Highlights
-
•
Leptospira from Eidolon helvum bats in Zambia was detected by flaB-nested PCR.
-
•
The Leptospira flaB was detected in 72 of 529 E. helvum bats.
-
•
The Leptospira rrs was also detected in E. helvum bats in Zambia.
-
•
Most of the Leptospira sequences belong to a unique cluster in the phylogeny.
-
•
E. helvum bats are a candidate natural reservoir to pathogenic Leptospira in Zambia.
Keywords: Leptospira, Fruit bat, Eidolon helvum, Zambia, Pathogenic
Abstract
The role played by bats as a potential source of transmission of Leptospira spp. to humans is poorly understood, despite various pathogenic Leptospira spp. being identified in these mammals. Here, we investigated the prevalence and diversity of pathogenic Leptospira spp. that infect the straw-colored fruit bat (Eidolon helvum). We captured this bat species, which is widely distributed in Africa, in Zambia during 2008–2013. We detected the flagellin B gene (flaB) from pathogenic Leptospira spp. in kidney samples from 79 of 529 E. helvum (14.9%) bats. Phylogenetic analysis of 70 flaB fragments amplified from E. helvum samples and previously reported sequences, revealed that 12 of the fragments grouped with Leptospira borgpetersenii and Leptospira kirschneri; however, the remaining 58 flaB fragments appeared not to be associated with any reported species. Additionally, the 16S ribosomal RNA gene (rrs) amplified from 27 randomly chosen flaB-positive samples was compared with previously reported sequences, including bat-derived Leptospira spp. All 27 rrs fragments clustered into a pathogenic group. Eight fragments were located in unique branches, the other 19 fragments were closely related to Leptospira spp. detected in bats. These results show that rrs sequences in bats are genetically related to each other without regional variation, suggesting that Leptospira are evolutionarily well-adapted to bats and have uniquely evolved in the bat population. Our study indicates that pathogenic Leptospira spp. in E. helvum in Zambia have unique genotypes.
1. Introduction
Leptospirosis is an important reemerging zoonotic disease caused by pathogenic spirochetes of the genus Leptospira. The disease is found worldwide, especially in tropical regions. Human leptospirosis presents with a variety of signs and symptoms, including general febrile disease an influenza-like illness, and results in liver or kidney failure. As a result, this disease is often confused with other diseases, such as dengue fever, hemorrhagic fever and malaria, all of which are common in tropical and subtropical regions of the world (World Health Organization, 2003). Pathogenic Leptospira spp. can infect the renal tubules of most animals and are excreted in their urine, resulting in contaminated environments (e.g., soil and water) (Adler and de la Peña Moctezuma, 2010). Humans become infected mainly through Leptospira-contaminated water or soil, or from contact with urine from animals infected with this bacterium (Adler and de la Peña Moctezuma, 2010). Rodents are the most important reservoir of Leptospira among a variety of wildlife reservoirs.
Over the past decade, there have been many reports of bats being an important reservoir and vector of emerging infectious diseases, such as Ebola and Marburg viral diseases, severe acute respiratory syndrome (known as SARS), Nipah and Hendra viral infections, and rabies (Calisher et al., 2006). Bats (order Chiroptera) are the second largest order in mammals after rodents (order Rodentia) and are geographically widespread. Loss of habitat for bats, caused by recent anthropogenic activities, may increase contact between bats and humans, resulting in transmission of various pathogens from peridomestic bats to humans (de Jong et al., 2011). Transmission of viral pathogens from bats to humans has been the main focus of studies in this area; however, there have not been many studies on pathogenic bacteria in bats (Mühldorfer, 2013).
A variety of pathogenic Leptospira spp. have been identified in bats worldwide (Bessa et al., 2010, Bunnell et al., 2000, Cox et al., 2005, Fennestad and Borg-Petersen, 1972, Harkin et al., 2014, Lagadec et al., 2012, Matthias et al., 2005, Tulsiani et al., 2011); however, little is known about the role of bats in the transmission of leptospirosis.
In this study, we performed a molecular epidemiological investigation of Leptospira spp. in straw-colored fruit bats (Eidolon helvum) captured from 2008 to 2013, which were migrating from the Democratic Republic of Congo to Zambia (Richter and Cumming, 2008).
2. Materials and methods
A total of 529 kidney samples were collected from captured E. helvum that were roosting in trees (Muleya et al., 2014, Ogawa et al., 2015) in Kasanka National Park in Central Province and in Ndola in Copperbelt Province of Zambia (Table 1 ). This research was performed under the research project “Molecular epidemiology of bacterial zoonoses in Zambia” approved by the Zambia Wildlife Authority, in the Republic of Zambia.
Table 1.
Summary of the kidney samples analyzed from fruit bats and Leptospira flaB prevalence.
| Year | Sample ID | Location | No. of samples |
No. of positives |
Positive rate (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | M | F | Total | M | F | Total | M | F | |||
| 2008 | ZFB08-01 – ZFB08-104 | Kasanka National Park | 104 | 38 | 66 | 28 | 10 | 18 | 26.9 | 26.3 | 27.3 |
| 2009 | ZFB09-01 – ZFB09-60 | Kasanka National Park | 60 | 15 | 45 | 7 | 2 | 5 | 11.7 | 13.3 | 11.1 |
| 2010 | ZFB10-01 – ZFB10-47 | Kasanka National Park | 47 | 13 | 34 | 4 | 1 | 3 | 8.5 | 7.7 | 8.8 |
| ZFB10-48 – ZFB10-52 | Ndola | 4 | 3 | 1 | 1 | 1 | 0 | 25.0 | 33.3 | 0 | |
| 2011 | ZFB11-01 – ZFB11-38 | Ndola | 38 | 18 | 20 | 3 | 0 | 3 | 7.9 | 0 | 15.0 |
| ZFB11-39 – ZFB11-95 | Kasanka National Park | 57 | 24 | 33 | 7 | 4 | 3 | 12.3 | 12.5 | 9.1 | |
| 2012 | ZFB12-01 – ZFB12-60 | Ndola | 60 | 22 | 38 | 4 | 2 | 2 | 6.7 | 9.1 | 5.3 |
| ZFB12-61 – ZFB12-110a | Kasanka National Park | 49 | 15 | 34 | 18 | 7 | 11 | 36.7 | 46.7 | 32.4 | |
| 2013 | ZFB13-01 – ZFB13-76 | Ndola | 76 | 23 | 53 | 0 | 0 | 0 | 0 | 0 | 0 |
| ZFB13-77 – ZFB13-111b | Kasanka National Park | 34 | 9 | 25 | 7 | 2 | 5 | 20.6 | 22.2 | 20.0 | |
| Total | 529 | 180 | 349 | 79 | 28 | 50 | 14.9 | 15.6 | 14.3 | ||
Kidney sample from ZFB12-97 was not available for PCR screening.
Kidney sample from ZFB13-93 was not available for PCR screening.
The kidney samples collected from E. helvum were placed directly in Korthof or Ellinghausen–McCullough–Johnson–Harris (EMJH) media (World Health Organization, 2003) and homogenized for DNA extraction and Leptospira isolation by crushing with beads. DNA was extracted from 10% (w/v) kidney homogenates using a DNA Isolation Kit for Mammalian Blood (Roche Diagnostics, Indianapolis, IN, USA) according to the manufacturer’s instructions with minor modifications. A nested PCR based on the flagellin B gene (flaB) sequence was used to amplify the extracted DNA samples (n = 529) to detect the flaB gene of pathogenic Leptospira spp. (Koizumi et al., 2008). Some of the flaB-nested PCR-positive samples (n = 27) were examined further. To identify Leptospira species, we also performed a nested PCR based on the 16S ribosomal RNA gene (rrs) and the preprotein translocase gene (secY) using the primer sets shown in Supplementary Tables 1 and 2.
The PCR products from the flaB-nested PCR (732 bp including the 41 bp primer sequence), the rrs-nested PCR (∼642 bp including the 48 bp primer sequence) and the secY-nested PCR (∼329 bp including the primer sequence) were purified and subjected to direct sequencing using a BigDye Terminator v3.1 Cycle Sequencing a Kit (Life Technologies, Waltham, MA, USA) according to the manufacturer’s instructions, and a 3130xl Genetic Analyzer (Life Technologies). The sequence data were aligned using the Clustal W software, and a maximum-likelihood phylogenetic tree was generated with 1,000 bootstrap replications using MEGA 5.2.2 software (Tamura et al., 2011).
The DDBJ accession numbers for the flaB and rrs sequences from the uncultured Leptospira spp. detected in E. helvum comprised LC005103 to LC005172 and LC005173 to LC005199, respectively (Supplementary Table 3).
3. Results and discussion
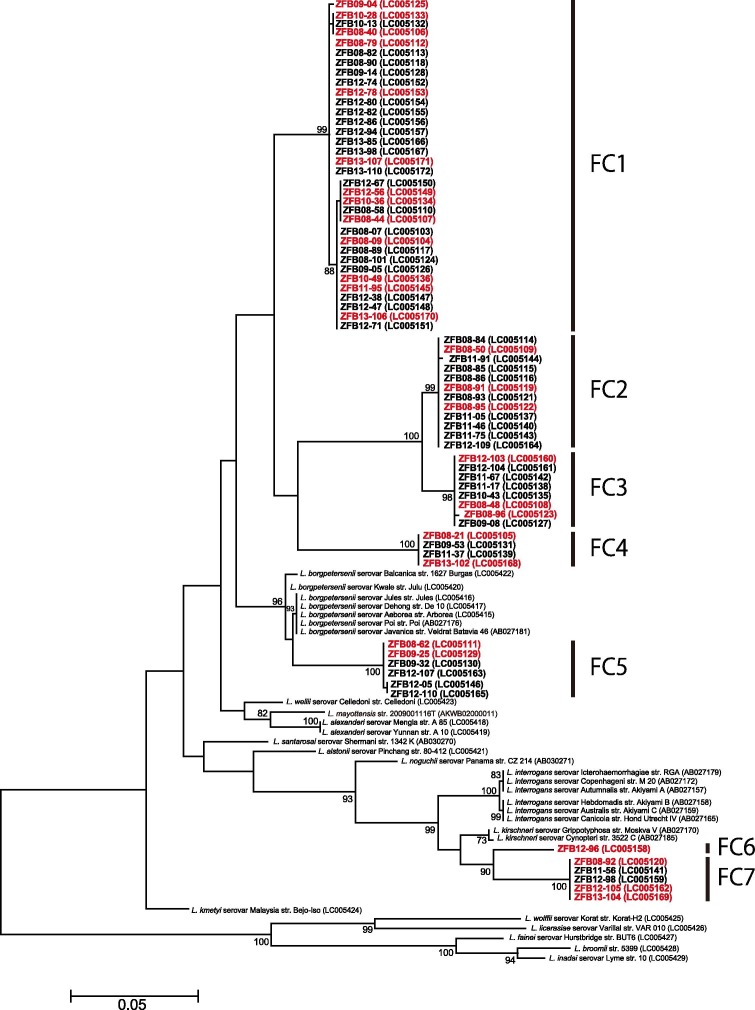
A 732 bp fragment of the Leptospira flaB gene was detected in 79 out of 529 E. helvum kidney samples (14.9%, Table 1). Among the 79 flaB-nested PCR-positive samples, 70 were used for direct sequencing and nine samples were not able to be sequenced because of insufficient DNA. Phylogenetic analysis (Fig. 1 ) revealed that the flaB sequences fell into seven clusters (FC1–FC7). Six flaB fragments (ZFB08-62, ZFB09-25, ZFB09-32, ZFB12-05, ZFB12-107 and ZFB12-110) in the FC5 cluster were related to the corresponding gene sequences, all of which were identical to Leptospira borgpetersenii strains including Jules, De 10, Arborea, Poi, and Veldrat Batavia 46. The six fragments shared sequence identities ranging from 96.2% to 96.4% with the L. borgpetersenii strains described above. The nucleotide identity of the flaB fragment for ZFB12-96 in the FC6 cluster with the Leptospira kirschneri strains Moskva V and 3522C was 95.5% and 95.4%, respectively. The nucleotide sequence identities of five flaB fragments (ZFB08-92, ZFB11-56, ZFB12-98, ZFB12-105 and ZFB13-104) in the FC7 cluster with Moskva V and 3522C L. kirschneri strains were 95.3% and 95.1%, respectively. The nucleotide sequence identities of the remaining 58 flaB fragments belonging to FC1 to FC4 with that of the closest species, L. borgpetersenii, were from 91.2% to 94.5%. In a previous report, L. borgpetersenii, L. kirschneri, and Leptospira interrogans were isolated predominantly from rodents in Africa (Ahmed et al., 2006, Nalam et al., 2010). A novel species, Leptospira mayottensis, for which strains were isolated in Mayotte located in the Comoros archipelago, was reported to be genetically similar to L. borgpetersenii (Bourhy et al., 2012, Bourhy et al., 2014). This new species was included in phylogenetic analysis; however, the 58 flaB fragments belonging to the FC1–FC4 clusters were also distantly related to L. mayottensis. The 12 flaB fragments belonging to the three clusters FC5–FC7 appear to group with those of L. borgpetersenii and L. kirschneri; however, the remaining 58 flaB fragments belonging to the FC1–FC4 clusters appear not to be associated with those of any reported species. Accordingly, the Leptospira flaB sequence data from kidney samples of captured E. helvum bats indicates that leptospires from E. helvum in Zambia have genotypes distinct from those previously reported. The results of phylogenetic analysis of the secY gene, which has been used as a valuable tool for discriminating between Leptospira spp. (Gravekamp et al., 1993, Rahelinirina et al., 2010, Victoria et al., 2008), were in accordance with the flaB-based phylogenetic tree (Supplementary Fig. 1), also supporting the hypothesis that leptospires from E. helvum in Zambia have unique genotypes.
Fig. 1.
Maximum-likelihood phylogenetic tree based on the nucleotide sequences of Leptospira spp. flaB in E. helvum bats. The dendrogram was constructed with the JC69 model, and with 1,000 replications using MEGA 5.2.2 software (Tamura et al., 2011). Numbers at nodes indicate bootstrap supports >70%. The sequences determined in this study are shown in bold. The samples colored red were also used in the phylogenetic analysis of rrs (Fig. 2). GenBank accession numbers are indicated in parentheses. Scale bar indicates the number of nucleotide substitutions per site.
Subsequently, we examined another gene from Leptospira, rrs, which has been used before to identify Leptospira spp. (Matthias et al., 2005, Postic et al., 2000). Fragments of the rrs gene (each ∼593 bp) were amplified and sequenced from 27 samples randomly selected from the seven clusters (FC1–FC7) (Fig. 2 ). Phylogenetic analysis showed that all 27 rrs fragments from E. helvum kidney samples grouped into a pathogenic group. ZFB08-92, ZFB12-105 and ZFB13-104, belonging to the FC7 cluster, were associated with the L. kirschneri strain, Kambale (FJ154562), and uncultured Leptospira sp. (JQ288731) from Triaenops menamena bats captured in Madagascar (Lagadec et al., 2012) (Fig. 2). ZFB09-25 (FC5) and ZFB12-96 (FC6) were closely related to L. borgpetersenii and uncultured Leptospira sp. (AY995720) from bats captured in Peru, respectively (Matthias et al., 2005) (Fig. 2). The rrs fragments of ZFB08-21 (FC4), ZFB08-91 (FC2), ZFB12-103 (FC3) and ZFB13-102 (FC4), all of which were identical, as well as those of ZFB08-50 (FC2), ZFB08-95 (FC2), ZFB08-48 (FC3) and ZFB08-96 (FC3), were located on unique branches (Fig. 2). The other 14 rrs fragments were closely related to an uncultured Leptospira sp. (JQ288732) from Rousettus obliviosus bats captured in Comoros; the latter sequence was closely related to L. borgpetersenii (Lagadec et al., 2012) (Fig. 2). Non-significant coevolutionary congruence was reported between the rrs sequence from Leptospira spp. and that of bats at the bat species level (Lei and Olival, 2014). However, the rrs sequences from bats are genetically related to each other and show no regional variations in phylogenetic analysis of the rrs sequences from various kinds of hosts (Fig. 2), suggesting that Leptospira have evolved uniquely in this bat population. Dietrich et al. reported that the host is an important factor in Leptospira diversification (Dietrich et al., 2014), also supporting our findings.
Fig. 2.
Maximum-likelihood phylogenetic tree based on the nucleotide sequences of Leptospira spp. rrs in E. helvum bats. The dendrogram was constructed with the general time reversible model with gamma distribution and invariable sites, and with 1000 replications using MEGA 5.2.2 software (Tamura et al., 2011). Numbers at nodes indicate bootstrap supports >70%. The sequences determined in this study are shown in red. The sequences from bats are shown in bold. The FC clusters shown in Fig. 1 and GenBank accession numbers are indicated in brackets and parentheses, respectively. Scale bar indicates the number of nucleotide substitutions per site.
Zambia is bordered by eight countries. Epidemiological studies in these countries have been reported; however, almost all of these reports were serological surveys using L. interrogans as the antigen, and most data originated from Tanzania and Zimbabwe (de Vries et al., 2014). In Zambia, data regarding circulating Leptospira spp. are limited. Serosurveys of Leptospira spp. in rodents and Leptospira weilii in pigs have been reported (de Vries et al., 2014). Although E. helvum examined in this study were migrating to Zambia from the Democratic Republic of Congo (Richter and Cumming, 2008), data in this country are also lacking and there are no previous reports on L. borgpetersenii that may be related to the Leptospira spp. detected in this study. E. helvum captured in Kasanka National Park were more frequently infected than those captured in Nodla (x2 = 23.0, df = 1, p < 0.01). The roosting environment and colony size may influence this difference. No significant difference in the prevalence of the Leptospira flaB gene was found between males and females.
The phylogenetic analyses of flaB and rrs infer that genes from potentially pathogenic Leptospira spp. were present in the kidney samples of E. helvum in Zambia. To the best of our knowledge, this is the first report of PCR detection of Leptospira spp. in fruit bats from the African continent. In addition, the nested PCR-positive rate for Leptospira (14.9%) in E. helvum in Zambia was relatively higher than that of previous reports (Mühldorfer, 2013). Although isolation of Leptospira directly from bat kidney samples using Korthof and EMJH media was not successful, the relatively high infection rate in the kidneys of E. helvum is likely to result in excretion of Leptospira via the urine. Contaminated urine has therefore been proposed as the potential transmission pathway of Leptospira spp. from fruit bats to rodents (Tulsiani et al., 2011). It is suggested, therefore, that E. helvum might be a candidate natural reservoir for Leptospira in Zambia. Continued surveillance in E. helvum, as well as in humans and rodents, is required to gain a better understanding of how Leptospira is maintained in, and transmitted by, E. helvum bats in Zambia.
Acknowledgments
We thank Ms. Yuka Thomas, Drs. Emiko Nakagawa, Akihiro Ishii, Reiko Yoshida, Yasuko Orba, Ichiro Nakamura, Kimihito Ito, and many PhD students and postdoctoral fellows at the Research Center for Zoonosis Control, Hokkaido University for technical assistance. We also thank Drs. Katendi Chabgula, Edgar Simulundu and Musso Munyeme of the University of Zambia, Dr. Frank Willems of the Kasanka Trust, the Ministry Agriculture and Livestock, the Zambia Wildlife Authority, and Mr. Bwalya Chisha. This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) and the Science and Technology Research Partnership for Sustainable Development (SATREPS) by the Japan Science and Technology Agency (JST) and Japan International Cooperation Agency (JICA).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2015.03.013.
Appendix A. Supplementary data
Supplementary Table.
Neighbor-joining phylogenetic tree based on the nucleotide sequences of Leptospira spp. secY genes in E. helvum bats. The dendrogram (corresponding to the G1–G2 region) was constructed using Tanura-Nei distances, with 1000 replications, using MEGA 5.2.2 software (Tamura et al., 2011). Numbers at nodes indicate bootstrap supports >70%. The sequences determined in this study are shown in red. The FC clusters shown in Fig. 1 and GenBank accession numbers are indicated in brackets and parentheses, respectively. Scale bar indicates the number of nucleotide substitutions per site.
References
- Adler B., de la Peña Moctezuma A. Leptospira and leptospirosis. Vet. Microbiol. 2010;140:287–296. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Ahmed N., Devi S.M., Valverde Mde L., Vijayachari P., Machang’u R.S., Ellis W.A., Hartskeerl R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrobe. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa T.Á., Spichler A., Chapola É.G., Husch A.C., de Almeida M.F., Sodré M.M., Savani E.S., Sacramento D.R., Vinetz J.M. The contribution of bats to leptospirosis transmission in São Paulo City. Braz. Am. J. Trop. Med. Hyg. 2010;82:315–317. doi: 10.4269/ajtmh.2010.09-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P., Collet L., Lernout T., Zinini F., Hartskeerl R.A., van der Linden H., Thiberge J.M., Diancourt L., Brisse S., Giry C., Pettinelli F., Picardeau M. Human leptospira isolates circulating in Mayotte (Indian Ocean) have unique serological and molecular features. J. Clin. Microbiol. 2012;50:307–311. doi: 10.1128/JCM.05931-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourhy P., Collet L., Brisse S., Picardeau M. Leptospira mayottensis sp. nov., a pathogenic species of the genus Leptospira isolated from humans. Int. J. Syst. Evol. Microbiol. 2014;64:4061–4067. doi: 10.1099/ijs.0.066597-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell J.E., Hice C.L., Watts D.M., Montrueil V., Tesh R.B., Vinetz J.M. Detection of pathogenic Leptospira spp. infections among mammals captured in the Peruvian Amazon basin region. Am. J. Trop. Med. Hyg. 2000;63:255–258. [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox T.E., Smythe L.D., Leung L.K. Flying foxes as carriers of pathogenic Leptospira species. J. Wildl. Dis. 2005;41:753–757. doi: 10.7589/0090-3558-41.4.753. [DOI] [PubMed] [Google Scholar]
- de Jong C., Field H., Newman S.H., Epstein J.H. Emerging infectious dieses. In: Newman S.H., Field H.E., de Jong C.E., Epstein J.H., editors. Investigating the Role of Bats in Emerging Zoonoses: Balancing Ecology, Conservation and Public Health Interests. Food and Agriculture Organization of the United Nations; Rome: 2011. pp. 1–13. [Google Scholar]
- de Vries S.G., Visser B.J., Nagel I.M., Goris M.G., Hartskeerl R.A., Grobusch M.P. Leptospirosis in Sub-Saharan Africa: a systematic review. Int. J. Infect. Dis. 2014;28:47–64. doi: 10.1016/j.ijid.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Dietrich M., Wilkinson D.A., Soarimalala V., Goodman S.M., Dellagi K., Tortosa P. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol. Ecol. 2014;23:2783–2796. doi: 10.1111/mec.12777. [DOI] [PubMed] [Google Scholar]
- Fennestad K.L., Borg-Petersen C. Leptospirosis in Danish wild mammals. J. Wildl. Dis. 1972;8:343–351. doi: 10.7589/0090-3558-8.4.343. [DOI] [PubMed] [Google Scholar]
- Gravekamp C., Van de Kemp H., Franzen M., Carrington D., Schoone G.J., Van Eys G.J., Everard C.O., Hartskeerl R.A., Terpstra W.J. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J. Gen. Microbiol. 1993;139:1691–1700. doi: 10.1099/00221287-139-8-1691. [DOI] [PubMed] [Google Scholar]
- Harkin K.R., Hays M., Davis R., Moore M. Use of PCR to identify Leptospira in kidneys of big brown bats (Eptesicus fuscus) in Kansas and Nebraska, USA. J. Wildl. Dis. 2014;50:651–654. doi: 10.7589/2013-08-201. [DOI] [PubMed] [Google Scholar]
- Koizumi N., Muto M., Yamamoto S., Baba Y., Kudo M., Tamae Y., Shimomura K., Takatori I., Iwakiri A., Ishikawa K., Soma H., Watanabe H. Investigation of reservoir animals of Leptospira in the northern part of Miyazaki Prefecture. Jpn. J. Infect. Dis. 2008;61:465–468. [PubMed] [Google Scholar]
- Lagadec E., Gomard Y., Guernier V., Dietrich M., Pascalis H., Temmam S., Ramasindrazana B., Goodman S.M., Tortosa P., Dellagi K. Pathogenic Leptospira spp. in bats, Madagascar and Union of the Comoros. Emerg. Infect. Dis. 2012;18:1696–1698. doi: 10.3201/eid1810.111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B.R., Olival K.J. Contrasting patterns in mammal-bacteria coevolution: Bartonella and Leptospira in bats and rodents. PLOS Negl. Trop. Dis. 2014;8:e2738. doi: 10.1371/journal.pntd.0002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthias M.A., Díaz M.M., Campos K.J., Calderon M., Willig M.R., Pacheco V., Gotuzzo E., Gilman R.H., Vinetz J.M. Diversity of bat-associated Leptospira in the Peruvian Amazon inferred by bayesian phylogenetic analysis of 16S ribosomal DNA sequences. Am. J. Trop. Med. Hyg. 2005;73:964–974. [PMC free article] [PubMed] [Google Scholar]
- Mühldorfer K. Bats and bacterial pathogens: a review. Zoonoses Publ. Health. 2013;60:93–103. doi: 10.1111/j.1863-2378.2012.01536.x. [DOI] [PubMed] [Google Scholar]
- Muleya W., Sasaki M., Orba Y., Ishii A., Thomas Y., Nakagawa E., Ogawa H., Hang’ombe B., Namangala B., Mweene A., Takada A., Kimura T., Sawa H. Molecular epidemiology of paramyxoviruses in frugivorous Eidolon helvum bats in Zambia. J. Vet. Med. Sci. 2014;76:611–614. doi: 10.1292/jvms.13-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam K., Ahmed A., Devi S.M., Francalacci P., Baig M., Sechi L.A., Hartskeerl R.A., Ahmed N. Genetic affinities within a large global collection of pathogenic Leptospira: implications for strain identification and molecular epidemiology. PLoS One. 2010;5:e12637. doi: 10.1371/journal.pone.0012637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa, H., Miyamoto, H., Nakayama, E., Yoshida, R., Nakamura, I., Sawa, H., Ishii, A., Thomas, Y., Nakagawa, E., Matsuno, K., Kajihara, M., Maruyama, J., Nao, N., Muramatsu, M., Kuroda, M., Simulundu, E., Changula, K., Hang’ombe, B., Namangala, B., Nambota, A., Katampi, J., Igarashi, M., Ito, K., Feldmann, H., Sugimoto, C., Moonga, L., Mweene, A., Takada, A., 2015. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J. Infect. Dis. (in press) http://jid.oxfordjournals.org/content/early/2015/03/17/infdis.jiv063.full.pdf+html?sid=7cd1608c-7e3b-49e3-84fe-bc4d0f857a4b. [DOI] [PubMed]
- Postic D., Riquelme-Sertour N., Merien F., Perolat P., Baranton G. Interest of partial 16S rDNA gene sequences to resolve heterogeneities between Leptospira collections: application to L. meyeri. Res. Microbiol. 2000;151:333–341. doi: 10.1016/s0923-2508(00)00156-x. [DOI] [PubMed] [Google Scholar]
- Rahelinirina S., Leon A., Harstskeerl R.A., Sertour N., Ahmed A., Raharimanana C., Ferquel E., Garnier M., Chartier L., Duplantier J.M., Rahalison L., Cornet M. First isolation and direct evidence for the existence of large small-mammal reservoirs of Leptospira sp. in Madagascar. PLoS One. 2010;5:e14111. doi: 10.1371/journal.pone.0014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter H.V., Cumming G.S. First application of satellite telemetry to track African straw-coloured fruit bat migration. J. Zool. 2008;275:172–176. [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulsiani S.M., Cobbold R.N., Graham G.C., Dohnt M.F., Burns M.A., Leung L.K., Field H.E., Smythe L.D., Craig S.B. The role of fruit bats in the transmission of pathogenic leptospires in Australia. Ann. Trop. Med. Parasitol. 2011;105:71–84. doi: 10.1179/136485911X12899838413501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria B., Ahmed A., Zuerner R.L., Ahmed N., Bulach D.M., Quinteiro J., Hartskeerl R.A. Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS One. 2008;3:e2752. doi: 10.1371/journal.pone.0002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2003. Human Leptospirosis: Guidelines for Diagnosis, Surveillance and Control. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table.
Neighbor-joining phylogenetic tree based on the nucleotide sequences of Leptospira spp. secY genes in E. helvum bats. The dendrogram (corresponding to the G1–G2 region) was constructed using Tanura-Nei distances, with 1000 replications, using MEGA 5.2.2 software (Tamura et al., 2011). Numbers at nodes indicate bootstrap supports >70%. The sequences determined in this study are shown in red. The FC clusters shown in Fig. 1 and GenBank accession numbers are indicated in brackets and parentheses, respectively. Scale bar indicates the number of nucleotide substitutions per site.