Abstract
In recent years, the outbreaks of porcine epidemic diarrhea (PED) caused by the highly virulent porcine epidemic diarrhea virus (PEDV) variants occurred frequently in China, resulting in severe economic impacts to the pork industry. In this study, we selected and analyzed the genetic evolution of 15 PEDV representative strains that were identified in fecal samples of diarrheic piglets in 10 provinces and cities during 2011–2017. The phylogenetic analysis indicated that all the 15 PEDV isolates clustered into G2 genotype associated with the current circulating strains. Compared with the genome of the prototype strain CV777, these strains had 103–120 amino acid mutations in their S proteins, most of which were in the N terminal domain of S1 (S1-NTD). We also found 37 common mutations in all these 15 strains, although these strains shared 96.9–99.7% nucleotide homology and 96.3–99.8% amino acid homology in the S protein compared with the other original pandemic strains. Computational analysis showed that these mutations may lead to remarkable changes in the conformational structure and asparagine (N)-linked glycosylation sites of S1-NTD, which may be associated with the altered pathogenicity of these variant PEDV strains. We evaluated the pathogenicity of the PEDV strain FJzz1 in piglets through oral and intramuscular infection routes. Compared with oral infection, intramuscular infection could also cause typical clinical signs but with a slightly delayed onset, confirming that the variant PEDV isolate FJzz1 was highly pathogenic to suckling piglets. In conclusion, we analyzed the genetic variation and pathogenicity of the emerging PEDV isolates of China, indicating that G2 variant PEDV strains as the main prevalent strains that may mutate continually. This study shows the necessity of monitoring the molecular epidemiology and the etiological characteristics of the epidemic PEDV isolates, which may help better control the PED outbreaks.
Keywords: PEDV, S gene, Variation, FJzz1, Pathogenicity
Highlights
-
•
G2 variant PEDV strains as the main prevalent strains continue to mutate along with time.
-
•
The isolated strains were prone to mutate in the S protein with many common amino acid mutations.
-
•
These mutations may lead to remarkable changes in the conformational structure of S1-NTD and the pathogenicity of PEDV.
-
•
Both oral and intramuscular routes caused typical clinical signs to piglets.
1. Introduction
Porcine epidemic diarrhea virus (PEDV) is a contagious enteric virus with high genetic variability. PEDV infects pigs of all ages, especially the newborn piglets, causing severe acute watery diarrhea, vomiting and dehydration with the morbidity of up to 100% (Coussement et al., 1982; Shibata et al., 2000; Wood, 1977). The highly contagious enteric disease of swine associated with PEDV was first reported in England in 1971 and then it spread to another 13 countries of Europe and Asia (Ben Salem et al., 2010; Horvath and Mocsari, 1981; Kusanagi et al., 1992; Martelli et al., 2008; Pensaert and de Bouck, 1978; Pijpers et al., 1993; Pospischil et al., 1981; Smid et al., 1993; Takahashi et al., 1983). Later, with the advent of prototype CV777-derived vaccine, PED outbreaks in Asia countries became endemic. From 2007 to 2010, however, PED re-emerged in Thailand, Vietnam and especially China, caused by new variants of PEDV that have been emerged repeatedly since the October 2010, for which neither CV777 inactivated vaccine nor attenuated live vaccine could provide effective protection (Puranaveja et al., 2009; Sun et al., 2012; Zhou et al., 2012). Later, a similar variant PEDV was reported for the first time in April 2013 in the United States, which quickly spread to 36 states across the United States and its neighboring countries such as Canada and Mexico (Lee and Lee, 2014; Stevenson et al., 2013; Vlasova et al., 2014). Subsequently, the outbreak of PED occurred in Okinawa, Japan in October 2013, followed by Korea and Taiwan province of China,resulting in huge economic losses to the pig industry of the world (Park et al., 2014; Sasaki et al., 2016). In 2014, a newly variant PEDV strain (described as “S-INDEL”) with insertions and deletions in the spike gene was reported in the US, which was different from the original highly virulent PEDV strains (Vlasova et al., 2014; Wang et al., 2014). Soon afterwards, S-INDEL was also reported in Germany and Portugal (Stadler et al., 2015; Wang et al., 2014). It has been proved that the major difference between the original variant strain and the newly emerging S-INDEL strain locates in the S protein, which might influence the antigenicity and pathogenicity of the two types of strains (Mesquita et al., 2015). Also, studies have indicated that PEDV strains are undergoing continuous evolution to adapt to the host environment.
PEDV is an enveloped, single-stranded, positive-sense RNA virus, which was thought to mutate easily under the environmental selective pressure. It is a member of the genus Alphacoronavirus, which also includes transmissible gastroenteritis virus (TGEV), human coronavirus 229E (HCoV-229E) and human coronavirus NL63 (HCoV-NL63)(Brian and Baric, 2005). PEDV consists of about 28 Kb PEDV genome including 7 open reading frames (ORF), coding polyprotein 1a and 1b, spike protein (S), accessary protein 3 (ORF3), envelop protein (E), membrane protein (M) and nucleocapsid protein (N). S protein contains two functional subunits: S1, responsible for the cellular receptor binding, and S2, responsible for the membrane fusion (Liu et al., 2015). Though the viral receptor binding domain (RBD) on S1 remains controversial, N-terminal of S1 subunit (S1-NTD) has been demonstrated to be able to recognize the cellular carbohydrate N-acetyl neuraminic acid (Neu5Ac), while the C-terminal of S1 subunit (S1-CTD) - binds to the amino peptidase N (APN)(Deng et al., 2016; Li et al., 2016; Liu et al., 2015). Studies have shown that, compared with the genome of prototype CV777, the mutations in the emerging PEDV variants are mainly concentrated in S1-NTD, reflecting the genetic correlation between different PEDV strains (Lin et al., 2017). Therefore, the analysis of S1-NTD is of vital importance in tracking the origin of the variant PEDV strains. Since 2010, the highly virulent PEDV variants have emerged in different parts of the world, resulting in significantly economic losses to the pork producers (Chen et al., 2012; Li et al., 2012b). Thus far, the classical CV777-derived vaccine has been widely used in the clinical practice in many Asian countries, which could not induce adequate protection against the disease induced by the emerged PEDV variants. Meanwhile, the application of vaccines in pigs may lead to continuous variation for PEDV to escape immune pressures. This study will provide more insights for predicting the prevalence as well as preventing the outbreaks of PED in the future by analyzing the genetic variation characteristics and the pathogenicity of PEDV isolates.
2. Material and methods
2.1. Sample preparation and detection
A total of 528 diarrheic samples including feces, fecal swabs and small intestine were collected from 43 pig farms of 10 provinces and cities during 2011–2017, where nearly 100% of the non-weaning neonates died from severe diarrhea (Table S1). And according to our survey, except for several pig farms with no PEDV vaccination, most of the pig farms were immunized by commercial inactivated PEDV vaccines. The total RNA of each sample was extracted by RNeasy Mini Kit (Qiagen, Hilden, Germany) and reverse transcribed into cDNA by Revert Aid First Stranded cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) as described in the handbooks. Then the cDNA was used for PCR with specific primers designed based on the conserved regions of PEDV, TGEV, porcine deltacoronavirus (PDCoV) and Group A Rotavirus (RoV-A) as described previously(Wang et al., 2016). PCR products were analyzed by electrophoresis in 1.0% agarose gels.
2.2. Amplification and sequence analysis of the S gene
All the positive samples from 43 pig farms were amplified and sequenced based on specific primers designed by our lab as described previously (Table 1 ) (Zhou et al., 2012). And 15 PEDV representative strains were selected from 43 acquired sequences of all the positive PEDV samples. Then a phylogenetic tree was constructed by the neighbor-joining method (NJ) using MEGA 7.0 based on these representative strains and another 60 representative PEDV strains with complete S gene sequences available in GenBank. Also, the complete amino acid sequence of these strains were aligned using Clustal W method of MegAlign (DNAStar Lasergene) (Table 2 ).
Table 1.
Primers used in this study.
| Names of primer | Sequence 5′~3′) | Sites | Length |
|---|---|---|---|
| S1-U | AAGTTACCTGATGGCATTATG | 19,835–21,850 | 2034 bp |
| S1-L | AATAGCCAAACCCATTGAC | ||
| S2-U | GCCATCTTTGCCATACCTCT | 21,700–24,185 | 2504 bp |
| S2-L | CATGCGTAAACAAGACTAAGC | ||
| S3-U | GCCCAAACCCTCACTAAGT | 23,923–25,969 | 2067 bp |
| S3-L | CACAACCGAATGCTATTGACA |
Table 2.
35 PEDV reference strains and 15 strains of our lab used in this study.
| Virus strain | Countries | Time | GenBank accession no. | Virus strain | Countries | Time | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| CV777 | Belgium | 1988 | AF353511 | MN | USA | 2013 | KF468752 |
| LZC | China | 2006 | EF185992 | IA1 USA | USA | 2013 | KF468753 |
| CH/S | China | 1986 | JN547228 | USA Colorado | USA | 2013 | KF272920 |
| BJ-2011-1 | China | 2011 | JN825712 | USA Indiana 17,846 | USA | 2013 | KF452323 |
| CHGD-01 | China | 2011 | JN980698 | USA/IA/2013/19321 | USA | 2013 | KM975738 |
| ZJCZ4 | China | 2011 | JX524137 | USA/Iowa107/2013 | USA | 2013 | KJ645696 |
| GD-1 | China | 2011 | JX647847 | USA/Minnesota52/2013 | USA | 2013 | KJ645704 |
| AJ1102 | China | 2012 | JX188454 | OH851 | USA | 2014 | KJ399978 |
| LC | China | 2012 | JX489155 | OH1414 | USA | 2014 | KJ408801 |
| CH/GDGZ | China | 2012 | KF384500 | PC21A | USA | 2014 | KR078299 |
| GD-A | China | 2012 | JX112709 | FJzz1 | China | 2011 | MK288006 |
| AH2012 | China | 2012 | KC210145 | SDxy | China | 2012 | Not submitted |
| CH/YNKM-8 | China | 2013 | KF761675 | ZJhz | China | 2012 | Not submitted |
| KPEDV-9 | Korea | 1997 | KF898124 | SHdt3 | China | 2013 | Not submitted |
| DR13 | Korea | 1999 | DQ862099 | HeN | China | 2013 | Not submitted |
| attenuated DR13 | Korea | 2002 | JQ023162 | HB10 | China | 2014 | Not submitted |
| KNU-0802 | Korea | 2008 | GU180143 | ZJ3 | China | 2014 | Not submitted |
| KNU-0902 | Korea | 2009 | GU180145 | SD1 | China | 2015 | Not submitted |
| CNU-091222-01 | Korea | 2009 | JN184634 | JXsc3 | China | 2015 | Not submitted |
| KNU-1303 | Korea | 2013 | KJ451038 | BJ4 | China | 2016 | Not submitted |
| K13JA12 | Korea | 2013 | KJ539151 | HBap7 | China | 2016 | Not submitted |
| KNU-1401 | Korea | 2014 | KJ451047 | JSqd3 | China | 2016 | Not submitted |
| KNU-1402 | Korea | 2014 | KJ451048 | ZJ9 | China | 2017 | Not submitted |
| K14JB01 | Korea | 2014 | KJ539154 | LNLY | China | 2017 | Not submitted |
| SM98 | Korea | 2010 | KF779469 | SHpd8 | China | 2017 | Not submitted |
2.3. Software prediction
The S glycoprotein (residues 30–1255) 3D structure models of FJzz1 and CV777 were generated based on the template with PDB accession code of 5SZS using SWISS-MEDEL server (https://swissmodel.expasy.org) (Arnold et al., 2006; Benkert et al., 2011; Biasini et al., 2014). In brief, the target sequences were first searched against SWISS-MODEL template library and the template with the highest quality (the spike protein of HCoV-NL63 employing PDB number of 5SZS and sequence similarity of 41% to both the two targets) was selected for model building based on the target-template alignment using the algorithms embedded in ProMod3 and PROMOD-II. Then after the overall model quality estimation, ligand modeling and oligomeric state conservation prediction, the final models obtained were utilized for our following structure variation analysis. In addition, the N-linked glycosylation sites were predicted by NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/).
2.4. Virus isolation and propagation
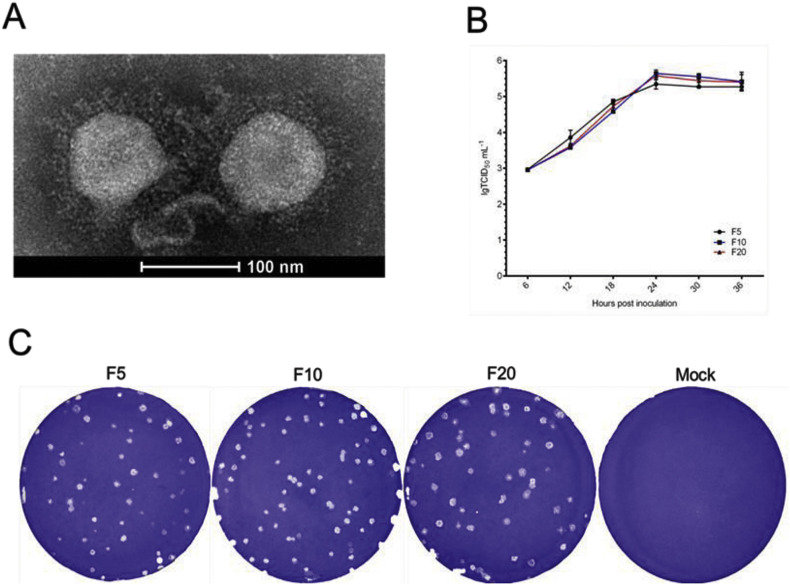
Some typical PEDV-positive samples were diluted by serum-free Dulbecco's Modified Eagle Medium (DMEM), then centrifuged at 4000 ×g for 4 min at 4 °C and the supernatants were collected and filtered through the syringe filter with a 0.22 μm pore size. Then 200 μl aliquots were mixed with 300 μl DMEM including 10 μg/ml trypsin (Sigma-Aldrich, St. Louis, MO, USA) and inoculated onto the monolayers of Vero cells rinsed twice in advance with phosphate-buffered saline as described by Hofmann et al. and Chen et al. (Chen et al., 2014; Hofmann and Wyler, 1988). After incubation at 37 °C for one hour with constant shaking, 1.4 ml maintenance media with 10 μg/ml trypsin were added and the plates were placed back at 37 °C incubator for several days. When 80% of the cells developed visible cytopathic effect (CPE), the plates were treated with freeze-thaw cycle twice to obtain the first passage (F1) viruses. After centrifuged at 4000 ×g for 1 min at 4 °C, the supernatant was inoculated onto the monolayers of Vero cells grown in T25 cell culture flask for further propagation until 20th passage (F20).
2.5. Identification of PEDV by immunofluorescent staining and electron microscope
The Vero cells inoculated with viruses from F1, F5, F10 or F20 were fixed with 80% ice-cold ethanol overnight at 4 °C. After washing with PBS twice, they were added with monoclonal antibody (MAb) against PEDV S protein, MAb against PEDV N protein, PEDV pig antisera or TGEV pig antisera as the primary anybody and incubated at 37 °C for 1 h. Then the cells were rinsed with PBS three times and added with 1:800 diluted fluorescein (FITC)-conjugated donkey anti-mouse IgG (H + L) antibody (Life Technologies, Carlsbad, CA, USA) onto MAb-treated cells or 1:500 diluted Anti-Pig IgG (whole molecule) − FITC antibody produced in rabbit (Sigma-Aldrich, St. Louis, MO, USA) onto pig antisera-treated cells as the secondary antibody. After incubated at 37 °C for 1 h, the cells were rinsed by PBS three times and subjected to epifluorescence microscopy analysis. Meanwhile, we purified and concentrated 1 ml of FJzz1 F5 virus as previously described (Wang et al., 2016). 5 μl of the purified viral suspension was first placed onto a carbon-coated copper grid one minute, which was followed by absorbing the remaining liquid by filter paper. Then 5 μl of 2% phosphotungstic acid (PTA) was applied on the grid and 30 s later the additional PTA was also absorbed. After drying the grid under natural ventilation, the specimen was subjected to observation by electron microscope (EM).
2.6. Plaque assay and growth curve
Viruses from F5, F10 and F20 were inoculated onto the Vero cell monolayers seeded in 6-well plates at a multiplicity of infection (MOI) of 0.01. After incubation at 37 °C for 1 h, the plates were washed with PBS three times and then covered with the prepared liquid agarose solution (3 ml/well) supplemented with 10 μg/ml trypsin. Two hours later, when the agar solidified completely at room temperature, the plates were placed back into the 37 °C cell incubator. As the plaques became large enough and clearly visible by naked eyes, the cells were fixed in 4% paraformaldehyde for 2 h at room temperature, and the plaques were visualized by staining with 2.5% crystal violet. Then the growth curves of viruses from F5, F10 and F20 were determined. First, the virus was inoculated onto a 96-well plate at a multiplicity of infection (MOI) of 0.01 and the cell culture supernatant was collected at 6 h post-inoculation (hpi), 12 hpi, 18 hpi, 24 hpi, 30 hpi and 36 hpi, respectively. Then the viral titers of these samples were measured by 50% tissue culture infectious dose (TCID50) assay according to the Reed-Muench method as below(Reed and Muench, 1938). After washing with PBS two times, 10-fold serial dilutions (10−1 to 10 −8) were prepared by adding 100 μl of the test supernatant to 900 μl of DMEM and each dilution was added to confluent monolayers of Vero cells in 8 vertical wells of a 96-well plate. Following this, the plates were placed in the cell incubator for 5 days at 37 °C and the viral titers were determined by observing CPE microscopically. Finally the growth curve of each virus was established based on the viral titers at different time points post infection.
2.7. Animal experiment
Fifteen 6-day-old conventional piglets were purchased from a commercial pig farm with no previous herd history of PED outbreak or PEDV vaccination. What's more, these piglets were diagnosed as negative for many viruses such as PEDV, TGEV, RoV-A, porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus (PCV), classical swine fever virus (CSFV) and pseudorabies virus (PRV). All animal experiments were approved by the Ethical Committee of the Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Shvri-pi-2,017,100,710). The piglets were assigned randomly into three groups, − A, B and C. Piglets in Group A and Group B were each infected with 1.0 × 105 TCID50 in 2 ml orally and intramuscularly, respectively. All the piglets in Group C were treated with the same amount of DMEM. During the experiment, the piglets were artificially fed milk replacer every 4 h; when anorexia occurred in some piglets, they were fed milk replacer by gavage every 4 h with 15–30 ml each time. The clinical signs, such as diarrhea, vomiting, anorexia, depression and changes in body temperature and weight were recorded daily. The fecal consistency was scored every day based on the standard including 0 for solid, 1 for pasty, 2 for semiliquid (mild diarrhea) and 3 for liquid (severe diarrhea). And the rectal swabs were collected at 1, 3, 5, 7, 9, 11 and 13 days post-inoculation (dpi) for virus shedding detection by TaqMan real-time RT-PCR (Hou et al., 2016; Jung et al., 2014). Intestinal segments including duodenum, jejunum, ileum, cecum, colon and rectum were collected after euthanasia of all experimental piglets at 14 days post infection for histopathological and immunohistochemical examinations as well as quantitative analysis.
2.8. Statistical analysis
GraphPad Prism 6 (GraphPad, La Jolla, CA, USA) was used to perform the statistical analysis, and statistical significance was assessed using Student's t-test. The p-value <.05 was considered significant.
3. Results
3.1. PEDV detection and sequence analysis of the S gene
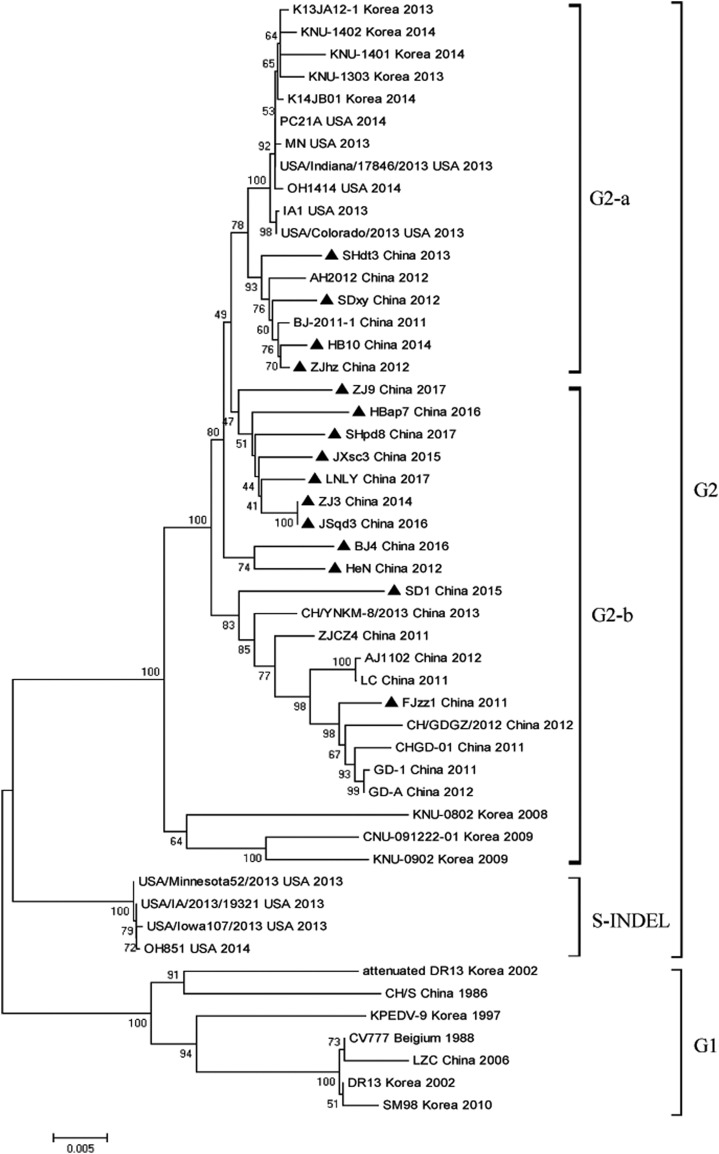
Of all the 528 diarrheic samples being tested, 385 samples were diagnosed as PEDV positive with the positive rate of 72.9%, whereas TGEV, porcine deltacoronavirus (PDCoV) and PRoV were detected to be lower positive rate (Table S1). Phylogenetic analysis showed that all PEDV strains in this study could be divided into two genogroups, namely the classical genotype (G1) represented by CV777 and the variant genotype (G2) of pandemic strains including 15 strains of this study. The G2 clade could be further divided into G2-a, G2-b and S-INDEL three subgroups. Four strains of this study namely ZJhz, SDxy, SHdt3 and HB10 belonged to G2-a along with the strains identified after 2013 in the United States and Korea. G2-b consisted in 11 strains of this study and parts of Chinese pandemic strains as well as some strains identified during 2008–2009 in the Korea (KNU-0802: GU180143, KNU-0902: GU180145 and CNU-091222-01: JN184634); While S-INDEL mainly referred to the strains emerging after 2013 in the United States represented by OH851 (Fig. 1 ).
Fig. 1.
Phylogenetic analysis. The phylogenetic tree was generated based on the complete S gene of 60 PEDV reference strains and 15 isolates in our study by neighbor-joining method of MEGA 6.0 with 1000 bootstrap replicates. The PEDV strains studied in this research were marked with black triangle.
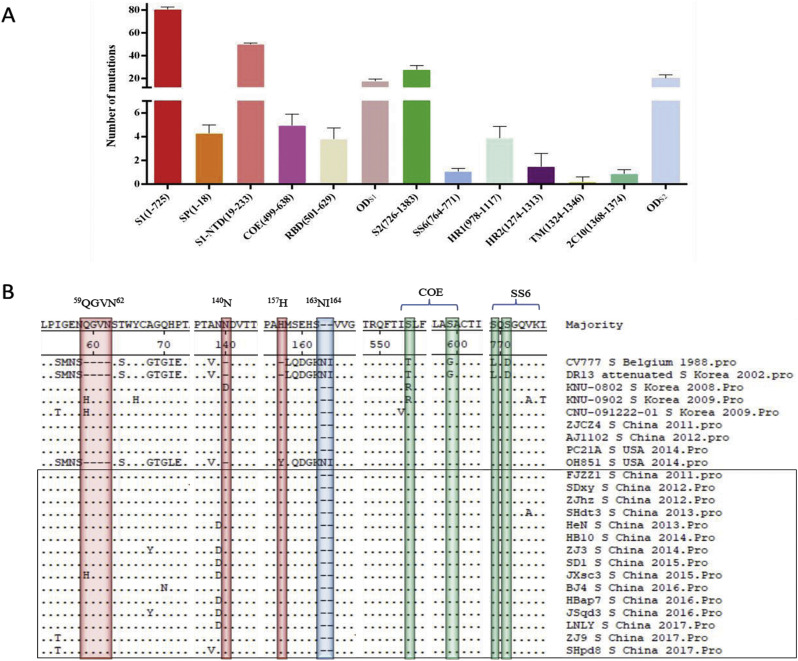
Sequence alignment of the deduced amino acids of the S gene showed that, compared with CV777, 103–120 amino acid mutations were observed in all the 15 strains of this study. S1 protein exhibited 78–83 mutations distributed in different regions including S1-NTD, COE, SP, RBD and other domains of S1 (ODS1) with most of the 49–52 amino acid mutations concentrating in S1-NTD, more than that of S2 protein with most of the 23–36 amino acid substitutions distributing in ODS2 while few amino acid substitutions distributing in SS6, HR1, HR2, TM and 2C10 (Fig. 2A and Table S2). Noticeably, 37 amino acid mutations were identified in the S1-NTD of all the 15 strains (Table 3 ). Among these mutations, 6 insertions (59QGVN62, 140N and 157H) and 2 deletions (163NI164) were found to be consistent with all the other variant strains except OH851. Notably, these mutations were also observed in another three South Korean PEDV strains (KNU-0802: GU180143, KNU-0902: GU180145 and CNU-091222-01: JN184634), which circulated in Korea in 2008 and 2009, the time earlier than the advent of the first so called “variant strains” in China in 2010 (Fig. 2B). Besides, sequence alignment revealed that the nucleotide and amino acid homology between our 15 PEDV strains and the three South Korean PEDV strains were 95.8–97.2% and 95.1–96.9% respectively, higher than that of 93.3–93.9% and 91.7–92.6% between the 15 PEDV strains and the prototype CV777 strain. What's more, our 15 PEDV strains were observed to share 96.9–99.7% nucleotide homology and 96.3–99.8% amino acid homology in the S protein compared with the other original pandemic strains, indicating that variant PEDV strains continue to mutate along with time to adapt the complex external pressure. In addition, all these 15 strains had four common amino acid substitutions including T554 - S, G599 - S, L769 - S and D771 - S in the COE (499-638aa) domain and SS6 (764-771aa) domain (Fig. 2B), and whether these mutations could affect the virus neutralizing activity remains unclear up till now.
Fig. 2.
Analysis of amino acid mutations in the S protein. (A) The number of amino acid mutations in the S protein of different domains including the S1 subunit (residue 1–725), the S2 subunit (residue 726–1383), the signal peptide (SP, residues1–18), the N-terminal domain of S1 (S1-NTD, residue 19–233), the neutralizing epitopes (COE, residues 499–638; SS6, residues 764–771; 2C10, residues 1368–1374), two heptad repeat regions (HR1, residues 978–1117 and HR2, residues 1274–1313), the transmembrane domain (TM, residues 1324–1346) and the other domains of both S1 and S2 (ODS1, ODS2)were counted. (B)The S protein amino acid sequences of 24 PEDV strains were aligned using Clustal W method. The insertion and deletion areas were highlighted in red and blue, respectively. The substitutions in neutralizing epitopes were highlighted in green. The PEDV strains studied in this research were marked with black box. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Statistics of common amino acid mutations in the S protein of the 15 strains in comparison with CV777.
| Domain | Mutations | Domain | Mutations | ||
|---|---|---|---|---|---|
| S1 | SP | 2TP3 → 2KS3 | S1 | COE | 548T → 548S |
| 5I → 5T | 593G → 593S | ||||
| 15L → 15S | Other domain(S1) | 281W → 281L | |||
| S1-NTD | 28S → 28A | 323F → 323S | |||
| 30I → 30T | 356I → 356T | ||||
| 56MNS58 → 56GEN58 | 364E → 364Q | ||||
| ΔΔΔΔ→59QGVN62 | 377K → 377N | ||||
| 60S → 60T | 392K → 392R | ||||
| 65T → 65G | 437V → 437I | ||||
| 67IE68 → 67HP68 | 473S → 473A | ||||
| 80Y → 80H | S2 | HR1 | 1043S → 1043A | ||
| 82DS83 → 82RG83 | 1050I → 1050V | ||||
| 85Q → 85H | HR2 | 1297R → 1297Q | |||
| 114S → 114N | Other domain(S2) | 763L → 763S | |||
| 116I → 116T | 765D → 765S | ||||
| 127N → 127I | 890G → 890R | ||||
| Δ → 136N | 958A → 958V | ||||
| 146S → 146F | 962L → 962F | ||||
| Δ → 153H | 972H → 972Y | ||||
| 154LQDGK158 → 154MSEHS158 | 1166D → 1166A | ||||
| 159NI160 → 159ΔΔ160 | 1172GD1173 → 1172DE1173 | ||||
| 190R → 190K | 1193Y → 1193H | ||||
| 195KRS197 → 195SGG197 | 1199L → 1199F | ||||
| 205T → 205E | 1231S → 1231R | ||||
| 222Y → 222S | 1259I → 1259T | ||||
| 224E → 224Q | |||||
3.2. Structural analysis of S glycoprotein
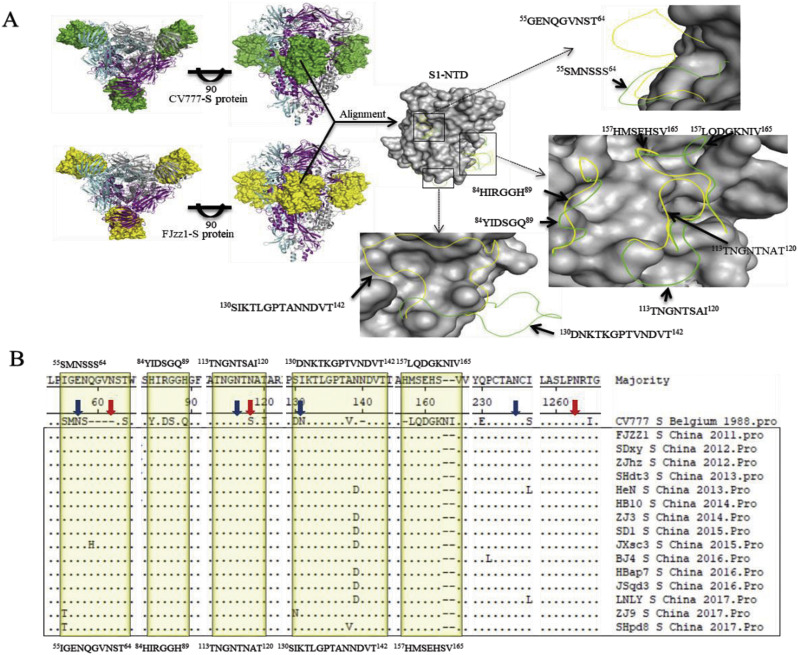
The S glycoprotein (residues 30-1255) 3D structure models of FJzz1 and CV777 were generated based on the template with PDB accession code of 5SZS using SWISS-MEDEL server. The structures of these two proteins were all predicted as homotrimers with S1-NTDs exposed on the surface of the proteins. When aligned with the structure of S1-NTD of CV777, S1-NTD of FJzz1 appeared five major conformational changes on residues 55–64, 84–89, 113–120, 130–142 and 157–165, which all resided on the surface of S1-NTD (Fig. 3A). Asparagine (N)-linked glycosylation sites prediction of S glycoprotein showed there existed 21 potential N-linked glycosylation sites on the S glycoprotein of CV777 (13 sites on S1 region, and another 8 sites on S2 region), while 20–23 N-linked glycosylation sites were predicted on S glycoprotein of the 15 strains obtained in this study. Remarkably, these 15stains lose four N-linked glycosylation sites at N57 (NSSS), N116 (NTSA), N131 (NKTL) and N235 (NCSG) predicted on the S1-NTD of CV777, but gained three additional putative sites at N62 (NSTW), N118 (NATA) and N1263 (NRTG) on the corresponding regions (Fig. 3B). Besides, 10 of our strains (SDxy, ZJhz, SHtd3, ZJ3, JXsc3, BJ4, HBap7, JSqd3, ZJ9, SHpd8) gained N728 (NSTR) and N1189 (NHTA) two sites on S1 and S2 regions, respectively (Table S3 and S4). And SHtd3 gained another one site N1310 (NNTL) on S2 regions. We speculated that these changes in structure and N-linked glycosylation sites of S glycoprotein may influence the antigenicity of PEDV.
Fig. 3.
Analysis of the 3D structural models and the predicted N-linked glycosylation sites of CV777 and FJzz1 in the S protein. (A) The predicted S protein 3D structural models of CV777 and FJzz1 were generated and observed on the top and front sides. The three monomers that form the glycoprotein trimer were colored in grey, magenta and cyan. The S1-NTDs of CV777 and FJzz1 were shown as ‘surface’ in green and yellow, respectively. Also, the structural alignment of S1-NTDs of these two proteins was carried out. The residues with no obvious structural variations were displayed as ‘surface’ in grey, while the residues with most obvious structural variations were shown as ‘ribbon’ in green for CV777 and yellow for FJzz1. (B) The S protein amino acid sequences of 15 PEDV strains in this study and the classical CV777 strain were aligned as described above. The residues with most obvious structural variations described in Fig. 3A were highlighted in yellow, while the predicted N-linked glycosylation sites of CV777 and our 15 strains were marked by blue and red arrows, respectively. The PEDV strains studied in this research were marked with black box. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Isolation and identification of PEDV
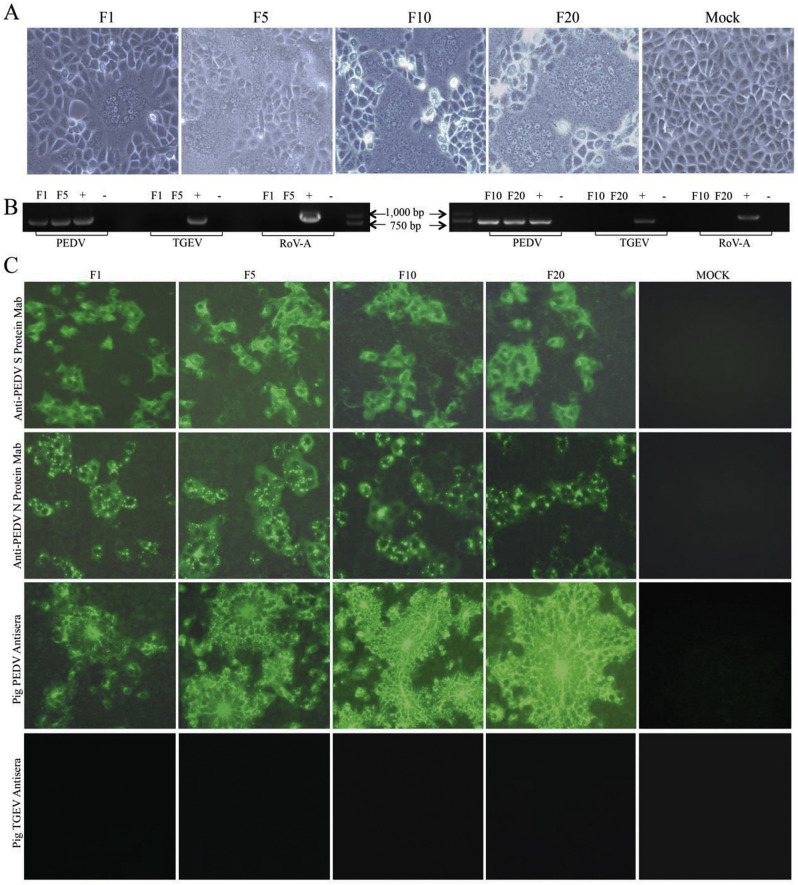
We successfully isolated six PEDV strains including FJzz1, SHdt3, HB10, SD1, BJ4 and ZJ9 from 344 samples of different provinces and cities in China during 2011–2017, all of which shared 97.3–98.4% amino acid sequence similarity for S protein. When infected with our isolated strain FJzz1, Vero cells developed visible CPE which was characterized by multiple regional cell fusion and syncytium formation within 24 hpi (Fig. 4A). Then, viruses from F1, F5, F10 and F20 were tested by RT-PCR and all these samples were positive for PEDV but negative for TGEV and RoV-A (Fig. 4B). Meanwhile, the cells inoculated with F1, F5, F10 or F20 viruses were fixed and tested by immunofluorescence assay (IFA). All these cells treated with MAb against PEDV S protein, MAb against Nucleocapsid (N) protein and pig PEDV antisera displayed bright fluorescence, while the cells emitted no fluorescence when treated with the pig antisera against TGEV as the primary antibody (Fig. 4C). The highly purified and condensed virus particles from the FJzz1 F5 supernatant were shown as roughly spherical virions around 100 nm in diameter with surface projections by electron microscopy (Fig. 5A).
Fig. 4.
Detection of FJzz1 in Vero cells. (A) The CPE of FJzz1 F1, F5, F10 and F20 at 24hpi was observed. (B) The cell culture supernatants of FJzz1 F1, F5, F10 and F20 were examined by RT-PCR using the primers specific for PEDV, TGEV and RoV-A. (C) Also, the monolayers of Vero cells inoculated with FJzz1 F1, F5, F10 and F20 were tested by IFA using MAb against PEDV S protein, MAb against PEDV N protein, pig PEDV antisera and pig TGEV antisera.
Fig. 5.
In vitro characterization of FJzz1. (A) The negative-staining EM image of FJzz1 F5 virus particles. (B) Multi-step growth curve of FJzz1 from different passages (F5, F10 and F20) on Vero cells at an MOI of 0.01. (C) The crystal violet stained plaques formed in the monolayers of Vero cells inoculated with FJzz1 from different passages (F5, F10 and F20) at 3 days post infection. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. In vitro characterization of FJzz1
Then, growth dynamics and plaque size were characterized. The growth curves of F5, F10 and F20 based on the TCID50 at different hpi were visualized. They showed that the titers of different virus propagations rose rapidly from 6 hpi to 24 hpi when the viral titers reached the peak ranging from 5.44 × 105 to 5.71 × 105 TCID50/mL (Fig. 5B). The plaques of each generation were mostly close to round with diameters of 1–2 mm (mm) (Fig. 5C).
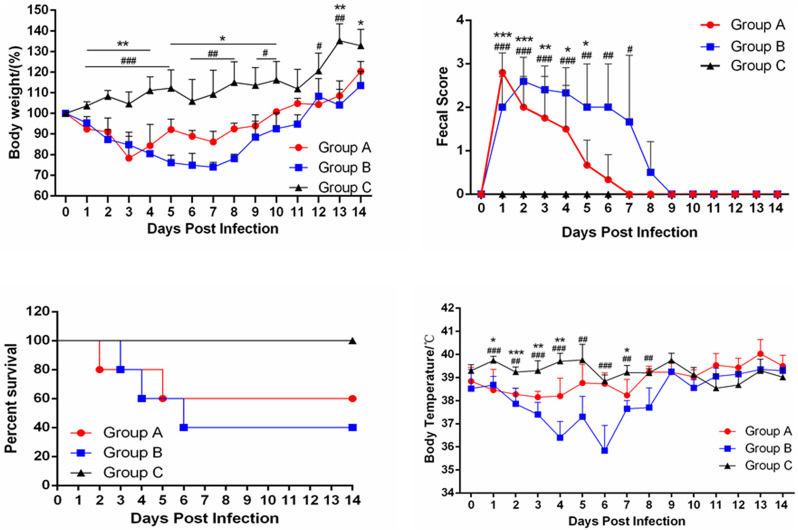
3.5. Pathogenic evaluation of piglets infected with FJzz1
After inoculation with PEDV strain FJzz1 orally (Group A) or intramuscularly (Group B), all the 10 pigs developed obvious clinical signs including diarrhea, vomiting and loss of weight (Fig. 6A). Notably, pig A2 in group A began to show diarrhea at 12 hpi, while pig B4 in Group B developed diarrhea at 20 hpi, later than that of pigs in group A, and all the 10 pigs inoculated with FJzz1 developed diarrhea within 24 h (Fig. 6B). One pig in Group A firstly died at 40 hpi, and the clinical signs became alleviated after 7 dpi, while the first pig in Group B died at 80 hpi. Finally, the mortality of Group A and Group B reached 40% (2/5) and 60% (3/5), respectively (Fig. 6C). Also, the daily body temperature and body weight of each piglet throughout the whole experiment were monitored and analyzed. No remarkable difference was found in body temperature between the three groups except for the initial infection, during which the body temperature of moribund piglets dropped significantly. And the two challenge groups lost significantly greater body weight than the control group (Fig. 6D). Also, the fecal scores for group A and group B is much higher than that for group C at 7–9 dpi (Fig. 6B), and no clinical signs were developed and no pig died in the negative control C group.
Fig. 6.
Pathogenicity analysis of FJzz1. (A) The average body weight changes in each group. (B) Fecal scores of piglets in different groups. Evaluation standard: 0 = normal; 1 = soft; 2 = semi-fluid; 3 = watery diarrhea. (C) The survival rate of piglets in each group. The survival curves of piglets infected with FJzz1 by oral inoculation or muscular injection are shown respectively. (D) The changes in average body temperature of each group within the first 14 dpi. Asterisk (*) indicates a significant difference between Group A and Group C (* p < .05; ** p < .01; *** p < .001). Pound (#) indicates a significant difference between Group B and Group C (# p < .05; ## p < .01; ### p < .001).
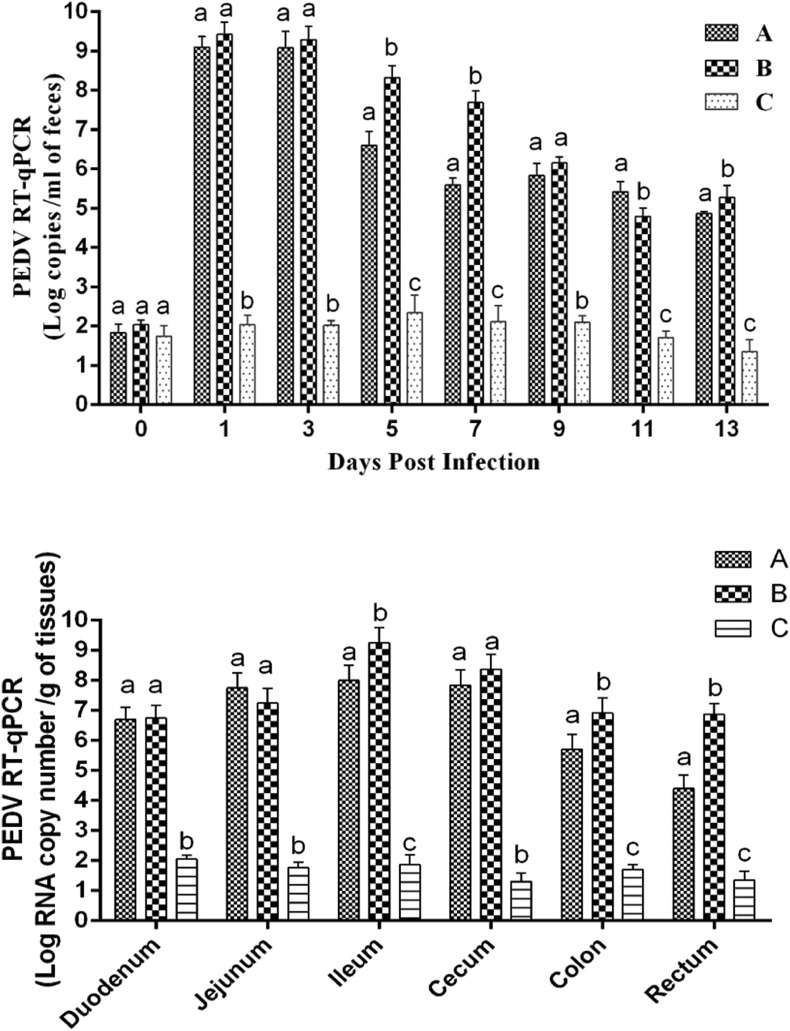
The viral RNA shedding of feces was investigated by TaqMan real-time RT-PCR. We found that piglets in both A and B groups shed virus at the highest level of shedding titers, with - 1.26 × 109 copies/ml and 2.63 × 109 copies/ml respectively within 24 h post infection (Fig. 7A). Then from 5 to 7 days post infection, the virus shedding of intramuscular injection group reduced slightly ranging from 4.92 × 107copies/ml to 2.08 × 108copies/ml, significantly higher than that of 3.85 × 105copies/ml to 3.96 × 106copies/ml of oral group. Subsequently, the virus shedding reduced continually with no significant difference between intramuscular injection group and oral group ranging from 7.19 × 104copies/ml to 1.89 × 105copies/ml. Meanwhile, we also examined the viral load of different segments of their intestines including duodenum, ileum, jejunum, cecum, colon and rectum. As shown in Fig. 7B, all segments of intestine exhibited high viral load in both of the infection groups ranging from 9.94 × 107copies/g to 1.78 × 109copies/g. During the whole experiment, neither the virus shedding in the feces nor the viral load in the intestine in group C was detected.
Fig. 7.
Virus shedding in feces and quantification of viral load in different intestine segments. (A) Virus shedding in feces. Daily virus shedding in feces of different groups as measured by real-time RT-PCR detection of virus genome in fecal swabs. (B) Quantification of viral load in different parts of intestine by TaqMan real-time RT-PCR targeting PEDV N gene. Labels without the same letters indicate significant differences.
.
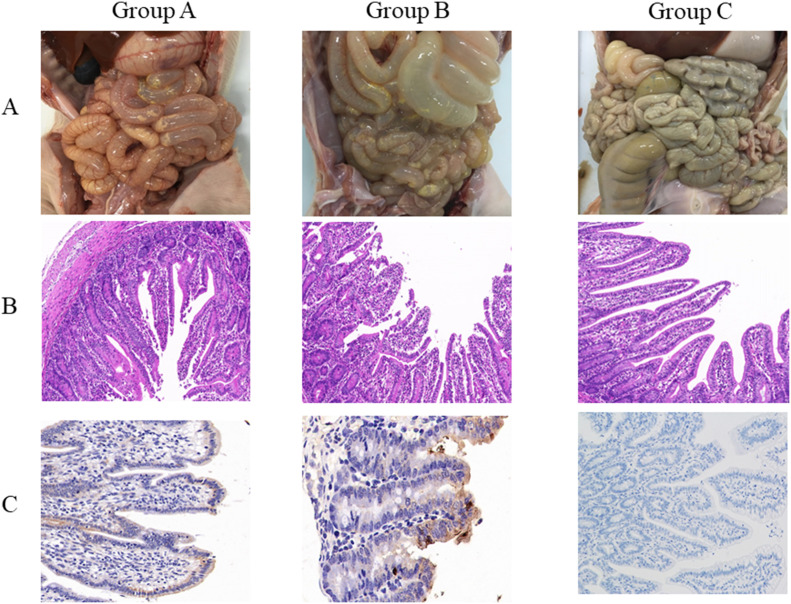
The pathological examination revealed that the intestinal walls of dead piglets of both A and B groups were thin or even transparent, with a large amount of yellowish fluid in the intestinal cavity (Fig. 8A). Results of HE staining showed that infection by PEDV both orally and intramuscularly caused severe damage to all intestinal segments characterized by shorten, atrophic or even shedding intestinal villus, especially jejunum and ileum. The epithelial layer of cecum and colon was destroyed with vacuolization and subcutaneous edema. In contrast, no pathological damage could be found in the mock group (Fig. 8B). Results of immunohistochemical staining indicated that the virus distributed extensively in the cytoplasm of the villous epithelial cells of jejunum and ileum (Fig. 8C). Therefore, we concluded that FJzz1 is an epidemic strain with strong pathogenicity to piglets, and both oral inoculation and muscular injection could cause the typical clinical signs and pathological damage to newborn piglets.
Fig. 8.
Gross lesions of piglets and histopathologic examination of intestines. (A) Gross lesions of piglets of each group. The intestinal lesions were examined at the day of death or at the final time points. (B) Histopathologic examination of jejunum. Jejunums collected from each group were processed for HE staining. (C) Immunohistochemical detection. Jejunum of each groups was stained with PEDV monoclonal antibody against spike protein (1:100 dilution).
4. Discussion
The entry of coronavirus is mainly mediated by the glycosylated S protein, which is also a key determinant for the tissue tropism. Previous studies demonstrated that S gene is prone to mutate, which could change the pathogenicity of the virus (Hou et al., 2017; Sun et al., 2012; Wang et al., 2014; Wu et al., 2012). In this study, a total of 528 diarrheic samples including feces, fecal swabs, and small intestine were collected from 43 pig farms of 10 provinces and cities during 2011–2017, and then detected by RT-PCR targeting N gene of PEDV. Results showed that 385 samples were detected as PEDV positive with a positive rate as high as 72.9%, whereas very few samples were diagnosed as TGEV, PDCoV or PRoV positive. A phylogenetic tree was constructed to investigate the genetic variation of PEDV strains with different time span. The results showed that all the 15 strains identified in this study were clustered into G2 genotype, the homology for nucleotide and amino acid sequence between all the 15 PEDV strains and the currently circulating variant isolates were 96.9–99.7% and 97.0–99.8% respectively, higher than that of 93.3–93.9% and 91.7–92.6% between the 15 PEDV strains and the G1 genotype strains represented by CV777. Interestingly, some typical mutations on the S protein of Chinese prevalent variant strains were also found on three South Korean PEDV strains (KNU-0802: GU180143, KNU-0902: GU180145 and CNU-091222-01: JN184634) that were circulating in 2008–2009 (Fig. 2B), which suggested that the early variant PEDV strains of China which emerged in the late 2010 associated with the circulating strains in South Korea. Up to now, the variant PEDV strains are still the main pathogen of enteric disease in neonatal piglets of China. Meanwhile, it cannot be ignored that these variant PEDV strains continue to mutate along with time.
Previous studies showed that the immunity induced by the prototype strain CV777-derived vaccine is effective to prevent the PED outbreaks caused by classical strains but poorly protects against the disease induced by variant strains (Li et al., 2012a; Puranaveja et al., 2009). Also the mutations, especially some deletions and insertions in the S protein may change the antigenicity, pathogenicity and neutralization properties(Park et al., 2014; Zhang et al., 2015), and S1-NTD (19-233aa) may be the potential region relevant to the virulence of PEDV (Hou et al., 2017; Su et al., 2018; Su et al., 2019; Suzuki et al., 2016). In this study, 49–53 mutations were found in the S1-NTD of all the 15 strains including FJzz1, which might change the major conformations of S1-NTD. Up to now, at least four neutralizing epitopes of PEDV have been identified in the S glycoprotein, namely the COE (499–638), SS2 (748–755), SS6 (764–771) and 2C10 (1368–1374) (Chang et al., 2002; Cruz et al., 2008; Sun et al., 2008). Coincidentally, four amino acid substitutions including T554 – S, G599 – S, L769 - S and D771 - S were observed in these neutralizing epitopes. Whether these amino acid mutations will affect the neutralization activity of the above epitopes warrant further exploration. What's more, compared with CV777, the 15 stains showed four amino acid insertions (59QGVN62) and three amino acid substitutions (S118 → N118, 130DN131 → 130SI131), lost three N-linked glycosylation sites at N57 (NSSS), N116 (NTSA), N131 (NKTL) predicted on the S1-NTD of CV777, but gained two additional putative sites at N62 (NSTW) and N118 (NATA) on the corresponding regions. Previously, researchers have demonstrated the N-linked glycosylation sites on the S protein of severe acute respiratory syndrome coronavirus (SARS-CoV) play a critical role in the viral entry (Han et al., 2007). Therefore, we speculate that these variations might result in more efficient recognition and stronger affinity with the specific cell receptor, sugar co-receptor or even other unknown co-receptors, thus better aiding the viral entry. This could be partially supported by the fact that the variant strain exhibited stronger sugar-binding activities than the prototype strain CV777 (Deng et al., 2016). Meanwhile, the changes of N-linked glycosylation sites may also have an influence on antigenicity of certain important epitopes, which may partly explain why CV777-derived vaccine provided poor protection against variant strains. This adaptive variation, however, may also be an evolutionary strategy for immune evasion of the variant PEDV strains, but further experiments such as reverse genetics are required to confirm all these assumptions(Beall et al., 2016; Hou et al., 2019; Wang et al., 2018).
In order to investigate the pathogenicity of variant strains to piglets, fifteen unweaned neonates were infected with FJzz1. The results showed that the oral Group developed diarrhea clinical signs around 8 h earlier than that of the intramuscular Group, and the piglets in oral Group began to die around 40 h earlier than that of the intramuscular Group. Moreover, the morbidity of the oral Group reached 100% 24 h post infection. By contrast, all the piglets showed diarrheic disease within 48 h post infection in the intramuscular Group. Nevertheless, diarrhea clinical signs of the intramuscular Group last longer than that of the oral group with more severe degree of clinical signs including diarrhea and loss of weight. Previous studies reported that the intestinal proteases are critical for PEDV release rather and entry (Liu et al., 2016; Shirato et al., 2011). When infecting piglets by the oral route, the virus could target to intestine through the digestive tract directly, and the intestinal proteases might lead to the earlier appearance of clinical signs by better aiding the viral release (Wicht et al., 2014). It has been reported that PEDV could invade the blood stream, but it fails to replicate actively there, and the viraemia levels may not necessarily be connected to virulence or pathogenicity with mechanisms not fully understood (Chen et al., 2016). Moreover, a recent study demonstrated that the PEDV particles could be recognized and presented by DCs of nasal epithelial cells and then delivered to CD3+ T cells of cervical lymph node, reaching the intestine through the blood and lymph circulation to infect the intestinal epithelial cells(Li et al., 2018). This report may support our study that intramuscular injection of PEDV could also cause the typical clinical signs to piglets, although the specific molecular mechanisms remain to be elucidated. Therefore, when infecting piglets by the intramuscular injection, the virus could be delivered to the target site through the blood circulation system without intestinal proteases, resulting in later appearance of clinical signs. As is known to all, fecal-oral transmission is the main natural route of PEDV for infection, so it is much easier for the virus to simulate this progress by oral inoculation. However, compared with the intramuscular injection, the oral inoculation is supposed to be unstable and require stricter operation, because part of the virus would be lost during the infection due to the uncertain in vivo conditions of piglets. Therefore, intramuscular injection exhibited basically the same efficiency to cause obvious clinical signs, despite slight delay of clinical signs at the initial phase of infection. This finding supports the idea that intramuscular injection of live, attenuated PEDV may be an alternative vaccination route. In conclusion, we analyzed the genetic variation and pathogenicity of the emerging PEDV isolates of China, indicating that G2 variant PEDV strains as the main prevalent strains continue to mutate along with time. The variant strain FJzz1 with mutations in the S protein is highly pathogenic to the piglets, and the intramuscular injection may also be an alternative inoculation route. This study will provide a basis for understanding of the genetic variation and pathogenicity of PEDV.
Author contributions
Pengfei Chen, Yixuan Hou, Huichun Li and Xianbin Li participated in the collecting and sequencing of some of the analyzed samples. Pengfei Chen, Guangzhi Tong and Yanjun Zhou conceived and designed the study. Kang Wang, Lingxue Yu, and Yifeng Jiang performed the animal experiments. Fei Gao, Wu Tong, and Hai Yu did the analyses. Pengfei Chen and Kang Wang conceived the analyses and drafted the manuscript. Zhibiao Yang, Guangzhi Tong and Yanjun Zhou provided critical review and editing of the manuscript. All authors have seen and approved the paper.
Competing interest
The authors declare that they have no competing interests.
Acknowledgments
The study was supported by the National Natural Science Foundation of China (31472207), the National Program on Key Research Project of China (2016YFD0500100), the earmarked fund for Modern Agro-industry Technology Research System of China (CARS-36).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2019.01.022.
Contributor Information
Guangzhi Tong, Email: gztong@shvri.ac.cn.
Yanjun Zhou, Email: yjzhou@shvri.ac.cn.
Appendix A. Supplementary data
Supplementary material
References
- Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Beall A., Yount B., Lin C.M., Hou Y., Wang Q., Saif L., Baric R. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. MBio. 2016;7 doi: 10.1128/mBio.01451-15. (e01451–01415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Salem A.N., Chupin Sergei A., Bjadovskaya Olga P., Andreeva Olga G., Mahjoub A., Prokhvatilova Larissa B. Multiplex nested RT-PCR for the detection of porcine enteric viruses. J. Virol. Methods. 2010;165:283–293. doi: 10.1016/j.jviromet.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S.H., Bae J.L., Kang T.J., Kim J., Chung G.H., Lim C.W., Laude H., Yang M.S., Jang Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells. 2002;14:295–299. [PubMed] [Google Scholar]
- Chen X., Yang J., Yu F., Ge J., Lin T., Song T. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) samples from field cases in Fujian, China. Virus Genes. 2012;45:499–507. doi: 10.1007/s11262-012-0794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Li G., Stasko J., Thomas J.T., Stensland W.R., Pillatzki A.E., Gauger P.C., Schwartz K.J., Madson D., Yoon K.J., Stevenson G.W., Burrough E.R., Harmon K.M., Main R.G., Zhang J. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014;52:234–243. doi: 10.1128/JCM.02820-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Gauger P.C., Stafne M.R., Thomas J.T., Madson D.M., Huang H., Zheng Y., Li G., Zhang J. Pathogenesis comparison between the United States porcine epidemic diarrhoea virus prototype and S-INDEL-variant strains in conventional neonatal piglets. J. Gen. Virol. 2016;97:1107–1121. doi: 10.1099/jgv.0.000419. [DOI] [PubMed] [Google Scholar]
- Coussement W., Ducatelle R., Debouck P., Hoorens J. Pathology of experimental CV777 coronavirus enteritis in piglets. I. Histological and histochemical study. Vet. Pathol. 1982;19:46–56. doi: 10.1177/030098588201900108. [DOI] [PubMed] [Google Scholar]
- Cruz D.J., Kim C.J., Shin H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize Porcine epidemic diarrhea virus. Virus Res. 2008;132:192–196. doi: 10.1016/j.virusres.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng F., Ye G., Liu Q., Navid M.T., Zhong X., Li Y., Wan C., Xiao S., He Q., Fu Z.F., Peng G. Identification and comparison of receptor binding characteristics of the spike protein of two porcine epidemic diarrhea virus strains. Viruses. 2016;8:55. doi: 10.3390/v8030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D.P., Lohani M., Cho M.W. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J. Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Wyler R. Propagation of the virus of porcine epidemic diarrhea in cell culture. J. Clin. Microbiol. 1988;26:2235–2239. doi: 10.1128/jcm.26.11.2235-2239.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath I., Mocsari E. Ultrastructural changes in the small intestinal epithelium of suckling pigs affected with a transmissible gastroenteritis (TGE)-like disease. Arch. Virol. 1981;68:103–113. doi: 10.1007/BF01314440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.-X., Xie C., Wang K., Zhao Y.-T., Xie Y.-Y., Shi H.-Y., Chen J.-F., Feng L., Tong G.-Z., Hua X.-G., Yuan C.-L., Zhou Y.-J., Yang Z.-B. Development and application of a TaqMan-MGB real-time RT-PCR assay for the detection of porcine epidemic diarrhoea virus strains in China. J. Vet. Res. 2016;60:127–133. [Google Scholar]
- Hou Y., Lin C.M., Yokoyama M., Yount B.L., Marthaler D., Douglas A.L., Ghimire S., Qin Y., Baric R.S., Saif L.J., Wang Q. Deletion of a 197-amino-acid region in the N-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J. Virol. 2017:91. doi: 10.1128/JVI.00227-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y., Meulia T., Gao X., Saif L.J., Wang Q. The deletion of both tyrosine-based endocytosis signal and endoplasmic reticulum-retrieval signal in the cytoplasmic tail of spike protein attenuates pedv in pigs. J. Virol. 2019;93(2) doi: 10.1128/JVI.01758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., Wang Q., Scheuer K.A., Lu Z., Zhang Y., Saif L.J. Pathology of US porcine epidemic diarrhea virus strain PC21A in gnotobiotic pigs. Emerg. Infect. Dis. 2014;20:662–665. doi: 10.3201/eid2004.131685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusanagi K., Kuwahara H., Katoh T., Nunoya T., Ishikawa Y., Samejima T., Tajima M. Isolation and serial propagation of porcine epidemic diarrhea virus in cell cultures and partial characterization of the isolate. J. Vet. Med. Sci. 1992;54:313–318. doi: 10.1292/jvms.54.313. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee C. Outbreak-related porcine epidemic diarrhea virus strains similar to US strains, South Korea, 2013. Emerg. Infect. Dis. 2014;20:1223–1226. doi: 10.3201/eid2007.140294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Li H., Liu Y., Pan Y., Deng F., Song Y., Tang X., He Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012;18:1350–1353. doi: 10.3201/eid1808.120002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.L., Zhu L., Ma J.Y., Zhou Q.F., Song Y.H., Sun B.L., Chen R.A., Xie Q.M., Bee Y.Z. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in south China. Virus Genes. 2012;45:181–185. doi: 10.1007/s11262-012-0735-8. [DOI] [PubMed] [Google Scholar]
- Li W., van Kuppeveld F.J.M., He Q., Rottier P.J.M., Bosch B.J. Cellular entry of the porcine epidemic diarrhea virus. Virus Res. 2016;226:117–127. doi: 10.1016/j.virusres.2016.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wu Q., Huang L., Yuan C., Wang J., Yang Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018;9:3811. doi: 10.1038/s41467-018-06056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.M., Hou Y., Marthaler D.G., Gao X., Liu X., Zheng L., Saif L.J., Wang Q. Attenuation of an original US porcine epidemic diarrhea virus strain PC22A via serial cell culture passage. Vet. Microbiol. 2017;201:62–71. doi: 10.1016/j.vetmic.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Tang J., Ma Y., Liang X., Yang Y., Peng G., Qi Q., Jiang S., Li J., Du L., Li F. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J. Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Ma Y., Yang Y., Zheng Y., Shang J., Zhou Y., Jiang S., Du L., Li J., Li F. Cell entry of porcine epidemic diarrhea coronavirus is activated by lysosomal proteases. J. Biol. Chem. 2016;291:24779–24786. doi: 10.1074/jbc.M116.740746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli P., Lavazza A., Nigrelli A.D., Merialdi G., Alborali L.G., Pensaert M.B. Epidemic of diarrhoea caused by porcine epidemic diarrhoea virus in Italy. Vet. Rec. 2008;162:307–310. doi: 10.1136/vr.162.10.307. [DOI] [PubMed] [Google Scholar]
- Mesquita J.R., Hakze-van der Honing R., Almeida A., Lourenco M., van der Poel W.H., Nascimento M.S. Outbreak of porcine epidemic diarrhea virus in Portugal, 2015. Transboundary Emerg. Dis. 2015;62:586–588. doi: 10.1111/tbed.12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Kim S., Song D., Park B. Novel porcine epidemic diarrhea virus variant with large genomic deletion, South Korea. Emerg. Infect. Dis. 2014;20:2089–2092. doi: 10.3201/eid2012.131642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pensaert M.B., de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978;58:243–247. doi: 10.1007/BF01317606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijpers A., van Nieuwstadt A.P., Terpstra C., Verheijden J.H. Porcine epidemic diarrhoea virus as a cause of persistent diarrhoea in a herd of breeding and finishing pigs. Vet. Rec. 1993;132:129–131. doi: 10.1136/vr.132.6.129. [DOI] [PubMed] [Google Scholar]
- Pospischil A., Hess R.G., Bachmann P.A. Light microscopy and ultrahistology of intestinal changes in pigs infected with epizootic diarrhoea virus (EVD): comparison with transmissible gastroenteritis (TGE) virus and porcine rotavirus infections. J. Vet. Med. B. 1981;28:564–577. doi: 10.1111/j.1439-0450.1981.tb01774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranaveja S., Poolperm P., Lertwatcharasarakul P., Kesdaengsakonwut S., Boonsoongnern A., Urairong K., Kitikoon P., Choojai P., Kedkovid R., Teankum K., Thanawongnuwech R. Chinese-like strain of porcine epidemic diarrhea virus, Thailand. Emerg. Infect. Dis. 2009;15:1112–1115. doi: 10.3201/eid1507.081256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints 12. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Sasaki Y., Alvarez J., Sekiguchi S., Sueyoshi M., Otake S., Perez A. Epidemiological factors associated to spread of porcine epidemic diarrhea in Japan. Prevent. Vet. Med. 2016;123:161–167. doi: 10.1016/j.prevetmed.2015.11.002. [DOI] [PubMed] [Google Scholar]
- Shibata I., Tsuda T., Mori M., Ono M., Sueyoshi M., Uruno K. Isolation of porcine epidemic diarrhea virus in porcine cell cultures and experimental infection of pigs of different ages. Vet. Microbiol. 2000;72:173–182. doi: 10.1016/S0378-1135(99)00199-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Matsuyama S., Ujike M., Taguchi F. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J. Virol. 2011;85:7872–7880. doi: 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smid B., Valicek L., Rodak L., Kudrna J., Musilova J. Detection of porcine epidemic diarrhea virus using electron microscopy in the Czech Republic. Vet. Med. 1993;38:333–341. [PubMed] [Google Scholar]
- Stadler J., Zoels S., Fux R., Hanke D., Pohlmann A., Blome S., Weissenbock H., Weissenbacher-Lang C., Ritzmann M., Ladinig A. Emergence of porcine epidemic diarrhea virus in southern Germany. BMC Vet. Res. 2015;11:142. doi: 10.1186/s12917-015-0454-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G.W., Hoang H., Schwartz K.J., Burrough E.R., Sun D., Madson D., Cooper V.L., Pillatzki A., Gauger P., Schmitt B.J., Koster L.G., Killian M.L., Yoon K.J. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013;25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- Su Y., Hou Y., Prarat M., Zhang Y., Wang Q. New variants of porcine epidemic diarrhea virus with large deletions in the spike protein, identified in the United States, 2016–2017. Arch. Virol. 2018;163:2485–2489. doi: 10.1007/s00705-018-3874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Hou Y., Wang Q. The enhanced replication of an S-intact PEDV during coinfection with an S1 NTD-del PEDV in piglets. Vet. Microbiol. 2019;228:202–212. doi: 10.1016/j.vetmic.2018.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Feng L., Shi H., Chen J., Cui X., Chen H., Liu S., Tong Y., Wang Y., Tong G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008;131:73–81. doi: 10.1016/j.vetmic.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R.Q., Cai R.J., Chen Y.Q., Liang P.S., Chen D.K., Song C.X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012;18:161–163. doi: 10.3201/eid1801.111259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Shibahara T., Yamaguchi R., Nakade K., Yamamoto T., Miyazaki A., Ohashi S. Pig epidemic diarrhoea virus S gene variant with a large deletion non-lethal to colostrum-deprived newborn piglets. J. Gen. Virol. 2016;97:1823–1828. doi: 10.1099/jgv.0.000513. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Okada K., Ohshima K. An outbreak of swine diarrhea of a new-type associated with coronavirus-like particles in Japan. Japn. J. Vet. Sci. 1983;45:829–832. doi: 10.1292/jvms1939.45.829. [DOI] [PubMed] [Google Scholar]
- Vlasova A.N., Marthaler D., Wang Q., Culhane M.R., Rossow K.D., Rovira A., Collins J., Saif L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013-February 2014. Emerg. Infect. Dis. 2014;20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Byrum B., Zhang Y. New variant of porcine epidemic diarrhea virus, United States, 2014. Emerg. Infect. Dis. 2014;20:917–919. doi: 10.3201/eid2005.140195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Xie C., Zhang J., Zhang W., Yang D., Yu L., Jiang Y., Yang S., Gao F., Yang Z., Zhou Y., Tong G. The identification and characterization of two novel epitopes on the nucleocapsid protein of the porcine epidemic diarrhea virus. Sci. Rep. 2016;6 doi: 10.1038/srep39010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Ge X., Chen D., Li J., Cai Y., Deng J., Zhou L., Guo X., Han J., Yang H. The S gene is necessary but not sufficient for the virulence of porcine epidemic diarrhea virus novel variant strain BJ2011C. J. Virol. 2018:92. doi: 10.1128/JVI.00603-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicht O., Li W., Willems L., Meuleman T.J., Wubbolts R.W., van Kuppeveld F.J., Rottier P.J., Bosch B.J. Proteolytic activation of the porcine epidemic diarrhea coronavirus spike fusion protein by trypsin in cell culture. J. Virol. 2014;88:7952–7961. doi: 10.1128/JVI.00297-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E.N. An apparently new syndrome of porcine epidemic diarrhoea. Vet. Rec. 1977;100:243–244. doi: 10.1136/vr.100.12.243. [DOI] [PubMed] [Google Scholar]
- Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Pan Y., Wang D., Tian X., Song Y., Cao Y. Identification and pathogenicity of a variant porcine epidemic diarrhea virus field strain with reduced virulence. Virol. J. 2015;12:88. doi: 10.1186/s12985-015-0314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.J., Wu Y.L., Zhu J.P., Tong W., Yu H., Jiang Y.F., Tong G.Z. Complete genome sequence of a virulent porcine epidemic diarrhea virus strain. J. Virol. 2012;86:13862. doi: 10.1128/JVI.02635-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material