Abstract
The role of cell-mediated immunity in human SARS-CoV infection is still not well understood. In this study, we found that memory T-cell responses against the spike (S) protein were persistent for more than 1 year after SARS-CoV infection by detecting the production of IFN-γ using ELISA and ELISpot assays. Flow cytometric analysis showed that both CD4+ and CD8+ T cells were involved in cellular responses against SARS-CoV infection. Interestingly, most of SARS-CoV S-specific memory CD4+ T cells were central memory cells expressing CD45RO+ CCR7+ CD62L−. However, the majority of memory CD8+ T cells revealed effector memory phenotype expressing CD45RO− CCR7− CD62L−. Thus, our study provides the evidence that SARS-CoV infection in humans can induce cellular immune response that is persistent for a long period of time. These data may have an important implication in the possibility of designing effective vaccine against SARS-CoV infection, specifically in defining T-cell populations that are implicated in protective immunity.
Keywords: SARS, SARS-CoV, S protein, IFN-γ, Cellular immune response, Memory T cells
Introduction
Severe acute respiratory syndrome (SARS) is a newly emerged infectious disease that led to thousands of human infections and hundreds of deaths. The etiologic agent of the syndrome, a novel coronavirus termed SARS-associated coronavirus (SARS-CoV) has been identified [1], [2], [3]. Although the epidemic was eventually controlled, the high morbidity and mortality make it extremely urgent to develop effective means to prevent this disease. Therefore, a successful vaccine against SARS-CoV remains a high priority. The unique opportunity of studying survivors of the infection allows for delineation of the T-cell populations implicated in the protective immunity, and those T cells would be the target population for expansion via a vaccine.
The spike (S) glycoprotein is a major structural protein of SARS-CoV and a potential target for SARS-specific humoral immunity and/or cell-mediated immune responses. Recent studies in animal models have demonstrated that vaccines based on the S protein of SARS-CoV seem to induce a considerable neutralizing antibody response [4], [5], [6], [7], [8] and provide protection via an inhibition of viral replication after challenge with live SARS-CoV [6], [7]. In addition, SARS-CoV-specific IgG antibody can be detected at week 3 after the onset of syndrome in SARS patients and persistent for a long period of time [9], [10]. Polyclonal immune sera from convalescent SARS patients were passively transferred to treat SARS patients during the outbreak in 2003 [11]. These findings revealed that humoral immunity may provide immediate protection against SARS-CoV infection in mice and humans.
In addition to humoral immune responses, the cell-mediated response likely plays a major role in eliminating virus. Although it has been demonstrated that DNA vaccine encoding the S protein of SARS-CoV can induce antigen-specific T-cell responses in animal models [4], [12], the protective effect of T cells specific for the S protein in humans remains undefined [13]. Thus, it is essential to determine whether recovered SARS patients maintained cell-mediated immunity against SARS-CoV and how long this immunity will persist. In addition, the phenotypic characteristics of memory CD4+ or CD8+ T cells specific for SARS-CoV have not been extensively studied. In this study, we analyzed CD4+ and CD8+ T-cell responses in peripheral blood of recovered SARS patients to a pool of 169 overlapping SARS-CoV S peptides and characterized the phenotype of memory T-cell subpopulations. Our study demonstrated that SARS-CoV S-specific memory CD4+ and CD8+ T cells in peripheral blood of recovered SARS patients were persistent for more than 1 year after infection. These memory T cells revealed both effector and central memory phenotypes.
Materials and methods
Donors
Eight fully recovered SARS patients, 5 male and 3 female, age 25 to 34 years, worked at the Third Affiliated Hospital of the Sun Yat-sen University, Guangzhou, China. All were diagnosed with the infection of SARS-CoV between March and April, 2003. Controls for the study were consisted of five healthy donors, 3 male and 2 female, age 27 to 33 years, without any contact history with SARS-CoV patients, and had no detectable antibodies specific to SARS-CoV in their sera.
Reagents
A total of 169 peptides spanning the entire SARS-CoV S protein (each peptide was 15–17 amino acids in length and overlapped by 10 amino acids) were kindly provided by the Vaccine Research Center, NIAID, National Institutes of Health, USA. Anti-CD4 PerCP and anti-CD8 PerCP were purchased from BD Bioscience Immunocytometry Systems (San Jose, CA, USA). Mouse anti-human CD28, anti-human CD49d, anti-CD4 FITC, anti-CD8 PE-Cy7, anti-CD45RO FITC, anti-CCR7 PE, anti-CD62L PE, anti-IFN-γ PE, anti-IFN-γ APC, anti-IL-2 APC and isotypes matched control antibodies were obtained from BD Bioscience Pharmingen (San Jose, CA, USA). RPMI 1640 medium, penicillin–streptomycin and 2-mercaptoethanol were purchased from GIBCO (Grand Island, NY, USA).
Cell preparation
Peripheral blood mononuclear cells (PBMCs) were isolated from heparinized venous blood that was mixed with an equal volume of Hank's balanced salt solution using Ficoll-Hypaque density gradient centrifugation. The cells were washed twice in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS) (HyClone), 100 U penicillin/ml, 100 μg streptomycin/ml and 50 μM 2-mercaptoethanol. The cells were finally adjusted to a concentration of 2 × 106/ml in complete RPMI 1640 medium.
Cell culture conditions and IFN-γ detection by ELISA
Fresh PBMCs were isolated from recovered SARS patients and healthy donors, suspended in complete RPMI 1640 medium and stimulated with a pool of 169 overlapping SARS-CoV S peptides in the presence of 1 μg/ml anti-CD28 and 1 μg/ml anti-CD49d mAbs. The final concentration of each peptide was 1 μg/ml. PBMCs were plated into a flat-bottomed 96-well plate at a total of 2 × 105 cells/well in triplicate and incubated for 72 h at 37°C with 5% CO2. The supernatants were harvested and assayed for IFN-γ production by ELISA according to the manufacturer's protocol (BD Pharmingen, San Diego, CA). In each experiment, PBMCs were cultured with 1 μg/ml anti-CD28 plus 1 μg/ml anti-CD49d in the absence of SARS-CoV S peptides as a negative control. In addition, the cells were stimulated with 20 ng/ml phorbol-12-mristate-13-acetate (PMA) (Sigma, St. Louis, MO, USA) and 1 μg/ml ionomycin (Sigma) as a positive control.
IFN-γ ELISpot assay
Human IFN-γ ELISpot assays were performed using a commercially available set (BD Pharmingen) according to the manufacturer's instructions. Briefly, 96-well nitrocellulose plates (Millipore) were coated overnight at 4°C with 5 μg/ml anti-human IFN-γ capture antibody. The wells were then washed and blocked for 2 h at room temperature with 10% FCS–RPMI 1640 medium. A total of 2 × 105 PBMCs were added to microwell in triplicate and incubated for 20 h with a pool of SARS-CoV S peptides (1 μg/ml), 1 μg/ml anti-CD28 and 1 μg/ml anti-CD49d mAbs. Similarly, the cells supplemented with anti-CD28 (1 μg/ml) and anti-CD49d (1 μg/ml) mAbs in the absence of SARS-CoV S peptides were considered as negative controls, and the cells were cultured with polyclonal stimuli (20 ng/ml PMA plus 1 μg/ml ionomycin) as positive controls. The plates were washed and subsequently incubated with 2 μg/ml biotinylated anti-human IFN-γ detection antibody for 2 h at room temperature. After washing, wells were developed for another 1 h with streptavidin–HRP followed by addition with AEC substrate reagent. Spot development was monitored, and substrate reaction was stopped by washing wells with water. The spots were counted automatically using Immunospot Image Analyzer (Cellular Technology Ltd, USA). A spot represents an IFN-γ-producing cell. The average of spots in triplicate wells was calculated and considered as the number of specific spot-forming cells (SFC)/2 × 105 cultured PBMCs.
Cell surface, intracellular cytokine staining and flow cytometric analysis
PBMCs from recovered SARS patients and healthy individuals were stimulated with or without a mixture of 169 overlapping SARS-CoV S peptides in the presence of 1 μg/ml anti-CD28 and 1 μg/ml anti-CD49d mAbs in 15 ml tubes. After the first 1 h incubation, brefeldin A (Sigma) at a final concentration 10 μg/ml was added to cultures to enable intracellular protein to accumulate in all stimulations. After incubation for a total 5 h, the cells were washed, fixed, permeabilized using saponin (Sigma) and blocked with 25 μg/ml human IgG for 30 min at 4°C. The cells were then stained with anti-CD4, anti-CD8, anti-IFN-γ, anti-IL-2, anti-CD45RO, anti-CCR7 and anti-CD62L. After staining, all samples were washed twice in PBS buffer (containing 0.1% saponin, 0.1% BSA and 0.05% NaN3) and resuspended in 300 μl PBS for flow cytometry. More than 600,000 cells were acquired on a FACS Aria flow cytometer (Becton Dickinson, San Jose, CA), and FACS data were analyzed using CellQuest software (Becton Dickinson, San Jose, CA).
Statistical analyses
The levels of IFN-γ and numbers of IFN-γ-producing cells were compared respectively using Student's t test between recovered SARS patients and healthy donors in the same condition. Value of P < 0.05 was considered significant.
Results
Quantification of SARS-CoV S-peptide-specific memory T lymphocytes
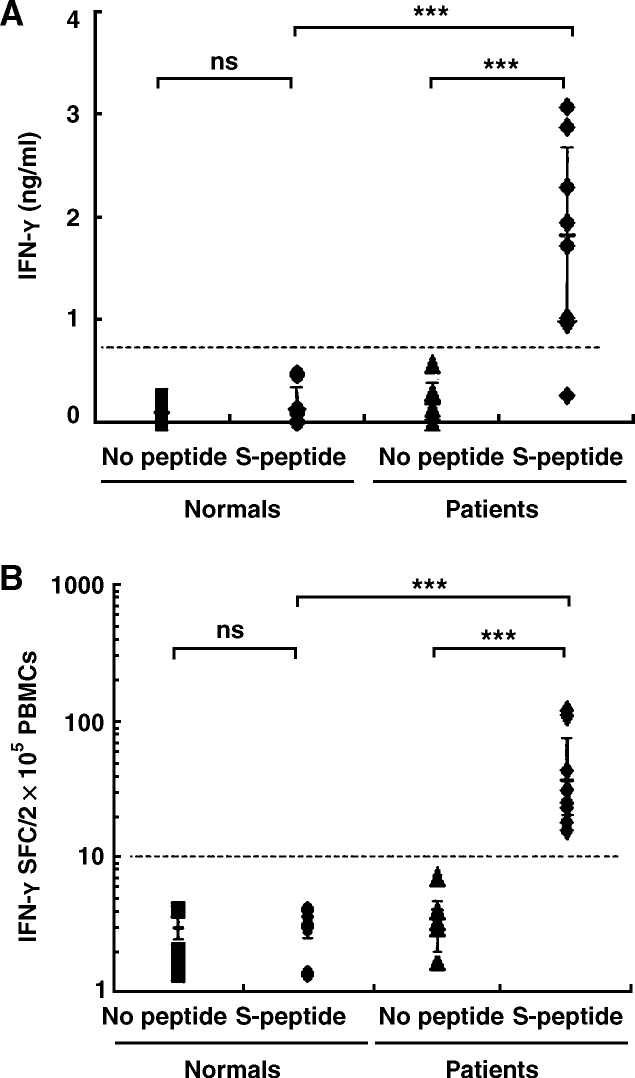
We first focused on understanding whether recovered SARS individuals could generate and maintain memory T-cell responses against the S protein of SARS-CoV more than 1 year after SARS-CoV infection. Fresh PBMCs were isolated from fully recovered SARS patients and healthy donors and cultured in vitro with or without a pool of 169 peptides spanning the entire S protein of SARS-CoV in the presence of 1 μg/ml anti-CD28 and 1 μg/ml anti-CD49d mAbs. The final concentration of each peptide was 1 μg/ml. The PBMCs were cultured with polyclonal stimuli (20 ng/ml PMA plus 1 μg/ml ionomycin) as a positive control. After stimulation for 72 h, cell-free culture supernatants were harvested and assayed for the production of IFN-γ by ELISA. The results showed that PBMCs from five healthy donors secreted very low levels of IFN-γ (0.10–0.13 ng/ml) when cultured either with or without the SARS-CoV S peptides. Statistic analysis indicated that the production of IFN-γ between these two culture conditions was not significantly different (P > 0.05) (Fig. 1A). In contrast, PBMCs from eight recovered SARS individuals produced significantly higher levels of IFN-γ (1.77 ± 0.98 ng/ml) when stimulated with SARS-CoV S peptides (P < 0.01) compared with the levels of IFN-γ from the cells cultured without SARS-CoV S peptides (0.19 ± 0.17 ng/ml) (Fig. 1A). In addition, no markedly difference in the production of IFN-γ between recovered patients and healthy individuals was observed when the PBMCs were stimulated with PMA plus ionomycin (P > 0.05) (data not shown).
Figure 1.
SARS-CoV S-peptide-specific IFN-γ production by PBMCs. Fresh PBMCs were isolated from fully recovered SARS patients and normal individuals and cultured in vitro with or without a pool of 169 SARS-CoV S peptides in the presence of anti-CD28 (1 μg/ml) and anti-CD49d mAbs (1 μg/ml). After incubation for 72 h, the level of IFN-γ in culture supernatants was assayed by ELISA (A). The frequency of IFN-γ-secreting cells was detected by ELISpot assay at the single cell level after stimulation for 20 h (B). Student's t test was used for statistical analysis. The significance was set at P < 0.01 (***). ns, not significant.
To determine the frequency of SARS-CoV S-specific T cells at the single cell level, fresh PBMCs isolated from recovered SARS patients and uninfected healthy individuals were cultured in the presence or absence of SARS-CoV S peptides. The frequency of IFN-γ-producing cells was detected by ELISpot assay after stimulation for 20 h. As shown in Fig. 1B, 2–5 IFN-γ-positive spots in 2 × 105 PBMCs were detected in cells without stimulation, either in healthy donors or recovered SARS individuals. The SARS-CoV S peptides, however, elicited a strong antigen-specific T-cell response in recovered SARS patients with an average number of spot-forming cells (SFC) 49/2 × 105 PBMCs (range 18 to 109) (Fig. 1B), which were significantly higher (P < 0.01) than spot-producing cells in healthy donors (1–6 spots in 2 × 105 PBMCs) when PBMCs were stimulated with SARS-CoV S peptides. Moreover, the PBMCs from recovered SARS patients revealed similar frequency of IFN-γ-forming cells (328/2 × 105 PBMCs) compared with the PBMCs from healthy individuals (299/2 × 105 PBMCs) when stimulated with PMA plus ionomycin (P > 0.05) (data not shown). Taken together, SARS-CoV S-specific memory T cells were persistent in peripheral blood of recovered SARS individuals and displayed the capacity to secrete IFN-γ upon subsequent stimulation with SARS-CoV S peptides in vitro.
Subpopulation of SARS-CoV S-peptide-specific memory T cells
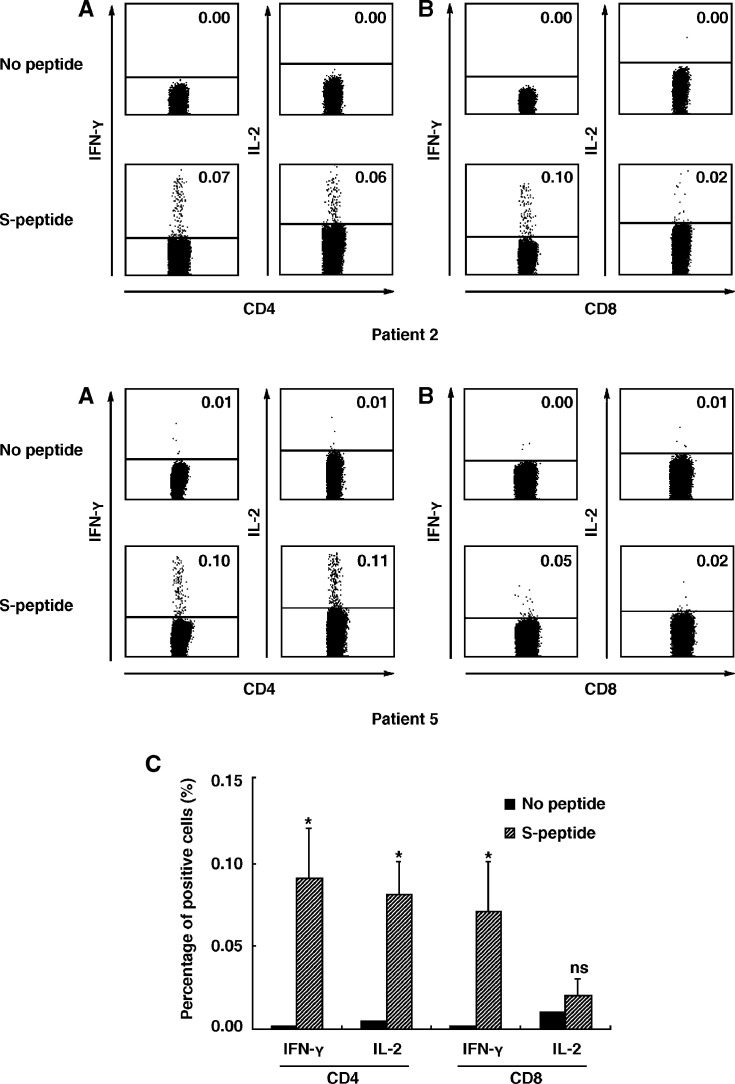
We next analyzed the subpopulation of IFN-γ-producing T cells by flow cytometry. PBMCs were isolated and stimulated with a pool of SARS-CoV S peptides (1 μg/ml), 1 μg/ml anti-CD28 and 1 μg/ml anti-CD49d mAbs in the presence of 10 μg/ml brefeldin A. After stimulation for 5 h, the cell surface markers and intracellular cytokines were stained with anti-CD4 FITC, anti-CD8 PerCP, anti-IFN-γ PE, anti-IL-2 APC and their corresponding isotype controls. The expression of IFN-γ and IL-2 was assessed when cells were gated on CD4+ or CD8+ T cells. The results showed that both CD4+ and CD8+ T cells from 5 recovered SARS patients failed to secret IFN-γ and IL-2 in the absence of SARS-CoV S peptides. However, addition of SARS-CoV S peptides to cultures resulted in the expression of IFN-γ and IL-2 in both CD4+ and CD8+ T-cell populations. Two representative examples, patient 2 and patient 5, are shown in Figs. 2A and B. The percentage of IFN-γ and IL-2-producing CD4+ and CD8+ T cells from 5 recovered SARS patients in SARS-CoV S-peptide-stimulated cultures was significantly higher than that from unstimulated cells (P < 0.05) (Fig. 2C), whereas no difference was observed in the proportion of IL-2-producing CD8 T cells between those two groups (P > 0.05). Taken together, the results demonstrated that both memory CD4+ and CD8+ T cells participated in cellular immune responses to SARS-CoV S antigen in humans.
Figure 2.
Expression of intracellular IFN-γ and IL-2 in PBMCs of recovered SARS patients after stimulation with SARS-CoV S peptides in vitro. Fresh PBMCs from recovered SARS patients were cultured for 5 h in vitro with a mixture of 169 SARS-CoV S peptides and stained with anti-CD4, CD8, IFN-γ and IL-2 mAbs. The expression of IFN-γ and IL-2 on CD4+ (A) and CD8+ (B) T cells was analyzed. The numbers in each quadrant were indicated as percentage. Data were representative of two of five recovered SARS individuals with similar results. The percentage (mean ± SE) of IFN-γ or IL-2-secreting cells from 5 independent experiments was shown within CD4 or CD8 T cells (C). Student's t test was used for statistical analysis. Statistical significance was set at P < 0.05 (*), ns, not significant.
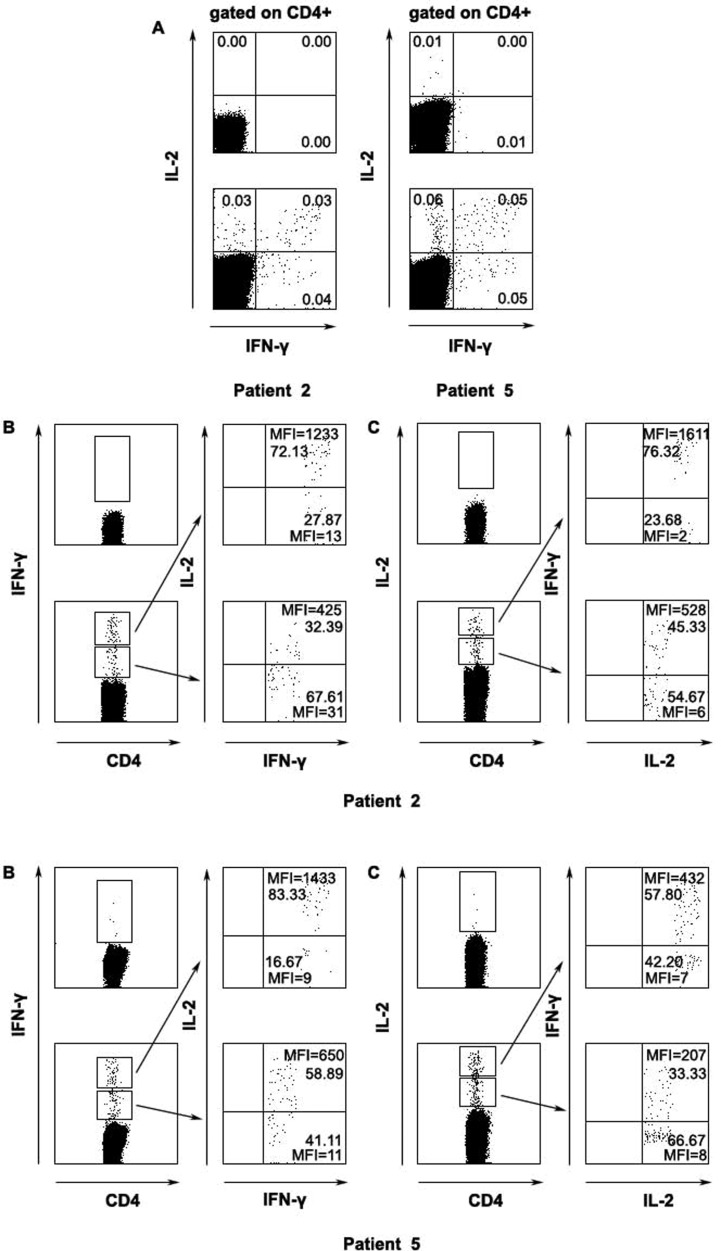
Correlation of the expression of IFN-γ and IL-2 by CD4 T lymphocytes
We next analyzed the expression of IFN-γ and IL-2 in CD4+ T cells following stimulation of PBMCs from SARS recovered patients with a pool of SARS-CoV S peptides. Based on the expression of IFN-γ and IL-2, antigen-specific CD4+ T cells could be divided into three distinct subpopulations: IFN-γ+ IL-2−, IFN-γ+ IL-2+ and IFN-γ− IL-2+ (two examples, patient 2 and patient 5, shown in Fig. 3A). We further analyzed the simultaneous expression of high or low levels of IFN-γ and IL-2 at the single cell (two examples, patient 2 and patient 5, shown in Figs. 3B, C). A total of four patients have been assessed for cytokine production. A representative of patient 2 where approximately 61–72% CD4+ IFN-γhigh cells showed high level expression of IL-2 and 67–81% CD4+ IFN-γlow cells was IL-2low. 64–76% of CD4+ IL-2high displayed phenotype of IFN-γhigh and 55–65% CD4+ IL-2low were IFN-γlow. Similar tendency was also found in the mean fluorescence intensity (MFI). However, in patient 5, IL-2low cells accounted for only 41% of CD4+ IFN-γlow but still showed similar results as seen in other 4 patients. These results suggested that the majority of IFN-γ+ IL-2+ T cells were confined to IFN-γhigh IL-2high subset.
Figure 3.
The correlation of intensity of IFN-γ and IL-2 within SARS-CoV S-specific CD4 T cells. The subsets of SARS-CoV S-specific CD4+ T cells based on the expression of IFN-γ and IL-2 (A), the expression of IL-2 in both IFN-γhigh or IFN-γlow subpopulations (B) and frequency as well as intensity of IFN-γ-producing cells in IL-2high and IL-2low subsets (C) (A, top panels, unstimulated cells; bottom panels, S-peptide-stimulated cells; B–C, left panels top quadrant, unstimulated cells; left panels bottom quadrant, S-peptide-stimulated cells). Percentage of indicated cells and the mean fluorescent intensity (MFI) were shown. Data were representative of two of five recovered SARS individuals.
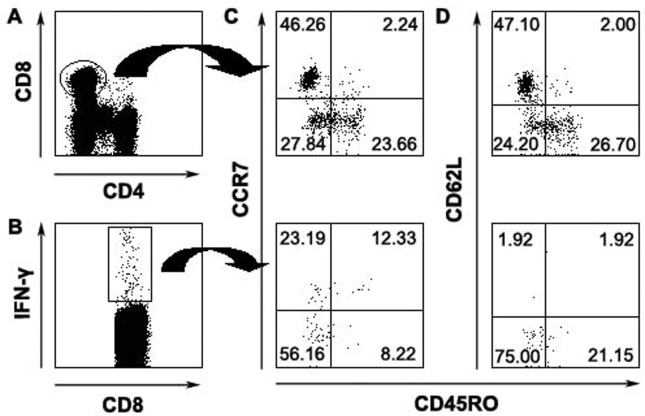
Characterization of memory CD4+ and CD8+ T cells specific for SARS-CoV S peptides
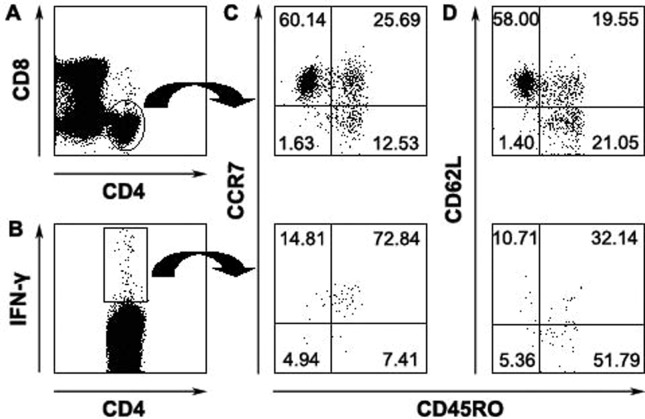
With regard to memory CD4+ and CD8+ T cells in humans, multiple phenotypes and a broad spectrum of functions have been observed in different viral infections. In this study, we performed phenotypic characterization of antiviral T-cell responses specific for SARS-CoV S peptides on the basis of their ability to secrete IFN-γ and the expression of CD45RO, CCR7 and CD62L. Consistent with previous reports, our results indicated that the majority of total CD4+ T cells (Fig. 4A) expressed high levels of CCR7 (85%) (Fig. 4C) and CD62L (77%) (Fig. 4D), whereas approximately 40% of CD4+ T cells expressed CD45RO (Figs. 4C, D). SARS-CoV S-specific IFN-γ-producing CD4+ T cells (Fig. 4B) had an increased expression of CD45RO (Fig. 4C) but had decreased expression of CD62L (Fig. 4D) compared with total CD4+ T cells. Furthermore, the majority of antigen-specific CD4+ IFN-γ+ cells were CD45RO+ CCR7+ (Fig. 4C) or CD45RO+ CD62L− (Fig. 4D). In contrast, a total of CD8+ T cells (Fig. 5A) expressed low level of CCR7 (Fig. 5C) and CD62L (Fig. 5D) compared with total CD4+ T cells. Moreover, IFN-γ-positive CD8+ T cells (Fig. 5B) were predominantly CD45RO− (78%) and mainly defined as CD45RO− CCR7− CD62L− (Figs. 5C, D). These results emphasized the heterogeneity of memory T cells and also demonstrated difference between memory CD4+ and CD8+ T-cell populations.
Figure 4.
Phenotypic analyses of memory CD4 T cells specific for SARS-CoV S peptides. Total CD4+ (A) or CD4+ IFN-γ+ cells (B) were analyzed for the expression of CD45RO and CCR7 (C) or CD45RO and CD62L (D). The numbers in each quadrant were indicated as percentage. Data were representative for 3 separate experiments with similar results.
Figure 5.
Characteristics of SARS-CoV S peptides-specific memory CD8 T cells. Total CD8+ (A) or CD8+ IFN-γ+ (B) were analyzed for the expression of CD45RO and CCR7 expression (C) or CD45RO and CD62L (D). Data were representative of 1 of 3 independent experiments with similar results.
Discussion
In the current study, we found that antigen-specific memory T cells were capable of secreting high levels of IFN-γ by ELISA assay upon stimulation in vitro with a pool of SARS-CoV S peptides. These memory T cells persisted for more than 1 year after recovery in peripheral blood of SARS individuals. These data were further confirmed by IFN-γ ELISpot assay showing a high frequency of SARS-CoV S-specific IFN-γ-producing T cells in recovered SARS patients. Our results are consistent with the report that two newly identified HLA-A*0201-restricted CD8+ T-cell epitopes from SARS-CoV S protein, S1203 and S978, elicit memory T-cell responses in PBMCs from HLA-A*0201+ recovered SARS patients up to 3 months post-infection [13], [14].
To date, most studies have been focused on mapping of CD8+ T-cell epitopes from patients who were fully recovered from SARS infection [13], [14], [15]. However, little is known about memory CD4+ T-cell responses to SARS-CoV. Consistent with observation in mouse model [4], we found that both memory CD4+ and CD8+ T cells participated in cellular responses to SARS-CoV. It is known that naive CD4+ T cells can be divided into two major subpopulations, Th1 and Th2, based on cytokine production and biological functions [16]. The Th1 cells are composed of IFN-γ+ and IFN-γ− cells. The IFN-γ− Th1 cells may differentiate into memory cells in vivo [17]. Based on the production of IFN-γ and IL-2, SARS-CoV S-specific CD4+ T cells could be divided into three distinct populations: single IFN-γ-secreting cells, double IFN-γ/IL-2-secreting cells and single IL-2-secreting cells. Interestingly, IFN-γ/IL-2 double positive CD4+ T cells were mainly restricted to IFN-γhigh IL-2high subset. These results further demonstrated the heterogeneity of memory Th1 cells and also indicated that distinct subpopulations of memory CD4+ T cells have different capacities to develop into short (IFN-γ+) and long-lived (IFN-γ+ IL-2+ and IL-2+) memory cells.
It is generally accepted that CD45RO expression defines to activated or memory T cells. Here, we found that most of SARS-CoV S-specific memory CD4+ T cells were CD45RO+, whereas the majority of memory CD8+ T cells displayed CD45RO− phenotype. Recently, SSp-1, an HLA-A*0201-restricted CD8+ T-cell epitope, was identified from SARS-CoV S protein. The SSp-1-specific CTL cells displayed the CD45RA+ CCR7− CD62L− phenotype, after PBMCs from recovered SARS patients were stimulated in vitro with heat-inactivated SARS-CoV for 7 days [18]. Consistent with these observations, we found that the majority of SARS-CoV S-specific memory CD8 T cells were confined to CD45RO− CCR7− CD62L− population.
Our results also demonstrated that the majority of memory CD8+ T cells exhibited effector memory phenotype of CD62L−. This observation is in agreement with recent studies showing that only central memory CD8+ T cells produce IL-2 following a short-peptide stimulation in lymphocytic choriomeningitis virus (LCMV)-infected mouse model [19]. Generally, the expression of CD62L is associated with the expression of CCR7 [20]. Surprisingly, SARS-CoV S-specific memory CD4+ T cells expressed a mixed CCR7+ CD62L− phenotype that differs from phenotypes of classical CCR7+ CD62L+ TCM and CCR7− CD62L− TEM cells. Some groups have also found memory CD4+ or CD8+ T cells expressing a mixed CCR7+ CD62L− phenotype in mouse model [21], [22]. These studies suggest that the expression of CCR7 and CD62L in memory CD4+ or CD8+ T cells may overlap only partially.
Based on the expression of chemokine receptor CCR7, memory T cells can be divided into two distinct subsets: central memory T cells (TCM) expressing CCR7 and effector memory T cells (TEM), which lack the expression of CCR7. In our study, the majority of IFN-γ-producing CD4+ T cells were CCR7+. This is supported by evidence that memory Th1 cells are mainly CCR7+ in mice [23]. Recent report has identified two functional distinct populations of memory CD8+ T cells: double IFN-γ/IL-2-secreting cells and single IFN-γ-secreting cells. Virus-specific IFN-γ/IL-2-secreting CD8+ T cells were CD45RA− CCR7−, whereas single IFN-γ-secreting CD8+ T cells were either CD45RA− CCR7− or CD45RA+ CCR7− [24]. Consistent with observations in HIV infection, we showed that SARS-CoV S-specific CD8+ T cells produced IFN-γ but not IL-2 and predominantly displayed CD45RO− CCR7− phenotype which might represent terminally differentiated effector memory T cells [25]. This may reflect difference in the inherent stability of CD4+ and CD8+ memory T cells that are CCR7− in normal steady-state conditions [26].
In conclusion, SARS-CoV infection elicits a cellular response mediated by both CD4+ and CD8+ T cells to the S protein. SARS-CoV S-specific memory CD4+ and CD8+ T cells are persistent for more than 1 year in peripheral blood of recovered SARS patients. This study provided an opportunity to delineate the memory T-cell response in survivors of a disease with high morbidity and mortality. The conclusions implicate a target population of T cells as a correlate for protection for effective vaccines.
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (2001CB510007), National Natural Science Foundation of China (No. 30340012), Guangdong Province of Science and Technology (No. 2003Z3-E0491) and Ministry of Education, Guangdong Province of China.
References
- 1.Peiris J.S.M., Lai S.T., Poon L.L.M., Guan Y., Yam L.Y.C., Lim W., Nicholls J., Yee W.K.S., Yan W.W., Cheung M.T., Chan K.H., Tsang D.N.C., Yung R.W.H., Ng T.K., Yuen K.Y. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349(25):2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z.Y., Kong W.P., Huang Y., Roberts A., Murphy B.R., Subbarao K., Nabel G.J. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature. 2004;428:561–564. doi: 10.1038/nature02463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisht H., Roberts A., Vogel L., Bukreyev A., Collins P.L., Murphy B.R., Subbarao K., Moss B. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. 2004;101(17):6641–6646. doi: 10.1073/pnas.0401939101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukreyev A., Lamirande E.W., Buchholz U.J., Vogel L.N., Elkins W.R., Claire M.S., Murphy B.R., Subbarao K., Collins P.L. Mucosal immunisation of African green monkeys (Cercopithecusaethiops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet. 2004;363:2122–2127. doi: 10.1016/S0140-6736(04)16501-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchholz U.J., Bukreyev A., Yang L.J., Lamirande E.W., Murphy B.R., Subbarao K., Collins P.L. Contributions of the structural proteins of severe acute respiratory syndrome coronavirus to protective immunity. Proc. Natl. Acad. Sci. 2004;101(26):9804–9809. doi: 10.1073/pnas.0403492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao W.T., Tamin A., Soloff A., D'Aluto L., Nwanegbo E., Robbins P.D., Bellini W.J., Barratt-Boyes S., Gambotto A. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362:1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li G., Chen X.J., Xu A.L. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 2003;349:508–509. doi: 10.1056/NEJM200307313490520. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y.D., Li Y., Xu G.B., Dong X.Y., Yang X.A., Feng Z.R., Tian C., Chen W.F. Detection of antibodies against SARS-CoV in serum from SARS-infected donors with ELISA and Western blot. Clin. Immunol. 2004;113:145–150. doi: 10.1016/j.clim.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soo Y.O.Y., Cheng Y., Wong R., Hui D.S., Lee C.K., Tsang K.K.S., Ng M.H.L., Chan P., Cheng G., Sung J.J.Y. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004;10(7):676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhi Y., Kobinger G.P., Jordan H., Suchma K., Weiss S.R., Shen H., Schumer G., Gao G.P., Boyer J.L., Crystal R.G., Wilson J.M. Identification of murine CD8 T cell epitopes in codon-optimized SARS-associated coronavirus spike protein. Virology. 2005;335:34–45. doi: 10.1016/j.virol.2005.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y.D., Chen W.F. Detecting specific cytotoxic T lymphocytes against SARS-coronavirus with DimerX HLA-A2:Ig fusion protein. Clin. Immunol. 2004;113:151–154. doi: 10.1016/j.clim.2004.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y.D., Fion Sin W.Y., Xu G.B., Yang H.H., Wong T.Y., Pang X.W., He X.Y., Zhang H.G., Ng J.N.L., Cheng C.S.S., Ju J., Meng L., Yang R.F., Lai S.T., Guo Z.H., Xie Y., Chen W.F. T-cell epitopes in severe acute respiratory syndrome (SARS) coronavirus spike protein elicit a specific T-cell immune response in patients who recovered from SARS. J. Virol. 2004;78(11):5612–5618. doi: 10.1128/JVI.78.11.5612-5618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B.M., Chen H.B., Jiang X.D., Zhang M.H., Wan T., Li N., Zhou X.Y., Wu Y.F., Yang F., Yu Y.Z., Wang X.N., Yang R.F., Cao X.T. Identification of an HLA-A*0201-restricted CD8+ T-cell epitope SSp-1 of SARS-CoV spike protein. Blood. 2004;104:200–206. doi: 10.1182/blood-2003-11-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abbas A.K., Murphy K.M., Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 17.Wu C.Y., Kirman J.R., Rotte M.J., Davey D.F., Perfetto S.P., Rhee E.G., Freidag B.L., Hill B.J., Douek D.C., Seder R.A. Distinct lineages of TH1 cells have differential capacities for memory cell generation in vivo. Nat. Immunol. 2002;3(9):852–858. doi: 10.1038/ni832. [DOI] [PubMed] [Google Scholar]
- 18.Chen H.B., Hou J.L., Jiang X.D., Ma S.W., Meng M.J., Wang B.M., Zhang M.H., Zhang M.X., Tang X.P., Zhang F.C., Wan T., Li N., Yu Y.Z., Hu H.B., Yang R.F., He W., Wang X.N., Cao X.T. Response of memory CD8+ T cells to severe acute respiratory syndrome (SARS) coronavirus in recovered SARS patients and healthy individuals. J. Immunol. 2005;175:591–598. doi: 10.4049/jimmunol.175.1.591. [DOI] [PubMed] [Google Scholar]
- 19.Wherry E.J., Teichgräber V., Becker T.C., Masopust D., Kaech S.M., Antia R., von Andrian U.H., Ahmed R. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 2003;4(3):225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 20.Sallusto F., Lenig D., Förster R., Lipp M., Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 21.Debes G.F., Bonhagen K., Wolff T., Kretschmer U., Krautwald S., Kamradt T., Hamann A. CC chemokine receptor 7 expression by effector/memory CD4+ T cells depends on antigen specificity and tissue localization during influenza A virus infection. J. Virol. 2004;78:7528–7535. doi: 10.1128/JVI.78.14.7528-7535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unsoeld H., Pircher H. Complex memory T-cell phenotypes revealed by coexpression of CD62L and CCR7. J. Virol. 2005;79:4510–4513. doi: 10.1128/JVI.79.7.4510-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unsoeld H., Krautwald S., Voehringer D., Kunzendorf U., Pircher H. Cutting edge: CCR7+ and CCR7− memory T cells do not differ in immediate effector cell function. J. Immunol. 2002;169:638–641. doi: 10.4049/jimmunol.169.2.638. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerli S.C., Harari A., Cellerai C., Vallelian F., Bart P.A., Pantaleo G. HIV-1-specific IFN-γ/IL-2-secreting CD8 T cells support CD4-independent proliferation of HIV-1-specific CD8 T cells. Proc. Natl. Acad. Sci. 2005;102(20):7239–7244. doi: 10.1073/pnas.0502393102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Champagne P., Ogg G.S., King A.S., Knabenhans C., Ellefsen K., Nobile M., Appay V., Rizzardi G.P., Fleury S., Lipp M., Förster R., Rowland-Jones S., Sekaly R.P., McMichael A.J., Pantaleo G. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature. 2001;410:106–111. doi: 10.1038/35065118. [DOI] [PubMed] [Google Scholar]
- 26.Seder R.A., Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat. Immunol. 2003;4(9):835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]