6996860. 2008. Available: [DOI]
- DeFries R.S., Rudel T., Uriarte M., Hansen M. Deforestation driven by urban population growth and agricultural trade in the twenty-first century. Nat. Geosci. 2010;3:178–181. [Google Scholar]
- Field H.E. Bats and emerging zoonoses: henipaviruses and SARS. Zoonoses Public Health. 2009;56:278–284. doi: 10.1111/j.1863-2378.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- Flemming T.H., Eby P. Ecology of bat migration. In: Kunz T.H., Fenton M.B., editors. Bat Ecology. University of Chicago Press; Chicago: 2003. pp. 156–208. [Google Scholar]
- Flint E.P. Changes in land use in South and Southeast Asia from 1880 to 1980: a data base prepared as part of a coordinated research program on carbon fluxes in the tropics. Chemosphere. 1994;29:1015–1062. [Google Scholar]
- Food and Agriculture Organization, World Bank Forest area (% of land area) 2016. http://data.worldbank.org/indicator/AG.LND.FRST.ZS?view=chart Available:
- Fortin M.J., Keitt T.H., Maurer B.A., Taper M.L., Kaufman D.M., Blackburn T.M. Species' geographic ranges and distributional limits: pattern analysis and statistical issues. Oikos. 2005;108:7–17. [Google Scholar]
- Gay N., Olival K.J., Bumrungsri S., Siriaroonrat B., Bourgarel M., Morand S. Parasite and viral species richness of Southeast Asian bats: fragmentation of area distribution matters. Int. J. Parasitol. 2014;3:161–170. doi: 10.1016/j.ijppaw.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.J., Wen H.L., Zhou C.M., Chen F.F., Luo L.M., Liu J.W., Yu X.J. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. doi: 10.1016/j.virusres.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle K., Davies K.F., Kleyer M., Margules C., Settele J. Predictors of species sensitivity to fragmentation. Biodivers. Conserv. 2004;13:207–251. [Google Scholar]
- Hutson A.M., Suyanto A., Kingston T., Bates P., Francis C., Molur S., Srinivasulu C. Macroglossus sobrinus. IUCN Red List Threat. Species 2008 ET12595A3363666. 2008. Available: [DOI]
- IUCN IUCN Red List of Threatened Species. Version 2.1 < www.iucnredlist.org >. 2017. http://www.iucn.org/fr Available:
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Threlfall C.G. Urbanisation and its effects on bats—a global meta-analysis. In: Voigt C.C., Kingston T., editors. Bats in the Anthropocene: Conservation of Bats in a Changing World. Springer International Publishing; 2016. pp. 13–33. [Google Scholar]
- Kingston T. Research priorities for bat conservation in Southeast Asia: a consensus approach. Biodivers. Conserv. 2010;19:471–484. [Google Scholar]
- Korstian J.M., Hale A.M., Bennett V.J., Williams D.A. Using DNA barcoding to improve bat carcass identification at wind farms in the United States. Conserv. Genet. Resour. 2016;8:27–34. [Google Scholar]
- Kunz T.H., Fenton M.B. University of Chicago Press; 2005. Bat Ecology. [Google Scholar]
- Lacroix A., Duong V., Hul V., San S., Davun H., Omaliss K., Chea S., Hassanin A., Theppangna W., Silithammavong S., Khammavong K., Singhalath S., Greatorex Z., Fine A.E., Goldstein T., Olson S., Joly D.O., Keatts L., Dussart P., Afelt A., Frutos R., Buchy P. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix A., Duong V., Hul V., San S., Davun H., Omaliss K., Chea S., Hassanin A., Theppangna W., Silithammavong S., Khammavong K., Singhalath S., Afelt A., Greatorex Z., Fine A.E., Goldstein T., Olson S., Joly D.O., Keatts L., Dussart P., Frutos R., Buchy P. Diversity of bat astroviruses in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;48:41–50. doi: 10.1016/j.meegid.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin W.I., Anthony S.J. Virus hunting. Virology. 2015;479:194–199. doi: 10.1016/j.virol.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Meyer C.F.J., Fründ J., Lizano W.P., Kalko E.K.V. Ecological correlates of vulnerability to fragmentation in Neotropical bats. J. Appl. Ecol. 2008;45:381–391. [Google Scholar]
- Mildenstein T., Tanshi I., Racey P.A. Exploitation of bats for bushmeat and medicine. In: Voigt C.C., Kingston T., editors. Bats in the Anthropocene: Conservation of Bats in a Changing World. Springer International Publishing; 2016. pp. 325–375. [Google Scholar]
- Morse S.S. Public health surveillance and infectious disease detection. Biosecurity Bioterrorism Biodefense Strategy Pract. Sci. 2012;10:6–16. doi: 10.1089/bsp.2011.0088. [DOI] [PubMed] [Google Scholar]
- National Geomatics Center of China NGCC 30 meter Global Land Cover Dataset, product description. 2014. http://www.globallandcover.com/GLC30Download/index.aspx
- Olson J.G., Rupprecht C., Rollin P.E., An U.S., Niezgoda M., Clemins T., Walston J., Ksiazek T.G. Antibodies to Nipah-like virus in bats (Pteropus lylei), Cambodia. Emerg. Infect. Dis. 2002;8:987–988. doi: 10.3201/eid0809.010515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A.S., Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV): animal to human interaction. Pathog. Glob. Health. 2015;109:354–362. doi: 10.1080/20477724.2015.1122852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne J.C., Rupprecht C.E., Olson J.G., Ksiazek T.G., Rollin P.E., Niezgoda M., Goldsmith C.S., An U.S., Nichol S.T. Isolation of Kaeng Khoi virus from dead Chaerephon plicata bats in Cambodia. J. Gen. Virol. 2003;8:2685–2689. doi: 10.1099/vir.0.19294-0. [DOI] [PubMed] [Google Scholar]
- Reynes J.M., Molia S., Audry L., Hout S., Ngin S., Walston J., Bourhy H. Serologic evidence of lyssavirus infection in bats, Cambodia. Emerg. Infect. Dis. 2004;10:2231–2234. doi: 10.3201/eid1012.040459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynes J.M., Counor D., Ong S., Faure C., Seng V., Molia S., Walston J., Georges-Courbot M.C., Deubel V., Sarthou J.L. Nipah virus in Lyle's flying foxes, Cambodia. Emerg. Infect. Dis. 2005;11:1042–1047. doi: 10.3201/eid1107.041350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosell-Ambal G., Tabaranza B., Heaney L., Gonzales J.C., Molur S., Srinivasulu C. Myotis horsfieldii. IUCN Red List Threat. Species 2008 ET14166A4413659. 2008. Available: [DOI]
- Salaün J.J., Klein J.M., Hebrad G. A new virus, Phnom-Penh bat virus, isolated in Cambodia from a short-nosed fruit bat, Cynopterus brachyotis angulatus. Ann. Microbiol. Inst. Pasteur. 1974;125A:485–495. [PubMed] [Google Scholar]
- Save Cambodia's Wildlife . Save Cambodia's Wildlife; Phnom Penh: 2014. Atlas of Cambodia. Maps on Socio-economic Development and Environment. [Google Scholar]
- Sisouphanthong B., Taillard C. St. Martin's Press; 2000. Atlas of Laos: The Spatial Structures of Economic and Social Development of the Lao People's Democratic Republic. [Google Scholar]
- Smith A.C., Fahrig L., Francis C.M. Landscape size affects the relative importance of habitat amount, habitat fragmentation, and matrix quality on forest birds. Ecography. 2011;34:103–113. [Google Scholar]
- Sodhi N.S., Koh L.P., Brook B.W., Ng P.K.L. Southeast Asian biodiversity: an impending disaster. Trends Ecol. Evol. 2004;19:654–660. doi: 10.1016/j.tree.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Song D., Moon H., Kang B. Porcine epidemic diarrhea: a review of current epidemiology and available vaccines. Clin. Exp. Vaccine Res. 2015;4:166–176. doi: 10.7774/cevr.2015.4.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibig H.J., Stolle F., Dennis R., Feldkotter C. JRC Scientific and Technical Reports, EUR, 22896. 2007. Forest cover change in Southeast Asia–the regional pattern. [Google Scholar]
- Walston, J., Kingston, T., Hutson, A.M., 2008. Rhinolophus affinis. Red List Threat. Species 2008 ET19522A8952553. Available: e.T19522A8952553. Accessed September 21, 2016.
- Wang L., Shi Z., Zhang S., Field H., Daszak P., Eaton B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006;12:18–34. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Masangkay J.S., Nagata N., Morikawa S., Mizutani T., Fukushi S., Alviola P., Omatsu T., Ueda N., Iha K., Taniguchi S., Fujii H., Tsuda S., Endoh M., Kato K., Tohya Y., Kyuwa S., Yoshikawa Y., Akashi H. Bat coronaviruses and experimental infection of bats, the Philippines. Emerg. Infect. Dis. 2010;16:1217–1223. doi: 10.3201/eid1608.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Development Indicators World Development Indicators: poverty & equity. 2016. http://povertydata.worldbank.org/poverty/home/ Available:
- WWF . World Wide Fund For Nature, Greater Mekong; Bangkok, Thailand: 2013. Ecosystems in the Greater Mekong: Past Trends, Current Status, Possible Futures. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correspondence between names of the areas of interest (AOI), corresponding to the 43 collection sites described in the related articles (Lacroix et al., 2017a, Lacroix et al., 2017b).