Abstract
Hantaviruses are emerging RNA viruses that cause human diseases predominantly in Asia, Europe, and the Americas. Besides rodents, insectivores and bats serve as hantavirus reservoirs. We report the detection and genome characterization of a novel bat-borne hantavirus isolated from insectivorous common noctule bat. The newfound virus was tentatively named as Brno virus.
Keywords: Hantavirus, Bat, Phylogenetic analysis, Emerging virus, Bat-borne virus
Highlights
-
•
Novel tentative Hantavirus species (Brno virus; BRNV) was identified in European bat species Nyctalus noctula
-
•
BRNV represents the first identified bat-borne hantavirus in Europe
-
•
Nearly complete sequence of all genomic segments of BRNV was determined
-
•
BRNV phylogenetically groups with other novel hantaviruses from bats
1. Introduction
Hantaviruses (genus Hantavirus, family Bunyaviridae) are responsible for life-threatening human diseases: hantavirus cardiopulmonary syndrome (HCPS) in the Americas and hemorrhagic fever with renal syndrome (HFRS) in Asia and Europe (Krüger et al., 2011). Rodents are natural reservoirs of hantaviruses; however, recent studies have demonstrated that insectivores and bats also represent hosts for divergent hantaviruses (Xu et al., 2015, Zhang, 2014, Witkowski et al., 2016). Bats are considered the natural reservoir of a large variety of zoonotic viruses causing serious human diseases, such as lyssaviruses, henipaviruses, severe acute respiratory syndrome coronavirus, and Ebola virus (Li et al., 2010). Genetically divergent bat-borne hantaviruses have been identified in Africa – Mouyassue virus (MOYV) in Cote d'Ivoire (Sumibcay et al., 2012), Magboi virus (MGBV) in Sierra Leone (Weiss et al., 2012), and Makokou virus (MAKV) in Gabon (Witkowski et al., 2016) and in Asia – Xuan Son virus (XSV) in Vietnam (Arai et al., 2013), Huangpi virus (HUPV), Longquan virus (LQUV), Laibin virus (LBV) in China (Xu et al., 2015, Guo et al., 2013), and Quezon virus (QZNV) in the Philippines (Arai et al., 2016). Here we report the detection of a novel hantavirus, tentatively named Brno virus (BRNV), in the European insectivorous bat species Nyctalus noctula collected in the Czech Republic, Central Europe. For genetic characterization, the three genome segments were sequenced by high-throughput sequencing (HTS).
2. The study
A total of 53 bats were collected during the years 2008–2013 in the South Moravia region in the Czech Republic. The sample collection contained bats that died accidentally or were found dead in the field. Bats represented 14 different species from two families: Eptesicus nilssonii (n = 1), E. serotinus (n = 1), Hypsugo savii (n = 4), Myotis bechsteinii (n = 1), M. daubentonii (n = 3), M. emarginatus (n = 1), M. mystacinus (n = 1), M. myotis (n = 1), Nyctalus noctula (n = 12), Pipistrellus pipistrellus (n = 15), Plecotus auritus (n = 2), Pl. austriacus (n = 2), Vespertilio murinus (n = 6) of the family Vespertilionidae and Rhinolophus hipposideros (n = 3) of the family Rhinolophidae.
Total RNA was extracted from the lungs, kidneys and livers of all animals using QIAamp viral RNA Mini Kit (QIAGEN) or QIAZOL/TRIZOL method and screened for the presence of hantavirus RNA by a broad-spectrum RT-PCR targeting the large (L) genome segment of hantaviruses (Klempa et al., 2006). Hantavirus RNA was identified in two noctule bats (N. noctula – the species was determined morphologically and confirmed by HTS-based results of three host genes) collected in the city of Brno. The obtained sequences (369 base pairs, bp) were designated as Brno 7/2012/CZE (amplified from liver tissue) and Brno 11/2013/CZE (identical sequences were obtained from liver and kidney samples) and deposited in GenBank (accession numbers KR920359 and KR920360, respectively).
Attempts to isolate the virus were done in Vero cells and in suckling mice, but were negative.
In order to determine the whole genome sequence of BRNV, IonTorrent HTS was conducted. The sample Brno 7/2012/CZE was selected as most suitable for sequencing based on high viral loads in a novel BRNV-specific real-time RT-qPCR (qScript XLT 1-step RT-PCR Kit (Quanta/VWR)). Briefly, RNA was extracted from the liver tissue (QIAamp RNeasy mini Kit, Qiagen) and its concentration was measured with a Nanodrop 1000 photometer (RNA concentrations in the two samples were of 1545.91 ng/μl and 896.98 ng/μl). A purification and concentration step with RNA clean beads (Beckman Coulter) followed. Synthesis of cDNA was performed with a cDNA Synthesis kit (Roche) and resulting cDNA was fragmented by M220 Focused-ultrasonicator (Corvaris). This fragmented cDNA with an average length of about 500 bp was used for library preparation (Gene Read Library Prep Kit, Qiagen). The library was purified and size selected using AMPure XP Beads (Beckman Coulter). Optimal size distribution and quality of the resulting library were verified on a Bioanalyzer in combination with a DNA High Sensitivity Chip (Agilent). The library was quantified with the Ion Library Taqman Quantitation Kit (ThermoFisher). Sequencing was performed on an Ion Personal Genome Machine (ThermoFisher) according to manufacturer's guidelines.
A metagenomics analysis of the complete dataset was conducted using RIEMS software (Scheuch et al., 2015). Reads classified as related to the family Bunyaviridae were extracted and de-novo assembled into contigs using 454 Sequencing System Software (version 3.0). Resulting contigs were subsequently analyzed using the BioEdit software 7.2.5 (Hall, 1999) and Geneious 9.0.5 (Kearse et al., 2012). The lengths of the sequences of S, M and L gene segments were 1441, 3575 and 6528 nt, respectively, encoding nucleocapsid protein (N), glycoprotein precursor (GPC) and viral RNA-dependent RNA polymerase (L) proteins of 423, 1136 and 2145 amino acids in length, respectively (GenBank accession numbers: KX845678, KX845679, KX845680). Sequence comparison revealed that the three genome segments and the encoded proteins of BRNV showed 54.7–78.3% nucleotide and 44.5–81.7% amino acid sequence identity with other bat-borne hantaviruses while sequence identity with hantaviruses from rodents, shrews and moles ranged between 50.1 and 64.8%at the nucleotide and between 38.9 and 64.1% at amino acid level (Table 1 ).
Table 1.
Nucleotide and amino acid sequence identities (%) of three genome segments and corresponding encoded nucleocapsid protein (N), glycoprotein precursor (GPC) and RNA-dependent RNA polymerase (RdRp) between Brno virus (BRNV) and other representative bat-, insectivore- and rodent-borne hantavirusesa.
| Host | Virus strain | Country | S segment/N |
M segment/GPC |
L segment/RdRp |
|||
|---|---|---|---|---|---|---|---|---|
| 1272 nt | 424 aa | 3411 nt | 1137 aa | 6435 nt | 2145 aa | |||
| Bats | Longquan virus | China | 65.9 | 65 | 66.3 | 62.5 | 78.3b | 80.5b |
| Laibin virus | China | 58.7 | 56.3 | 55.4 | 45.6 | 66 | 66.7 | |
| Huangpi virus | China | 65.6b | 64b | – | – | 71.7b | 81.7b | |
| Xuan Son virus | Vietnam | 58.5 | 54.3 | 61.2b | 54.4b | 70b | 75.1b | |
| Mouyassue virus | Côte d'Ivoire | – | – | – | – | 73.4b | 78.9b | |
| Magboi virus | Sierra Leone | – | – | – | – | 74.2b | 75.7b | |
| Makokou virus | Gabon | – | – | – | – | 66.8b | 67.6b | |
| Quezon virus | Philippines | 59.2 | 55.5 | 54.7 | 44.5 | 65.4 | 66.6 | |
| Shrews | Uluguru virus | Tanzania | 50.9 | 40.9 | 54.8b | 42.9b | 64.5 | 62.7 |
| Altai virus | Russia | 56.9b | 52.6b | 54.8b | 48.9b | 63.3 | 62.3 | |
| Cao Bang virus | Vietnam | 57.1 | 51.7 | 51.9 | 40.4 | 63.7 | 62.1 | |
| Seewis virus | Switzerland | 57.8 | 49.4 | 54.2b | 46.9b | 60.7b | 60.3b | |
| Thottapalayam virus | India | 54 | 46.5 | 50.1 | 38.9 | 62.8 | 62.2 | |
| Moles | Nova virus | Belgium | 57.5 | 51.7 | 54.6 | 44.2 | 64.5 | 63.3 |
| Asama virus | Japan | 55.6 | 51 | 52.2b | 40.3b | 64.8b | 64.1b | |
| Rodents | Puumala virus | Finland | 58.3 | 51.9 | 51 | 40.1 | 63.2 | 60.3 |
| Sin Nombre virus | USA | 58.8 | 53 | 52.5 | 41.6 | 63.4 | 61.3 | |
| Seoul virus | Korea | 55.4 | 48.8 | 51.6 | 39.1 | 62.2 | 60.7 | |
| Hantaan virus | Korea | 56.6 | 50.1 | 51 | 40 | 62.2 | 60.6 | |
| Dobrava-Belgrade virus | Greece | 55.7 | 49.8 | 50.4 | 40.1 | 62.5 | 61.1 | |
| Tula virus | Czech Republic | 57.2 | 52.9 | 51.9 | 40.9 | 63.1 | 61.3 | |
– no sequence available.
Viral sequences used to generate sequence identities: Bat-borne hantaviruses: Longquan virus (JX465415, JX465397, JX465381), Laibin virus (KM102247, KM102248, KM102249), Huangpi virus (JX473273, JX465369), Xuan Son virus (KF704710, KJ000539, KF704715), Mouyassue virus (JQ287716), Magboi virus (JN037851), Makokou virus (KT316176), Quezon virus (KU950713, KU950714, KU950715); Shrew-borne hantaviruses: Uluguru virus (JX193695, JX193696, JX193697), Altai virus (KM361048, KM361053, KM361061), Cao Bang virus (EF543524, EF543526, EF543525), Thottapalayam virus (NC_010704, NC_010708, NC_010707), Seewis virus (EF636024, EF636025, EF636026); Mole-borne hantaviruses: Nova virus (KT004445, KT004446, KT004447), Asama virus (EU929072, EU929075, EU929078); Rodent-borne hantaviruses: Puumala virus (NC_005224, NC_005223, NC_005225), Sin Nombre virus (NC_005216, NC_005215, NC_005217), Seoul virus (NC_005236, NC_005237, NC_005238), Hantaan virus (NC_005218, NC_005219, NC_005222), Dobrava-Belgrade virus (NC_005233, NC_005234, NC_005235), Tula virus (NC_005227, NC_005228, NC_005226).
Comparison based on shorter sequences.
The observed amino acid sequence differences clearly exceed one of the current species demarcation criteria of the current International Committee on Taxonomy of Viruses (ICTV) for the genus Hantavirus, 7% difference in amino acid sequences of the N and GPC proteins. The current ICTV criteria also include serological differences in virus neutralization, preventing virus classification without a cell culture isolate. However, a new proposal for species demarcation criteria (assigned code of the proposal: 2016.023a,bM; available for download at https://talk.ictvonline.org), submitted by the ICTV Bunyaviridae Study Group, is currently under consideration which allows the classification solely based on genetic data. It is based on a concatenated multiple sequence alignment of complete amino acid sequences of the N and GPC proteins which is used to calculate PED (pairwise evolutionary distances) values using WAG amino acid substitution matrix. A species of the genus Hantavirus is defined by a PED value greater than 0.1. According to this criterion, BRNV clearly represents a new hantavirus species because the lowest PED value, observed for the most closely related LQUV, is 0.5.
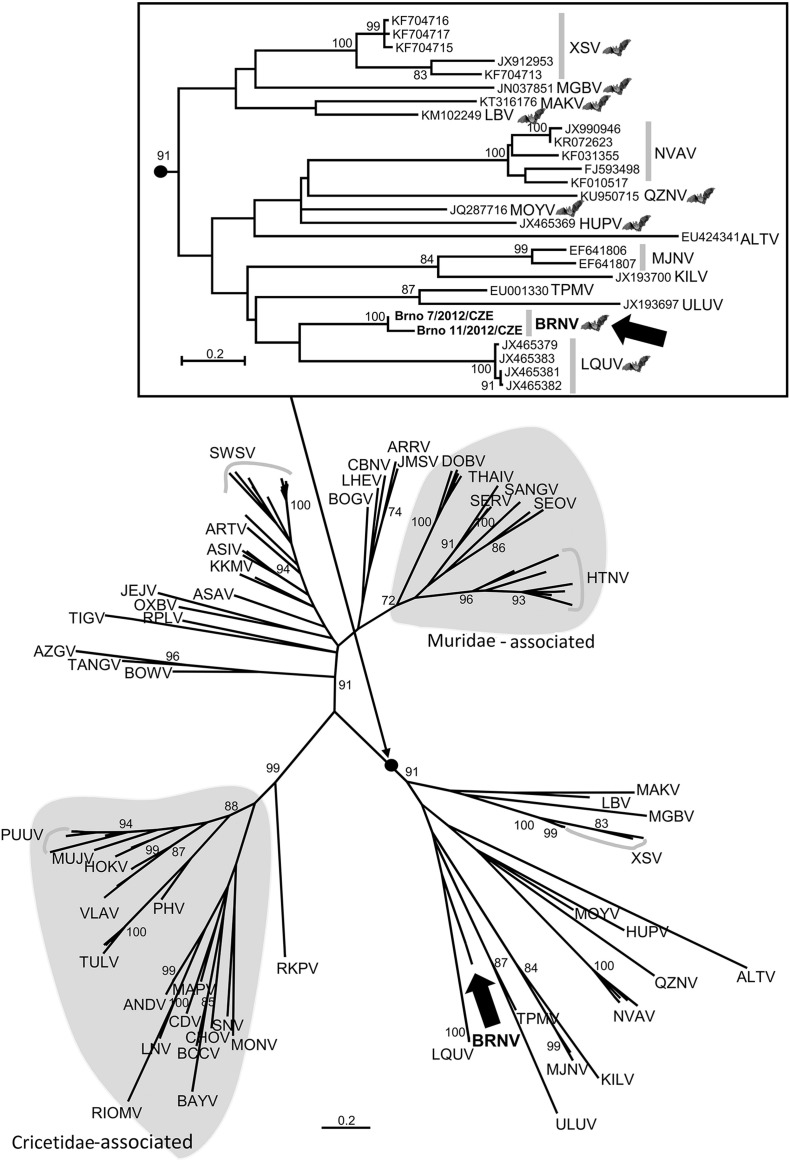
The obtained BRNV sequences were subjected to phylogenetic analyses within the Maximum Likelihood framework using MEGA7 (Kumar et al., 2016). Unfortunately, only partial L segment sequences are available for the majority of bat-borne hantaviruses. Therefore, we first based our analysis on a short L segment dataset (352 nt) which contains all currently recognized bat-borne hantaviruses (Fig. 1 ). BRNV clustered within the clade containing all bat-borne hantaviruses and shared the most recent common ancestor with LQUV found in insectivorous Rhinolophus sp. bats in China. This pattern has been consequently observed in all other phylogenetic analyses including complete amino acid sequences for all three segments (phylogenetic trees based on partial sequences of S and M segments are shown in Supplementary Figs. 1 and 2).
Fig. 1.
Maximum-Likelihood phylogenetic tree showing the phylogenetic position of Brno virus (BRNV; marked by black arrow and bold face) constructed on the basis of partial L segment nucleotide sequences (352 nt). Evolutionary analysis was conducted in MEGA7 (15). The evolutionary history was inferred by using the Maximum-Likelihood method based on the General Time Reversible (GTR) model with using a discrete Gamma distribution (+ G) with 5 rate categories and by assuming that a certain fraction of sites are evolutionarily invariable (+ I) which was estimated to be the Best-Fit substitution model according to Bayesian Information Criterion. The scale bars indicate an evolutionary distance in substitutions per position in the sequence. Bootstrap values ≥ 70%, calculated from 500 replicates, are shown at the tree branches. The insert shows the phylogenetic group containing all bat-borne hantaviruses (marked by bat pictogram) in greater detail. Rodent-borne hantaviruses associated with members of the families Muridae or Cricetidae are indicated by grey-shaded background. The list of the accession numbers used in the analysis is available from the authors upon request. Abbreviations: ALTV, Altai virus; ANDV, Andes virus; ARRV, Ash River virus; ARTV, Artybash virus; ASAV, Asama virus; ASIV, Asikkala virus; AZGV, Azagny virus; BAYV, Bayou virus; BCCV, Black Creek Canal virus; BOGV, Boginia virus; BOWV, Bowé virus; BRNV, Brno virus; CBNV, Cao Bang virus; CDV, Cano Delgadito virus; CHOV, Choclo virus; DOBV, Dobrava-Belgrade virus; HOKV, Hokkaido virus; HUPV, Huanqpi virus; HTNV, Hantaan virus; JEJV, Jeju virus; JMSV, Jemez Springs virus; KILV, Kilimanjaro virus; KKMV, Kenkeme virus; LAIV, Laibin virus; LHEV, Lianghe virus; LNV, Laguna Negra virus; LQUV, Longquan virus; MAKV, Makokou virus; MAPV, Maporal virus; MGBV, Magboi virus; MJNV, Imjin virus; MONV, Montano virus; MOUV, Mouyassué virus; MUJV, Muju virus; NVAV, Nova virus; OXBV, Oxbow virus; PHV, Prospect Hill virus; PUUV, Puumala virus; QDLV, Qiandao Lake virus; QZNV, Quezon virus; RIOMV, Rio Mamore virus; RKPV; Rockport virus; RPLV, Camp Ripley virus; SANGV, Sangassou virus; SEOV, Seoul virus; SERV, Serang virus; SNV, Sin Nombre virus; SWSV, Seewis virus; TANGV, Tanganya virus; TIGV, Tigray virus; THAIV, Thailand virus; TPMV, Thottapalayam virus; TULV, Tula virus; ULUV, Uluguru virus; VLAV, Vladivostok virus; XSV, Xuan Son virus. In cases of HTNV, PUUV, SWSV, and XSV, several sequences of the same virus were marked by grey curve to allow unambiguous designation of the taxa.
3. Conclusions
In the present study a novel bat-borne hantavirus has been identified and its complete sequence of all coding genomic regions of has been determined. The successful determination of the genome segment sequences of BRNV provide reference data for improving detection methods and determining the genome sequences of further European bat-borne hantaviruses.
Its phylogenetic relatedness to other bat-borne hantaviruses, high genetic distance to other known hantaviruses, and its independent detection in two distinct animals of the same species and in two organs led us to conclude that BRNV is an indigenous bat-borne hantavirus associated with common noctule bat (N. noctula). This study provided a missing molecular proof that bat-borne hantaviruses occur in Europe too, and need to be considered as putative public health threat.
Acknowledgments
This study was funded by grant no. LO1218 of the MEYS of the Czech Republic under the NPU I program (to D.R.), Ministry of Agriculture of the Czech Republic (RO0516 to D.R.), by the contract-research-project for the Bundeswehr Medical Service FV E/U2AD/CF512/DF557 META-InfRisk (to R.G.U.) and partially funded by the Slovak Scientific Grant Agency VEGA, grant number 2/0174/15 (to B.K.). The continuous support of Martin Beer is kindly acknowledged. Petra Straková acknowledges support by Erasmus + stipendium.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.meegid.2016.12.025.
Appendix A. Supplementary data
Supplementary figures: Maximum-Likelihood phylogenetic trees showing the phylogenetic position of BRNV (constructed on the basis of partial nucleotide sequences of the S and M genomic segments).
References
- Arai S., Nguyen S.T., Boldgiv B., Fukui D., Araki K., Dang C.N. Novel bat-borne hantavirus, Vietnam. Emerg. Infect. Dis. 2013;19:1159–1161. doi: 10.3201/eid1907.121549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S., Taniguchi S., Aoki K., Yoshikawa Y., Kyuwa S., Tanaka-Taya K. Molecular phylogeny of a genetically divergent hantavirus harbored by the Geoffroy's rousette (Rousettus amplexicaudatus), a frugivorous bat species in the Philippines. Infect. Genet. Evol. 2016;45:26–32. doi: 10.1016/j.meegid.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Guo W.P., Lin X.D., Wang W., Tian J.H., Cong M.L., Zhang H.L. Phylogeny and origins of hantaviruses harbored by bats, insectivores, and rodents. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;25:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempa B., Fichet-Calvet E., Lecompte E., Auste B., Aniskin V., Meisel H. Hantavirus in African wood mouse, Guinea. Emerg. Infect. Dis. 2006;12:838–840. doi: 10.3201/eid1205.051487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger D.H., Schönrich G., Klempa B. Human pathogenic hantaviruses and prevention of infection. Hum. Vaccin. 2011;7:685–693. doi: 10.4161/hv.7.6.15197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. http://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Victoria J.G., Wang C., Jones M., Fellers G.M., Kunz T.H. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010;84:6955–6965. doi: 10.1128/JVI.00501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuch M., Höper D., Beer M. RIEMS: a software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinf. 2015;16:69. doi: 10.1186/s12859-015-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumibcay L., Kadjo B., Gu S.H., Kang H.J., Lim B.K., Cook J.A. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d'Ivoire. Virol. J. 2012;9:34. doi: 10.1186/1743-422X-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S., Witkowski P.T., Auste B., Nowak K., Weber N., Fahr J. Hantavirus in bat, Sierra Leone. Emerg. Infect. Dis. 2012;18:159–161. doi: 10.3201/eid1801.111026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski P.T., Drexler J.F., Kallies R., Ličková M., Bokorová S., Mananga G.D. Phylogenetic analysis of a newfound bat-borne hantavirus supports a laurasiatherian host association for ancestral mammalian hantaviruses. Infect. Genet. Evol. 2016;41:113–119. doi: 10.1016/j.meegid.2016.03.036. [DOI] [PubMed] [Google Scholar]
- Xu L., Wu J., He B., Qin S., Xia L., Qin M. Novel hantavirus identified in black-bearded tomb bats, China. Infect. Genet. Evol. 2015;31:158–160. doi: 10.1016/j.meegid.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z. Discovery of hantaviruses in bats and insectivores and the evolution of the genus Hantavirus. Virus Res. 2014;187:15–21. doi: 10.1016/j.virusres.2013.12.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures: Maximum-Likelihood phylogenetic trees showing the phylogenetic position of BRNV (constructed on the basis of partial nucleotide sequences of the S and M genomic segments).