Abstract
Recently several infectious agents with a zoonotic potential have been detected in different bat species. However, there is still a lack of knowledge on the transmission dynamics within and between bat species, as well as from bats to other mammals. To better understand these processes, it is important to compare the phylogenetic relationships between different agents to that of their respective hosts. In this study, we analysed more than 950 urine, faeces and oral swab samples collected from 653 bats from mainly four species (Myotis nattereri, Myotis bechsteinii, Myotis daubentonii, and Plecotus auritus) for the presence of coronavirus, paramyxovirus and astrovirus related nucleic acids located in three different regions of Germany. Using hemi-nested reverse transcriptase (RT)-PCR amplification of fragments within the highly conserved regions of the respective RNA dependent RNA polymerase (RdRp) genes, we detected astrovirus sequences at an overall detection rate of 25.8% of the analysed animals, with a maximum of 65% in local populations. The detection rates for coronaviruses and paramyxoviruses were distinctly lower, ranging between 1.4% and 3.1%. Interestingly, the sequence similarities in samples collected from the same bat species in different geographical areas were distinctly larger than the sequence similarities between samples from different species sampled at the same location. This indicates that host specificity may be more important than host ecology for the presence of certain viruses in bats.
Keywords: Insectivore bats, Astroviruses, Coronaviruses, Paramyxoviruses, Host specificity
Highlights
-
•
we combined the spatio-temporal data for bat colonies with the virological data
-
•
high host specificity of astrovirus and coronavirus determined
-
•
host ecology and spatial proximity of colonies has low impact on virus occurrence
-
•
some bat astrovirus sequences closely related to those of other species, research gap
1. Introduction
Since the majority of emerging infectious diseases in humans have their origin in animals (Wolfe et al., 2007, Jones et al., 2008), improving our understanding of the mechanisms underlying interspecies transmissions is of major importance. Bats are of particular interest in this respect for a number of reasons: Firstly, they represent almost 20% of all recognised mammalian species, forming the second largest mammalian order after rodents, with more than 1300 different bat species distributed almost worldwide (Moratelli and Calisher, 2015). Secondly, bats harbour a large variety of viruses. Mostly in tropical regions, bats have been identified as reservoir hosts of some highly pathogenic and zoonotic viruses such as rabies virus, Ebola and Marburg viruses, Hendra and Nipah viruses, and coronaviruses including severe acute respiratory syndrome (SARS)-like and middle east respiratory syndrome (MERS)-like coronaviruses (Burns et al., 1958, O'Shea et al., 2014, Smith and Wang, 2013, Calisher et al., 2006). For European bat species, several studies revealed a broad variety of viruses by examining samples of various species, mostly using molecular detection methods, e.g. astroviruses, coronaviruses, herpes viruses, picornaviruses and paramyxoviruses in insectivorous bats from Germany, Hungary and Italy (Wibbelt et al., 2007, Drexler et al., 2011, Kurth et al., 2012, Kemenesi et al., 2014, Kemenesi et al., 2015, Lelli et al., 2012). Except for lyssaviruses and potentially orthoreoviruses, however, there is no proof to date of any pathogenicity of European bat viruses to humans (Kohl et al., 2012, Lelli et al., 2013, Steyer et al., 2013). Thirdly, bats are very gregarious and the vast majority of all species form colonies, which can reach enormous numbers and densities of individuals during certain periods of the year (Kerth, 2008). This could facilitate the spread of viruses within bat populations. Fourthly, due to their ability to fly, several bat species are potentially capable of spreading viruses over large distances (species specific dispersal distances range from dozens to hundreds of kilometres, Dietz et al., 2007). Finally, bats often share roosts, such as tree cavities, with other animals and/or settle close to humans, e.g. in roofs of houses. As a consequence bats may spread viruses to other animals and to humans (Steyer et al., 2013, Kemenesi et al., 2015).
Despite the large number of molecular virus records in temperate zone bats, hardly anything is known about the course of infection in the individual hosts and the factors that influence intra- and inter-species transmission of viruses. The particular life history and physiology of bats may reduce the pathogenic effects of virus infections, but at the same time increase the virus diversity in bats. For example, the elevated body temperature due to flight activity may mimic the fever reaction of other mammals. This increased body temperature activates the immune response (O'Shea et al., 2014). In contrast, the reduced body temperature during torpor bouts in summer and prolonged torpor during hibernation will reduce the immune response and therefore hamper the virus clearance. At the same time torpor has been suggested to facilitate the development of a persistent virus infection (Sulkin and Allen, 1974, George et al., 2011). It has even been postulated that with these mechanisms bats may have developed the preference of incomplete viral clearance in order to evade immunopathologically induced morbidity and mortality (O'Shea et al., 2014). Given these facts, we predicted to observe an intermittent virus detectability rather than a long-term presence of the same virus strain in individual bats sampled at different time points.
Even less is known to what degree ecological conditions as well as social structures influence virus transmission within and among bat species. In our study, we analysed the detection rates of nucleic acids related to astro-, corona- and paramyxoviruses in bat colonies in the German regions of Bavaria (BY), Mecklenburg Western Pomerania (MV) and North Rhine Westphalia (NRW). We focused on Bechstein's bat (Myotis bechsteinii), Natterer's bat (M. nattereri), Daubenton's bats (M. daubentonii) and brown long-eared bat (Plecotus auritus), which have a similar roosting ecology (Dietz et al., 2007) and where long-term data on the social structure and the individual movements among roosts are available (e.g. Kerth and Van Schaik, 2012, Fleischmann and Kerth, 2014). Some of the colonies studied in BY have been monitored for more than twenty years. In these colonies, subcutaneously implanted microchips, so called PIT-tags, allowed precise monitoring and documentation of social structures in the past (Kerth et al., 2011) and for this present study.
Combining ecological, social and spatial data on the bats with the virological data of the same host populations, our aim was to assess the factors influencing intra- and interspecies virus transmission in bats. We predicted that if host specificity matters more than host ecology (e.g. roost type used, habitat type used, colony size etc.) and host spatial proximity for virus transmission, virus strains should be associated with bat species regardless of the geographical location of the respective populations.
2. Material and methods
2.1. Study areas and sample collection
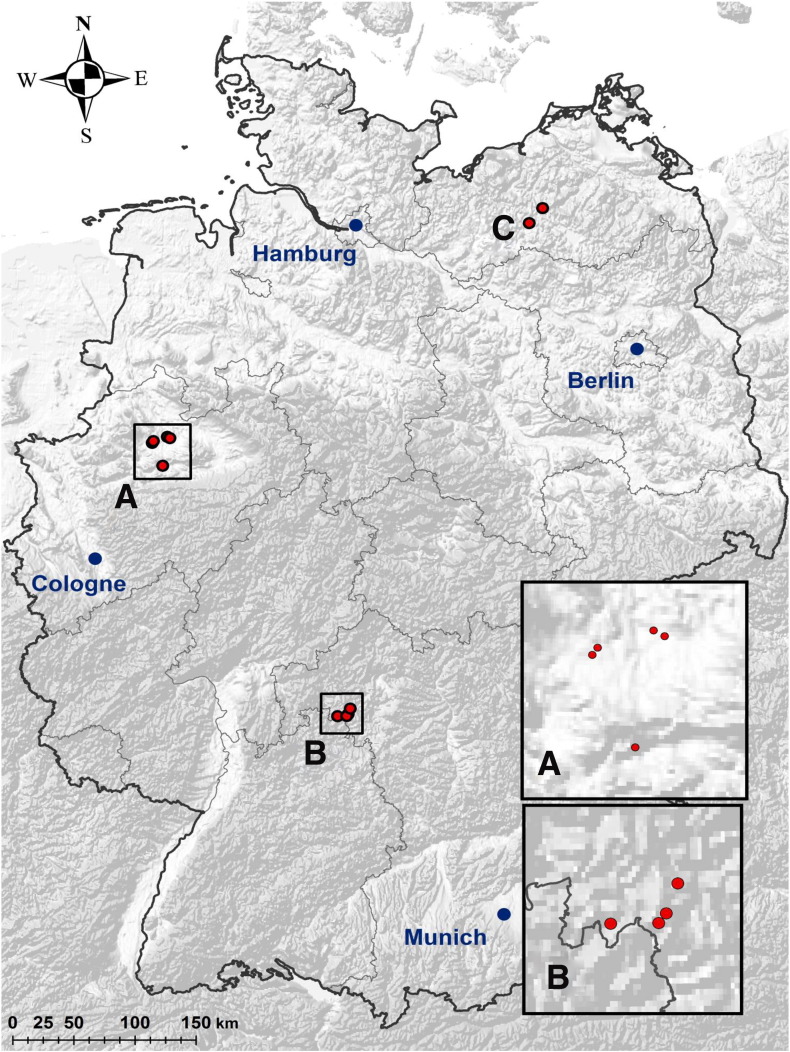
Sampling of bat saliva, faeces and urine was performed in three different regions of Germany (Fig. 1 ), namely in BY between May 2011 and May 2014, in MV between July 2012 and July 2013, and in NRW between July and September 2014. While bats were captured from bat boxes in forests in BY and MV, animals in NRW were captured in their nursery roosts in cow barns and during swarming at hibernacula in autumn. The urine and faeces samples were collected directly from the individual bats during regular capture events while being handled, in order to minimise the possibility of cross-contamination. Urine samples were collected as a swab sample. The permissions to capture and sample the bats were issued by the respective local conservation authorities. Adult bats in the sampled colonies in BY were individually marked with implanted passive integrated transponders (PIT-tags; Kerth et al., 2011). The last four digits of their PIT tags were used to identify individuals. In these colonies, samples were collected from the majority of individuals twice per year over four consecutive years (from 2011 until 2014), once before parturition and once after parturition. In the other areas, adult individuals were marked with rings and/or PIT-tags, and samples were collected once per year.
Fig. 1.
Map of Germany with red dots representing sampling locations. (A) sampling locations in North Rhine Westphalia (B) sampling locations in Bavaria, (C) sampling locations in Mecklenburg Western Pomerania.
Each faecal, urine or saliva sample was put directly either into 500 μl of cell culture medium (Minimal Essential Medium; Collection of Cell Lines in Veterinary Medicine, Friedrich-Loeffler-Institut, Germany) during the first two years of sampling. Starting in the second year, RNAlater RNA preservative solution (QIAGEN, Hilden, Germany) was used due to the increased thermal tolerance of nucleic acids stored in this solution. The samples were kept frozen until processed in the laboratory.
2.2. Preparation of samples and detection of viral RNA
RNA was extracted using the Viral RNA Mini Kit (QIAGEN) following the manufacturer's instructions and eluted in a volume of 60 μl AVE buffer per sample. In order to be able to detect as many different virus strains as possible, we applied conventional RT-PCR protocols, using degenerated primers. Samples were investigated for the presence of astrovirus (AstV) related RNA by hemi-nested RT-PCR targeting the highly conserved region of the RNA-dependent RNA polymerase (RdRp) gene (Chu et al., 2008). A synthetic RNA based on a bat AstV sequence detected in China (Bat astrovirus 1 isolate AFCD337 polyprotein GenBank: EU847155) was used as the positive control. To confirm our results, we have performed exemplary repeats (up to four times) of PCR and sequence analysis, and the results were 100% consistent in all cases. Samples were also analysed for the presence of paramyxovirus (PmV) and coronavirus (CoV) RNA as described before (Tong et al., 2008, de Souza Luna et al., 2007). Respective primer sequences and amplicon lengths are listed in Supplementary Table 1. As positive controls, we used bovine parainfluenza virus 3- and porcine transmissible gastroenteritis virus-derived RNA (both kindly provided from the FLI virus strain bank).
For detection of AstV related RNA, we applied the protocol by Chu et al. (2008), with some modifications. In detail, the first round of RT-PCR was carried out using the Super Script III One Step RT-PCR Kit (Invitrogen, Karlsruhe, Germany) and a mixture of two forward and one reverse primers (Supplementary Table 1) with a final concentration of 2 μM each. The total reaction volume of 50 μl contained 5 μl of purified RNA per investigated sample. Amplification involved 30 min at 50 °C for reverse transcription of RNA and 2 min at 94 °C, followed by 30 cycles of 15 s at 94 °C, 30 s at 50 °C for annealing and 40 s at 72 °C for extension, completed by a last step of 7 min at 72 °C for final extension. Hemi-nested PCR was conducted with two forward primers and the first-round PCR reverse primer at a final concentration of 2 μM each. Reactions were composed using the PWO DNA Polymerase Kit (Roche Diagnostics GmbH, Mannheim) and a 10 mM dNTP Mix (Roche Diagnostics GmbH, Mannheim). The final volume of 50 μl contained 2 μl of the first-round product. After initial denaturation of 2 min at 94 °C, thermal cycling involved 40 cycles of 15 s at 94 °C, 30 s at 50 °C and 40 s at 72 °C. The expected size of the second-round product was 422 nucleotides (nt). In-vitro-transcribed RNA of Bat astrovirus 1 isolate AFCD337 polyprotein 1AB and polyprotein 1a genes (Chu et al., 2008) was included as a positive control in each reaction, nuclease-free water was used as a negative control. PCR products were analysed by agarose gel electrophoresis, purified by using the QIAquick Gel Extraction kit (QIAGEN) according to the manufacturer's instructions and further verified by forward and reverse sequencing.
2.3. Phylogenetic analyses of the RdRp gene sequences
Sequences were assembled and proofread in Geneious 8.1.3 (Biomatters Limited, Auckland, New Zealand). We only included sequences in the phylogenetic analyses of which the quality was satisfactory, i.e. where the electropherogram signals were readable and where forward and reverse sequencing resulted in the same sequence, and if the contig sequence generated from the forward and reverse sequencing results comprised more than 250 nt. Fragment lengths from 279 to 389 nt were included in the phylogenetic analysis of AstV sequences, while the fragments used for the phylogenetic analysis of CoV comprised 381–419 nt, and of 382–459 nt for the PmV analysis. According to BLAST analyses against the GenBank database, sequences were allocated to the host species of the most similar hit. For phylogenetic analyses, our sequences and selected reference sequences from GenBank were aligned using MAFFT 7.017 (Katoh and Standley, 2013). As the AstV and PmV sequences exhibited indels, these alignments had to be corrected manually in a few positions. Base frequencies of the AstV and PmV sequences were inhomogeneous as indicated by a Χ2-test implemented in PAUP* 4b10 (Swofford, 2003) [χ2 = 193.123, df = 159, p = 0.034; χ2 = 100.055, df = 63, p = 0.002; after exclusion of constant sites as proxies for invariant sites (Lockhart et al., 1994): χ2 = 240.429, df = 159, p < 0.001; χ2 = 124.562, df = 63, p < 0.001]. Therefore, the phylogeny of the AstV and PmV sequences were not only reconstructed using maximum likelihood (ML) and Bayesian (BA) methods as the CoV sequences, but also by neighbour-joining trees based on LogDet distances, which are insensitive to the effects potentially caused by inhomogeneous base frequencies (Lockhart et al., 1994). The neighbour-joining analyses were conducted in PAUP* 4b10 and invariant sites were removed in proportion to the frequencies estimated from constant sites (Waddel and Steel, 1997). For ML and BA, jModeltest 2.1.4 (Darriba et al., 2012) was used to find the best fitting substitution models. These were TVM + I + Γ for the AstV sequences and TIM3 + Γ for the CoV and PmV sequences according to the Bayesian information criterion. ML and BA were conducted in GARLI 2.1 (Zwickl, 2006) and MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003). As MrBayes implements only a restricted number of models, the next similar to those identified by jModeltest, viz. GTR + I + Γ and GTR + Γ, were applied for AstV sequences and CoV as well as Paramyxo sequences, respectively. MrBayes was run for 2 Mio generations sampling every 100th with a burn-in of 10%, and otherwise default settings. Convergence of parameter estimates was monitored using the criteria implemented in MrBayes as well as in Tracer 1.6 available at http://tree.bio.ed.ac.uk/software/tracer/. The best ML tree was selected among 500 replicates. The analysis comprised 500 replicates in the case of ML bootstrap analyses, and 1000 replicates for the neighbour-joining trees. The PmV trees contained one long-branching sequence. Excluding it from the analyses did not change the topologies so that long-branch attraction (Felsenstein, 1978) can be ruled out.
3. Results
3.1. Sampling
In total, 653 individual bats were sampled, with 122 bats being sampled at least twice, resulting in 775 samples. More than one sample (e.g. urine and faeces) per bat were collected on several capture events, adding up to 957 samples altogether. We collected 47 (5%) oral swab samples, 430 (45%) urine samples and 480 (50%) faecal samples.
The majority (94.7%) of the samples (734 out of 775) were collected from Bechstein's bats (Myotis bechsteinii), Natterer's bats (Myotis nattereri), Daubenton's bats (M. daubentonii) and brown long-eared bats (Plecotus auritus). In addition, in MV, 41 samples were collected from Nathusius' pipistrelle (Pipistrellus nathusii), common pipistrelle (Pipistrellus pipistrellus) and soprano pipistrelle (Pipistrellus pygmaeus) bats (see Supplementary Tables 2–4).
3.2. Detection of CoV, PmV and AstV nucleic acid
While CoV RNA was only detectable in faecal samples, we were able to detect PmV and AstV RNA from oral swab samples, faeces and urine. Regarding AstV, the highest detection rate was found in faecal (29.8%) samples, followed by oral swab samples (25.5%), and urine samples (14%). PmV RNA was also mostly detected in faecal samples, albeit at a much lower rate (3.4%), followed by oral swab (2.1%) and urine (1.6%) samples.
3.3. Molecular detection of CoV and PmV
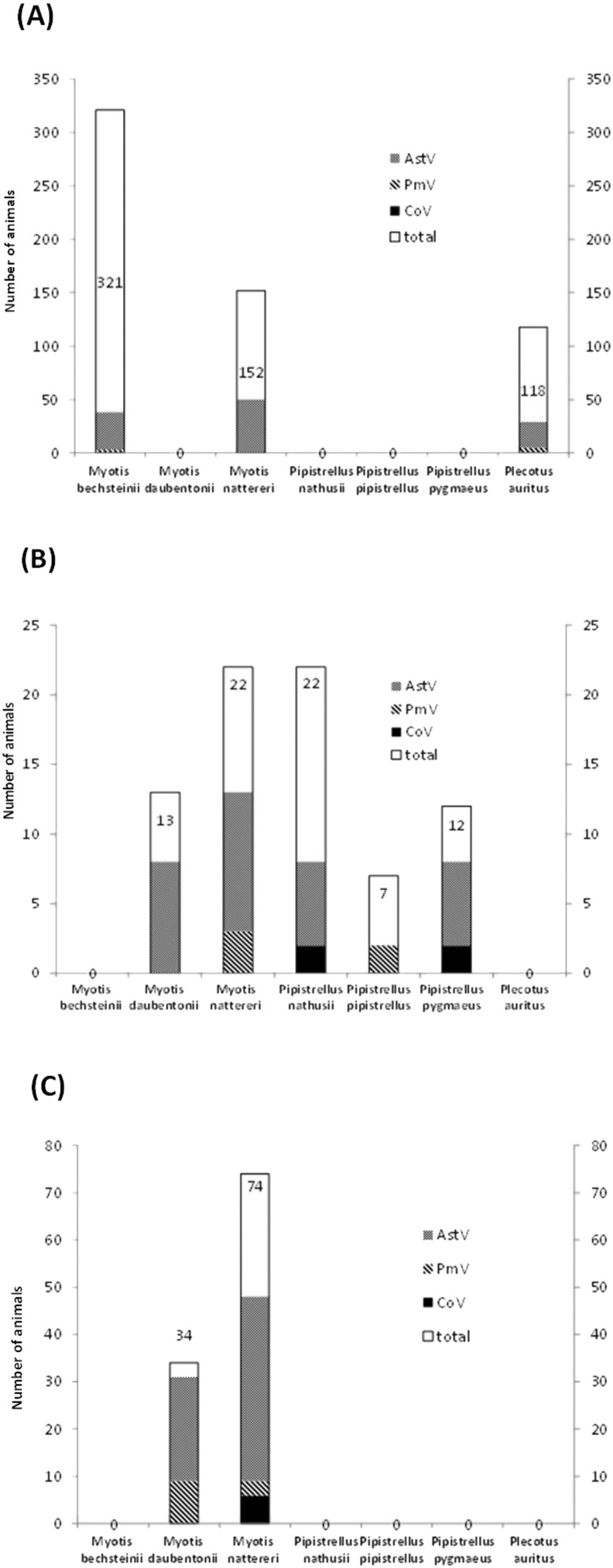
We analysed 957 samples collected from 653 individual bats belonging to seven species in three different geographical regions in Germany. CoV RNA was only detected in six M. nattereri in NRW, in two P. nathusii and two P. pygmaeus bats in MV, and in one sample collected from a M. bechsteinii bat in BY, adding up to 11 positive samples in total (Fig. 2A, B, Supplementary Table 2). The highest detection rates were determined for six out of 74 analysed individual samples from M. nattereri in NRW (8.1%) and two out of 22 analysed samples from P. nathusii in MV (9%), while the average detection rate for CoV was only 1.4%. Sequence analysis revealed a close similarity to alphacoronavirus sequences described from P. nathusii in Northern Germany (Gloza-Rausch et al., 2008) and greater mouse-eared bats (Myotis myotis) in Rhineland-Palatinate (RP) (Drexler et al., 2011) (Supplementary Fig. 1; Genbank accession numbers KT894920 - KT894926). Six of our CoV sequences were included in further phylogenetic analyses. These analyses indicated a species-specificity of CoV strains, i.e. sequences detected in samples from P. nathusii from MV and RP (Drexler et al., 2011), two sites, which are approx. 600 km apart from each other, showed a sequence similarity of 97.3%, while CoV strains from P. pygmaeus from MV and a sampling location approx. 100 km apart (Gloza-Rausch et al., 2008) displayed a similarity of 98.5%. Moreover, sequences amplified from M. nattereri from our different sampling locations showed a similarity of 98.9% with a sequence from M. nattereri sampled in Hungary (Kemenesi et al., 2014). The only CoV sequence we obtained from a M. bechsteinii sample also falls into this cluster of M. nattereri bat-associated sequences. However, the overall CoV detection rate was too low to allow for a more detailed analysis of the bat species-specificity of CoV.
Fig. 2.
Viral RNA detection of coronavirus (CoV)-, paramyxovirus (PmV)- and astrovirus (AstV)-related sequences in bats from three locations in Germany (2011–2014). Total numbers of tested animals are displayed in the columns and include recaptured and re-analysed animals; (A) bats tested in Bavaria from 2011 to 2014, (B) bats tested in Mecklenburg Western Pomerania (2012–2013), (C) bats tested in North Rhine Westphalia (2014). A “0” instead of a column for a given species indicates that this species was not sampled at the respective location.
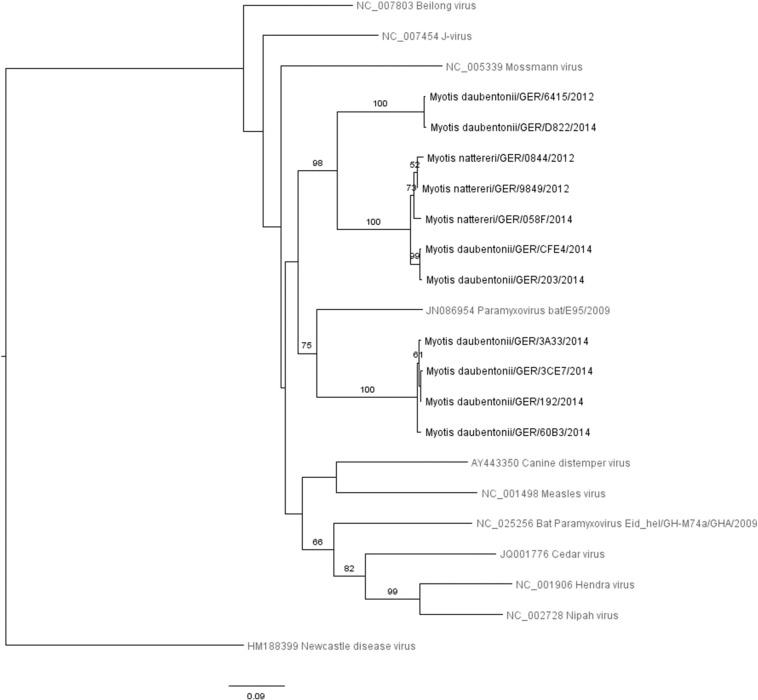
The overall PmV detection rate was 3.1%, adding up to 24 positive samples (Fig. 2, Supplementary Table 3). Phylogenetic analyses of PmV were very similar and mainly differed in the position of HM188399_NDV. Distance as well as character based analyses identified the same three well supported clades, however, the basal relationships were not robustly resolved. Our bat-derived paramyxovirus sequences clustered together with a published sequence from Germany (Kurth et al., 2012). These paramyxoviruses detected in German bats display some similarity to J-virus, Beilong virus and the Morbillivirus group, but they do not seem to be closely related to highly pathogenic Hendra and Nipah viruses carried by Old World fruit bats (Fig. 3 ; Genbank accession numbers KT894927 - KT894937). Since we detected only one sequence from a M. nattereri and six sequences from six M. daubentonii which were all sampled in NRW, these data did not allow us to analyse species-specificity of PmV.
Fig. 3.
Neighbour-joining tree based on LogDet distances of PmV related sequences. Sequences established by us are printed in black, sequences available in the literature (for bat PmV see Kurth et al., 2012) are printed in grey. Nomenclature for the newly generated sequences: bat species, SAMPLING LOCATION, PIT no, sequence number.
3.4. Molecular detection of AstV
Out of the 957 samples from 653 animals, we determined an overall AstV detection rate of 23.5% for the analysed samples and of 25.8% for the analysed animals (Fig. 2, Supplementary Table 4). The majority of the samples were collected in BY, mostly from M. bechsteinii, followed by M. nattereri and P. auritus. For M. nattereri, representing the only bat species that could be analysed in all three sampling areas, the AstV detection rate was lowest in BY (50 positives out of 152 samples, i.e. 32.9%) and highest in NRW (39 positives out of 74 samples, i.e. 52.7%). The detection ranges determined for BY, where sampling was performed in four consecutive years, varied between the years, resulting in a comparably broad range of determined detection rates (Supplementary Tab 4). We detected 47 different AstV sequences altogether, from 200 positive out of the 775 analysed bats. 16 different sequences alone were identified from 30 positive M. daubentonii samples (out of 47 analysed individuals of this species). On the other hand, M. bechsteinii, representing the species where the largest quantity of bats were sampled (321) only yielded 35 positive samples carrying 8 different sequences.
The marking of the animals with PIT-tags in BY allowed us to follow up the history of AstV infections for individual bats. 122 bats were sampled repeatedly. As it turned out, 82% of the animals that were tested positive at one sampling event were tested negative in the following event. However, in some rare instances (5 animals out of 28 recaptured positive bats, representing 18%), animals were identified as positive again after recapture. One M. bechsteinii (PIT-tag: C073) turned out to carry the same AstV sequence (B1, Table 1 ) in May 2011 as well as three months later in August. Another M. bechsteinii (1A0D) also carried the sequence B1 in May 2011, and carried a sequence assigned as avian AstV by GenBank (sequence B8) in August 2013. AstV sequences amplified from a M. nattereri (2ACE) at both sampling events (sequences N8 and N4) just showed an 80% similarity, suggesting a re-infection with a different strain. Sequence N8 was also detected again in four M. nattereri captured in the same sampling area in May 2013. Two P. auritus (AD8A and 1681) of the colony “BS2” carried the identical AstV sequences (P3) in May 2011. Recaptured in May 2013, both bats tested positive, and again carried again identical sequences (P2). Moreover, the amplified sequences showed 98% sequence similarity between the two years, with only silent mutations in the RdRp coding region. Noteworthy, one of the two bats was recaptured in 2012 and tested negative for AstV.
Table 1.
Astrovirus sequences determined for M. nattereri, M. daubentonii, M. bechsteinii and P. auritus captured in Bavaria, North Rhine Westphalia and Mecklenburg Western Pomerania.
| Bat species | Sequence number | Detected at sampling events | Detected at sampling location | Number of bats carrying this sequence |
|---|---|---|---|---|
| Myotis nattereri | N1 | Aug 14 | NRW | 1 |
| Myotis nattereri | N2 | Aug 14 | NRW | 1 |
| Myotis nattereri | N3 | May 13 | BY | 1 |
| N3 | May 14 | BY | 2 | |
| Myotis nattereri | N4 | Aug 11 | BY | 3 |
| Myotis nattereri | N5 | Aug 13 | BY | 10 |
| N5 | May 14 | BY | 3 | |
| Myotis nattereri | N6 | Sep 14 | NRW | 1 |
| Myotis nattereri | N7 | Jul 13 | MV | 2 |
| Myotis nattereri | N8 | May 11 | BY | 13 |
| N8 | May 13 | BY | 4 | |
| Myotis nattereri | N9 | Jul 14 | NRW | 4 |
| Myotis nattereri | N10 | Jul 13 | MV | 2 |
| Myotis nattereri | N11 | May 11 | BY | 1 |
| Myotis nattereri | N12 | Aug 14 | NRW | 11 |
| Myotis nattereri | N13 | Aug 14 | NRW | 1 |
| Myotis nattereri | N14 | Jul 14 | NRW | 3 |
| Myotis nattereri | N15 | Aug 12 | BY | 5 |
| N15 | May 2014 | BY | 1 | |
| Myotis nattereri | N16 | Aug 14 | NRW | 2 |
| Myotis nattereri | N17 | Jul12 | MV | 1 |
| Myotis nattereri | N18 | Oct14 | NRW | 1 |
| Myotis nattereri | N19 | Sep 14 | NRW | 1 |
| Myotis daubentonii | D1 | Sep 14 | NRW | 1 |
| Myotis daubentonii | D2 | Jul 13 | MV | 1 |
| Myotis daubentonii | D3 | Sep 14 | NRW | 1 |
| Myotis daubentonii | D4 | Sep 14 | NRW | 2 |
| Myotis daubentonii | D5 | Sep 14 | NRW | 2 |
| Myotis daubentonii | D6 | Jul 13 | MV | 1 |
| Myotis daubentonii | D7 | Jul 13 | MV | 1 |
| Myotis daubentonii | D8 | Sep 14 | NRW | 1 |
| Myotis daubentonii | D9 | Jul 13 | MV | 1 |
| Myotis daubentonii | D10 | Jul 13 | MV | 1 |
| Myotis daubentonii | D11 | Jul 13 | MV | 1 |
| Myotis daubentonii | D12 | Sep 14 | NRW | 2 |
| Myotis daubentonii | D13 | Sep 14 | NRW | 1 |
| Myotis daubentonii | D14 | Sep 14 | NRW | 1 |
| Myotis daubentonii | D15 | Sep 14 | NRW | 3 |
| Myotis daubentonii | D16 | Aug 14 | NRW | 1 |
| Myotis bechsteinii | B1 | May 11 | BY | 12 |
| B1 | Aug 12 | BY | 13 | |
| Myotis bechsteinii | B2 | Aug 13 | BY | 1 |
| Myotis bechsteinii | B3 | Aug 12 | BY | 1 |
| Myotis bechsteinii | B4 | Aug 11 | BY | 1 |
| Myotis bechsteinii | B5 | Aug 12 | BY | 1 |
| Myotis bechsteinii | B6 | Aug 12 | BY | 1 |
| Myotis bechsteinii | B7 | May 14 | BY | 1 |
| Myotis bechsteinii | B8 | Aug 13 | BY | 1 |
| Plecotus auritus | P1 | May 14 | BY | 2 |
| Plecotus auritus | P2 | May 13 | BY | 5 |
| P2 | May 14 | BY | 3 | |
| Plecotus auritus | P3 | May 11 | BY | 9 |
| Plecotus auritus | P4 | May 11 | BY | 1 |
Interestingly, in May 2014 members of another P. auritus colony “BS1” within the same area in BY carried AstV strains with the exact same sequence (P2) as the bats AD8A and 1681 caught in May 2013, although members of these two colonies do not mix (Zeus et al., unpublished data). Unfortunately, efforts to recapture these two bats AD8A and 1681 that tested positive in 2011 and 2013 failed in May 2014, thus the information, if two strictly separated colonies nevertheless shared identical viral sequences in 2014, is lacking. All the other members of colony BS2 that were captured in 2014 tested negative.
Furthermore, it became evident that certain AstV strains were dominant in a certain species and region at a given sampling event. For example, out of the 18 M. bechsteinii found positive in May 2011, 12 carried the identical sequence (B1), out of the 14 P. auritus samples collected in May 2013, five bats sampled in one colony carried an identical sequence (P2), and of 6 positives out of 23 M. nattereri from one colony, four carried one sequence (N8) and two carried another sequence (N3). All our findings regarding the detection of different sequences are summarised in Table 1.
3.5. Phylogenetic analysis of AstV sequences
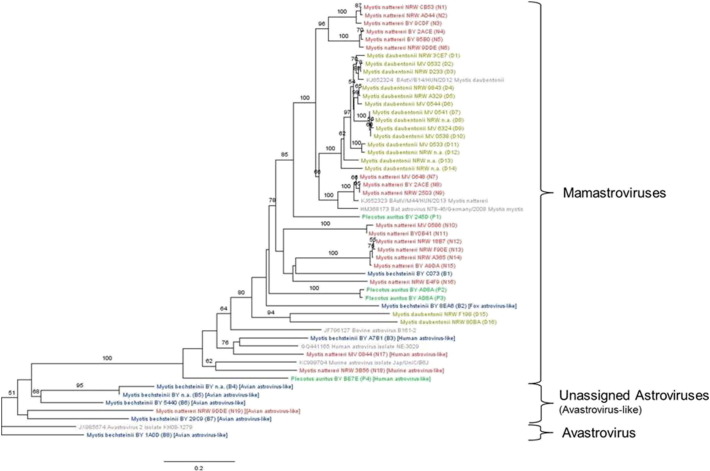
The topologies of the phylogenetic reconstructions of AstV sequences differed slightly between ML and Bayesian approaches on the one hand and the LogDet analyses on the other hand. As the latter is insensitive to the effects of the detected base composition heterogeneity, we only present the LogDet analyses in Fig. 4 (GenBank accession numbers KT894874 - KT894920). The differences among methods were not relevant for our questions and interpretations. The Bayesian reconstruction, practically identical to that obtained by ML, is presented in Supplementary Fig 2. In general, strains detected from particular species were not monophyletic and their relationships were also largely independent of the geography. The ingroup was not monophyletic either due to one avian-like sequence gained from a M. bechsteinii branching off the root level. The monophyly of the rest of the ingroup was only poorly supported. The earliest branches were also formed by strains related to avian AstV, five from M. bechsteinii and one from a M. nattereri. The monophyly of the remaining sequences received 100% support. Still, seven non-bat specific strains, four of which were detected in bats (for details see below), formed a group paraphyletic with respect to the majority of sequences detected almost exclusively in bats.
Fig. 4.
Neighbour-joining tree based on LogDet distances of AstV related sequences Sequences available in the literature are printed in grey (for bat AstV see Kemenesi et al., 2014). AstV related sequences identified from different bat species are indicated in different colours: Natterer's bat (Myotis nattereri): red; Daubenton's bat (Myotis daubentonii): ocher; Brown long-eared bat (Plecotus auritus): green; Bechstein's bat (Myotis bechsteinii): blue. Nomenclature for the newly generated sequences: bat species, SAMPLING LOCATION, PIT no, sequence number.
From the 30 positive results obtained from M. daubentonii in MV and NRW, we were able to identify 16 different AstV sequences. Of these 16 sequences, 14 clustered with an AstV sequence recently reported from M. daubentonii in Hungary (Kemenesi et al., 2014), while the remaining two sequences formed another more distant clade.
For M. nattereri, we identified 17 bat-related AstV sequences from 99 positive samples collected in all three sampling locations. These formed four clades. One of these clades contained a published sequence from Hungary (Kemenesi et al., 2014) as well as a sequence published from a mouse-eared bat colony in Rhineland Palatinate (Drexler et al., 2011). Another clade containing sequences from M. nattereri had one sequence from a M. bechsteinii (B1) as a sister group (Fig. 4). In addition to the bat-related sequences, we detected one sequence in a M. nattereri in NRW that was most similar to a murine AstV sequence. Two other sequences amplified from two M. nattereri sampled in in NRW (9DDE) and MV (0844) were closely related to an avian AstV and a human AstV, respectively.
From the 24 positive P. auritus identified in BY we determined three sequences (P1, P2, P3). P1 was closer related to strains obtained from M. daubentonii and M. nattereri than to P2 and P3, which were sister strains. A fourth isolate (P4) from the P. auritus BE7E was most similar to a human AstV.
Interestingly, from the 35 positive samples identified from M. bechsteinii from BY dividing into 8 different sequence types, only one type seemed to be clearly bat-specific (see above). One sequence was most similar to a fox AstV (bat 8EA6), while another sequence was closely related to a human AstV (bat A7B1). As already mentioned, five additional sequences generated from faecal samples collected from M. bechsteinii in BY were identified as avian AstV-like sequences and clustered with one avain AstV sequence amplified from a M. nattereri from NRW mentioned above (9DDE) and one additional sequence from GenBank.
4. Discussion
While several studies were focused on the screening of European bats for the presence of viral nucleic acids, predominantly RNA viruses, this is to our knowledge the first study to combine spatio-temporal data for different bat species over several consecutive years and the corresponding virological data. By doing so, our aim was to analyse the correlation between certain viruses and bat host species, in order to improve our understanding of the virus/host relationship and possibly the transmissibility within and between bat species.
4.1. CoV and PmV detection
CoV detection rates in our study were up to 9% in one site and species with an overall average of only 1.4% over all samples. This is lower than the CoV detection rates described in another German study (Gloza-Rausch et al., 2008), although the same RT-PCR protocol was applied. This may be due to the sampled bat species and the geographical region of the sampling, which were both different from the earlier studies. The CoV sequences identified in this study fall into the genus alphacoronavirus, and showed an overall high sequence similarity of 88.2% with the published bat CoV sequences from Germany (Gloza-Rausch et al., 2008) and Hungary (Kemenesi et al., 2014). We observed a bat species specificity of certain CoV strains, which had also been observed in earlier studies (Gloza-Rausch et al., 2008), even over distances of more than 600 km. However, one M. bechsteinii sequence is found in a cluster with four sequences related to M. nattereri. These five sequences display a similarity of 98.8%, indicating in this case a spillover infection between two Myotis species.
Our PmV detection rates reached 28.6% per study population, with an overall average of 3.1%, which is in the range of what has been described in the literature (Kurth et al., 2012). Since we detected PmV-related RNA in oral swabs as well as faeces and urine samples, these virus strains seem to be excreted by various routes. The restriction of our seven sequences to one sampling area and two species precluded a deeper analysis of the species-specificity of these viruses.
4.2. AstV detection and phylogeny
We determined the highest detection rates for AstV, with up to 63.8% of the analysed M. daubentonii being positive in the applied RT-PCR assay, and an overall average detection rate of 25.8%. We found the highest detection rates in the bat populations in NRW (56.5% on average), and the lowest rates in bat populations in BY (18.4% on average). The overall detection rates were lower than described in the literature, where a real-time RT-PCR protocol was applied that was designed for the specific detection of the strains that were present in that colony (Drexler et al., 2011). The phylogenetic analysis of the 47 sequences detected in four bat species revealed a good correlation between the sampled bat species and the AstV strain, almost completely irrespective of the sampling region in Germany and Hungary. This confirms a bat species-specificity of AstV, combined with an obviously low rate of interspecies transmission of these viruses.
Positive animals sampled from the same colony at the same respective sampling event displayed a high rate of sequence similarity of up to 100% especially in BY, indicating the circulation of a certain strain at a given time within a colony. Out of 122 bats that were sampled repeatedly, only five bats turned out to be positive at more than one sampling occasion. This may either mirror an oscillation in the virus shedding intensity as postulated before (Plowright et al., 2015), or a transient virus infection. It must however also be kept in mind that the applied RT-PCR assay was selected for its ability to detect a wide range of different virus strains, at the cost of a high detection sensitivity, therefore some false negative results cannot be completely ruled out. In order to increase the sensitivity of our assays, we considered establishing specific real-time PCR assays, but due to the variability of the sequence types, at least eight different real-time PCR assays would need to be established to be able to detect all sequence types known so far. This would, however, bear the risk of not detecting any new sequence type because it would not be covered by these assays. Confirmation of any positive results would still need to be done by sequence analysis.
We were nevertheless able to identify several bats that were positive for AstV when recaptured several months or up to two years after the initial positive result. We only detected the identical virus sequence in one occasion after three months, and in another occasion after two years, with negative results in between. In the three other repeatedly recaptured bats that had been found to be AstV positive on at least two sampling events, we detected different sequences, with either a few mutations (98% sequence similarity) or only 50% similarity to the original sequence. We therefore conclude that in these cases, the original virus strain was replaced by a different strain.
Our analysis revealed a number of AstV sequences that were identified to be closely related to human, murine, fox, or avian AstV by GenBank, although AstV originating from these species have never been introduced to our lab. These AstV sequences were predominantly found in M. bechsteinii, where we only detected one AstV sequence type that was directly allocated by GenBank as a bat AstV. In this species, we detected five AstV sequences that were assigned as avian AstV, which were closely related to a similar sequence from a M. Nattereri, suggesting the presence of a novel AstV group. Similar findings on species assignment of AstV sequences have been reported by others in AstV screening studies. For example, AstV surveys of bats (Zhu et al., 2009) and birds (Chu et al., 2012) in China also revealed a close relation of AstV sequences assigned as human and feline sequences with bat associated sequences. Furthermore, screening of a variety of small mammalian species in China revealed a surprisingly close phylogenetic relationship between some rodent AstV and ungulate AstV (Hu et al., 2014), that cannot be explained yet. It therefore cannot be completely ruled out that the AstV nomenclature is in some cases misleading, since the newly identified virus sequences are often simply nominated according to the species of first detection of virus related nucleic acid (Monroe et al., 2005, De Benedictis et al., 2011). In most cases, this is done without a successful virus isolation from this species as a proof of a productive infection herein, leaving the possibility of a passive virus transmission, or even an external contamination. From these findings we conclude that further studies regarding the occurrence of AstV in different mammalian species, especially bats, is required. Such studies need to include virus isolation attempts, in order to be able to assess the real species spectrum of AstV strains that have been assigned to a species based on phylogenetic analysis. Results of such studies would also shed more light on a possible zoonotic potential of some AstV strains.
In our study we did follow individual colonies throughout the summer within and between years, but it was impossible to follow colonies throughout the entire year, as the hibernacula are typically unknown. It is possible that different species meet at joint hibernacula, which thus provide opportunities for virus transmission. However, this should not affect our conclusions, because if bats of different species meet during winter and if viruses were not species specific we should find the same or at least very similar virus sequences in different bat species within a region where joint hibernation is possible. This was clearly not the case, supporting our conclusion of a strong species specificity of the detected virus sequences.
In conclusion, our study reveals a high degree of bat species specificity of AstV and also CoV strains, that is largely independent of the spatial proximity of the different hosts. The finding is interesting, as our four main study species are not migrating very far (typically less than 50 km; Dietz et al., 2007). As a consequence our study populations are very unlikely to exchange individuals between the sampled regions MV, BY, and NRW. Overall, our findings are valuable for the general understanding of virus distribution in habitats with mixed species, and also to improve our understanding of the spectrum of host species of AstV and CoV strains.
Acknowledgements
This work was partly funded by the Deutsche Forschungsgemeinschaft (DFG: GR 980/3-1 and KE 746/6-1) within the DFG priority programm “Ecology and species barriers in emerging viral diseases (SPP 1596)”. The following people provided samples: Lena Grosche, Markus Melber, Frauke Meyer. Patrick Wysocki prepared the map showing the sampling areas. We thank the local forestry and conservation departments as well as the Fledermausforschungsprojekt Wooster Teerofen e.V. for their support. We highly appreciate the constructive comments given the anonymous reviewers who helped us to improve this manuscript. We are indebted to Anna-Maria Hübener for her excellent technical assistance in the lab.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.meegid.2015.11.010.
Contributor Information
Kerstin Fischer, Email: Kerstin.fischer@fli.bund.de.
Veronika Zeus, Email: zeusv@uni-greifswald.de.
Gerald Kerth, Email: gerald.kerth@uni-greifswald.de.
Martin Haase, Email: mhaase@uni-greifswald.de.
Martin H. Groschup, Email: martin.groschup@fli.bund.de.
Anne Balkema-Buschmann, Email: anne.buschmann@fli.bund.de.
Appendix A. Supplementary data
Supplementary material
References
- Burns K.F., Shelton D.F., Grogan E.W. Bat rabies: experimental host transmission studies. Ann. N. Y. Acad. Sci. 1958;70(3):452–466. doi: 10.1111/j.1749-6632.1958.tb35403.x. (3) [DOI] [PubMed] [Google Scholar]
- Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19(3):531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Leung C.Y., Perera H.K., Ng E.M., Gilbert M., Joyner P.H., Grioni A., Ades G., Guan Y., Peiris J.S., Poon L.L. A novel group of avian astroviruses in wild aquatic birds. J. Virol. 2012;86(24):13772–13778. doi: 10.1128/JVI.02105-12. (Epub 2012 Oct 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Poon L.L., Guan Y., Peiris J.S. Novel astroviruses in insectivorous bats. J. Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. JModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedictis P., Schultz-Cherry S., Burnham A., Cattoli G. Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11(7):1529–1544. doi: 10.1016/j.meegid.2011.07.024. (Epub 2011 Aug 5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Luna L.K., Heiser V., Regamey N., Panning M., Drexler J.F., Mulangu S., Poon L., Baumgarte S., Haijema B.J., Kaiser L., Drosten C. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J. Clin. Microbiol. 2007;45(3):1049–1052. doi: 10.1128/JCM.02426-06. (Epub 2007 Jan 17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz C., Helversen O., Nill D. Kosmos Verlag (Franckh-Kosmos); Stuttgart: 2007. Handbuch der Fledermäuse Europas und Nordwestafrikas. [Google Scholar]
- Drexler J.F., Corman V.M., Wegner T., Tateno A.F., Zerbinati R.M., Gloza-Rausch F., Seebens A., Müller M.A., Drosten C. Amplification of emerging viruses in a bat colony. Emerg. Infect. Dis. 2011;17(3):449–456. doi: 10.3201/eid1703.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Biol. 1978;27(4):401–410. [Google Scholar]
- Fleischmann D., Kerth G. Roosting behavior and group decision-making in two syntopic bat species with fission-fusion dynamics. Behav. Ecol. 2014;25(5):1240–1247. [Google Scholar]
- George D.B., Webb C.T., Farnsworth M.L., O'Shea T.J., Bowen R.A., Smith D.L., Stanley T.R., Ellison L.E., Rupprecht C.E. Host and viral ecology determine bat rabies seasonality and maintenance. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10208–10213. doi: 10.1073/pnas.1010875108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloza-Rausch F., Ipsen A., Seebens A., Göttsche M., Panning M., Drexler J.F., Petersen N., Annan A., Grywna K., Müller M., Pfefferle S., Drosten C. Detection and prevalence patterns of group I coronaviruses in bats, northern Germany. Emerg. Infect. Dis. 2008;14(4):626–631. doi: 10.3201/eid1404.071439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Chmura A.A., Li J., Zhu G., JS2 D., Zhang Y., Zhang W., Epstein J.H., Daszak P., Shi Z. Detection of diverse novel astroviruses from small mammals in China. J. Gen. Virol. 2014;95:2442–2449. doi: 10.1099/vir.0.067686-0. [DOI] [PubMed] [Google Scholar]
- Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. (Epub 2013 Jan 16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemenesi G., Dallos B., Görföl T., Boldogh S., Estók P., Kurucz K., Kutas A., Földes F., Oldal M., Németh V., Martella V., Bányai K., Jakab Molecular survey of RNA viruses in Hungarian bats: discovering novel astroviruses, coronaviruses, and caliciviruses. Vector Borne Zoonotic Dis. 2014;14(12):846–855. doi: 10.1089/vbz.2014.1637. [DOI] [PubMed] [Google Scholar]
- Kemenesi G., Zhang D., Marton S., Dallos B., Görföl T., Estók P., Boldogh S., Kurucz K., Oldal M., Kutas A., Bányai K., Jakab F. Genetic characterization of a novel picornavirus detected in miniopterus schreibersii bats. J. Gen. Virol. 2015;96(Pt 4):815–821. doi: 10.1099/jgv.0.000028. (Apr 2015, Epub 2014 Dec 16) [DOI] [PubMed] [Google Scholar]
- Kerth G. Animal sociality: bat colonies are founded by relatives. Curr. Biol. 2008;18(17):R740–R742. doi: 10.1016/j.cub.2008.07.038. [DOI] [PubMed] [Google Scholar]
- Kerth G., Van Schaik J. Causes and consequences of living in closed societies: lessons from a long-term socio-genetic study on bechstein's bats. Mol. Ecol. 2012;21(3):633–646. doi: 10.1111/j.1365-294X.2011.05233.x. (Epub 2011 Aug 29) [DOI] [PubMed] [Google Scholar]
- Kerth G., Perony N., Schweitzer F. Bats are able to maintain long-term social relationships despite the high fission–fusion dynamics of their groups. Proc. R. Soc. B. 2011;278:2761–2767. doi: 10.1098/rspb.2010.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl C., Lesnik R., Brinkmann A., Ebinger A., Radonić A., Nitsche A., Mühldorfer K., Wibbelt G., Kurth A. Isolation and characterization of three mammalian orthoreoviruses from European bats. PLoS ONE. 2012;7(8):e43106. doi: 10.1371/journal.pone.0043106. (Epub 2012 Aug 14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth A., Kohl C., Brinkmann A., Ebinger A., Harper J.A., Wang L.F., Mühldorfer K., Wibbelt G. Novel paramyxoviruses in free-ranging European bats. PLoS ONE. 2012;7(6):e38688. doi: 10.1371/journal.pone.0038688. (Epub 2012 Jun 21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli D., Moreno A., Lavazza A., Bresaola M., Canelli E., Boniotti M.B., Cordioli P. Identification of Mammalian orthoreovirus type 3 in Italian bats. Zoonoses Public Health. 2012;60(1):84–92. doi: 10.1111/zph.12001. (Epub 2012 Aug 30) [DOI] [PubMed] [Google Scholar]
- Lelli D., Papetti A., Sabelli C., Rosti E., Moreno A., Boniotti M.B. Detection of coronaviruses in bats of various species in Italy. Viruses. 2013;5(11):2679–2689. doi: 10.3390/v5112679. (Oct 31 2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart P.J., Steel M.A., Hendy M.D., Penny D. Recovering evolutionary trees under a realistic model of sequence evolution. Mol. Biol. Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- Monroe S.S., Carter M.J., Herrmann J., Mitchel D.K., Sanchez Fauquier A. Astroviridae. In: Fauquet C.M., Mayo A., Maniloff J., Desselberger U., Ball L.A., editors. Virus Taxonomy. Classification and Nomenclature of Viruses: Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; California: 2005. pp. 859–864. [Google Scholar]
- Moratelli R., Calisher C.H. Bats and zoonotic viruses: can we confidently link bats with emerging deadly Viruses? Mem. Inst. Oswaldo Cruz. 2015;110(1):1–22. doi: 10.1590/0074-02760150048. (2015 Feb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea T.J., Cryan P.M., Cunningham A.A., Fooks A.R., Hayman D.T., Luis A.D., Peel A.J., Plowright R.K., Wood J.L. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014;20(5):741–745. doi: 10.3201/eid2005.130539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright R.K., Eby P., Hudson P.J., Smith I.L., Westcott D., Bryden W.L., Middleton D., Reid P.A., McFarlane R.A., Martin G., Tabor G.M., Skerratt L.F., Anderson D.L., Crameri G., Quammen D., Jordan D., Freeman P., Wang L.F., Epstein J.H., Marsh G.A., Kung N.Y., McCallum H. Ecological dynamics of emerging bat virus spillover. Proc. R. Soc. B. 2015;282:20142124. doi: 10.1098/rspb.2014.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J.P. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Smith I., Wang L.F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. 2013;3(1):84–91. doi: 10.1016/j.coviro.2012.11.006. (Epub 2012 Dec 21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer A., Gutiérrez-Aguire I., Kolenc M., Koren S., Kutnjak D., Pokorn M., Poljšak-Prijatelj M., Racki N., Ravnikar M., Sagadin M., Fratnik Steyer A., Toplak N. High similarity of novel orthoreovirus detected in a child hospitalized with acute gastroenteritis to mammalian orthoreoviruses found in bats in Europe. J. Clin. Microbiol. 2013;51(11):3818–3825. doi: 10.1128/JCM.01531-13. (2013 Nov, Epub 2013 Sep 11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulkin S.E., Allen R. Virus infections in bats. In: Melnick J.L., editor. Monographs in Virology. vol. 8. Karger; Basel: 1974. (103 pp.) [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates; Sunderland, MA: 2003. Paup*. (Phylogenetic Analysis Using Parsimony (*and other methods). Version 4). [Google Scholar]
- Tong S., Chern S.W., Li Y., Pallansch M.A., Anderson L.J. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J. Clin. Microbiol. 2008;46(8):2652–2658. doi: 10.1128/JCM.00192-08. (Epub 2008 Jun 25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddel P.J., Steel M.A. General time-reversible distances with unequal rates across sites: mixing G and inverse Gaussian distributions with invariant sites. Mol. Phylogenet. Evol. 1997;8:398–414. doi: 10.1006/mpev.1997.0452. [DOI] [PubMed] [Google Scholar]
- Wibbelt G., Kurth A., Yasmum N., Bannert M., Nagel S., Nitsche A., Ehlers B. Discovery of herpesviruses in bats. J. Gen. Virol. 2007;88(Pt 10):2651–2655. doi: 10.1099/vir.0.83045-0. [DOI] [PubMed] [Google Scholar]
- Wolfe N.D., Dunavan C.P., Diamond J. Origins of major human infectious diseases. Nature. 2007;447(7142):279–283. doi: 10.1038/nature05775. (17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.C., Chu D.K., Liu W., Dong B.Q., Zhang S.Y., Zhang J.X., Li L.F., Vijaykrishna D., Smith G.J., Chen H.L., Poon L.L., Peiris J.S., Guan Y. Detection of diverse astroviruses from bats in China. J. Gen. Virol. 2009;90(Pt 4):883–887. doi: 10.1099/vir.0.007732-0. (Epub 2009 Mar 4) [DOI] [PubMed] [Google Scholar]
- Zwickl D.J. The University of Texas at Austin; Austin, TX: 2006. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets under the Maximum Likelihood Criterion. (Ph.D. dissertation) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material