Highlights
► Mixed infections prevail among CCoV infections. ► CCoV triple (CCoV-I/CCoV-IIa/CCoV-IIb) infections are reported for the first time. ► The identification of a divergent strain is reported. ► CCoV-II is more conserved in the first part of the S gene compared to CCoV type I.
Keywords: Canine coronavirus, Diarrhea, Genotype, Subtype, Characterization
Abstract
Canine coronavirus (CCoV) is an etiologic agent of diarrhea in dogs and is known to have spread worldwide. Mild disease or asymptomatic carriage are probably in many cases common outcomes of infection. To date, two different genotypes of CCoV are known, CCoV type I (CCoV-I) and CCoV type II (CCoV-II). CCoV type II is divided in two subtypes, CCoV-IIa (classical strains) and CCoV-IIb, with CCoV-IIb emerging as a result of a putative recombination between CCoV-IIa and transmissible gastroenteritis virus (TGEV). The aim of the present study was to investigate the presence of CCoV in Greece and to genetically analyze the circulating strains. Between December 2007 and December 2009, 206 fecal samples were collected from dogs with diarrhea from kennels, pet shops and veterinary clinics of different country regions. RT-PCR and real time RT-PCR assays were used for CCoV detection and characterization. CCoV was identified in 65.1% of the dogs presenting diarrhea, being more frequently detected in animals younger than 3 months old and in animals housed in groups. In 47% of the positive samples more than one CCoV genotype/subtype were detected, with triple CCoV-I/CCoV-IIa/CCoV-IIb infections being identified for the first time. Molecular and phylogenetic analysis revealed that CCoV-I Greek strains share low genetic relatedness to each other and to the prototype CCoV-I strains in the 5’ end of the S gene. Moreover, a divergent CCoV-IIa strain was identified. The circulation of highly variable CCoV-I and CCoV-IIb emerging strains, as well as the detection of the divergent strain, raise concerns on the importance of these new strains as primary pathogens of diarrhoeic syndromes diagnosed in dogs.
1. Introduction
Coronaviruses (order Nidovirales, family Coronaviridae) are large enveloped, positive-stranded RNA viruses that infect a variety of animal and bird species, including humans. They are usually associated with enteric or respiratory infections and until recently they were divided into three groups, based on genetic and antigenic similarities (Enjuanes et al., 2000, Lai and Holmes, 2001). Canine coronavirus (CCoV), former member of group 1, is currently classified as a variant of the species Alphacoronavirus 1, genus Alphacoronavirus, along with the feline coronavirus (FCoV) and the transmissible gastroenteritis virus (TGEV), according to the International Committee of Taxonomy of Viruses (Carstens, 2010).
To date, two different genotypes of CCoV are known, CCoV type I (CCoV-I) and CCoV type II (CCoV-II). Typical reference strains of CCoV are classified as CCoV-II (Decaro and Buonavoglia, 2008, Decaro and Buonavoglia, 2011). CCoV-I strains were identified in the beginning of the 2000s, when Pratelli and coworkers highlighted that FCoV-like CCoV strains, that share high homology with FCoVs, are circulating in canine population (Pratelli et al., 2001a, Pratelli et al., 2002). Recently, CCoV-II genotype was divided in two subtypes, CCoV-IIa (classical strains) and CCoV-IIb (TGEV-like strains), with CCoV-IIb emerging as a result of a putative recombination between CCoV-II and TGEV (Decaro et al., 2009, Decaro et al., 2010).
The genome of CCoV consists of 4 ORFs, encoding spike (S), envelope (E), membrane (M) and nucleocapsid (N) structural proteins and several ORFs translated into non structural proteins (polyprotein-precursor of RNA-dependent RNA polymerase, 3a, 3b, 3c, 7a and 7b) (Lai and Holmes, 2001). S, E and M proteins are present in the viral envelope. Glycoprotein S forms club-shaped projections on the surface of the virus and it is responsible for binding to host cell membranes, membrane fusion and induction of neutralizing antibodies (Enjuanes et al., 2000).
CCoV was described for the first time in 1974, being associated with reports of diarrhea in dogs (Binn et al., 1974). It is considered to be an etiologic agent of gastroenteritis in dogs, usually manifested with anorexia, dullness, diarrhea and vomiting, lasting for up to 2 weeks. Feces are usually loose to watery and they may contain mucus or blood. Younger dogs are more severely affected. Mild disease or asymptomatic carriage are probably in many cases common outcomes of infection. Clinical signs are usually mild and the disease is highly contagious with low mortality rate (Appel et al., 1979, Carmichael and Binn, 1981).
CCoV is known to have spread worldwide, with the virus detected or isolated in Europe, U.S.A., Asia and Australia (Appel, 1987, Bandai et al., 1999, Carmichael and Binn, 1981, Naylor et al., 2001, Pollock and Carmichael, 1979). In Greece, CCoV detection was reported for the first time in 2010, and it was associated with a severe outbreak of enteritis in young dogs in a municipal kennel (Ntafis et al., 2010). The aim of the present study was the detection, molecular characterization and phylogenetic analysis of CCoV strains circulating in Greece.
2. Materials and methods
2.1. Samples
Between December 2007 and December 2009, 206 fecal samples were collected from dogs with diarrhea (from mild to severe), from northern, central and southern Greece. In detail, 103, 44, 56 and 3 samples came from the Administrative division of the Central Macedonia Region, the Thessaly Region, the Attica Region and the Crete Region, respectively. Dogs were housed in kennels or pet shops or were presented at veterinary practices at the time of the sampling. Signalment (including age, sex, breed, singly housed/housed in groups and vaccination history) was recorded.
Rectal swabs were placed in isothermal boxes using ice bags and transferred in the lab. Samples were kept frozen at −80 °C until tested.
2.2. RNA extraction, CCoV detection and characterization
Fecal samples were homogenized in phosphate buffered saline solution (PBS, Sigma–Aldrich, USA) and were clarified by centrifugation at 8000g for 5 min. Total RNA was extracted from the supernatant fluid with a commercial RNA extraction kit (QIAamp Viral RNA Mini Kit, Qiagen GmbH, Germany).
For the CCoV RNA detection and quantitation, both a reverse transcription nested PCR (RT-nPCR) and a real time RT-PCR assay were used, as previously described (Decaro et al., 2004, Pratelli et al., 1999).
For CCoV type I and II detection and quantitation, two RT-PCR assays (amplifying a part of the S gene) (Pratelli et al., 2004) and two real time RT-PCR assays (amplifying a part of the M gene) were used (Decaro et al., 2005).
For CCoV subtyping and discrimination of CCoV-IIa and CCoV-IIb strains, two RT-PCR assays with comparable levels of sensitivity were performed, as previously described (Decaro et al., 2010). A RT-PCR, able to discriminate CCoV and TGEV, was used in order to verify the absence of TGEV strains in the CCoV-IIb positive samples (Wesley, 1999).
2.3. Sequencing and sequence analysis
To determine strains’ sequence of each genotype/subtype, positive samples from all regions of sampling were chosen randomly. At least 10% of each category was subjected to sequencing.
To determine the sequence of CCoV-I strains, a 1-kb region of the 5’ end of the S gene was amplified from 15 samples, using SuperScript One-Step RT-PCR for Long Templates (Invitrogen S.R.L.) and primers PolEnd (5′-CTAAGGAAGGGTAAGTTGCTCA-3′) and 1060dR (5′-ATCAGCAGATGCTTGRGGACA-3′). The cDNA was synthesized at 50 °C for 30 min with denaturation at 94 °C for 2 min. The amplification reaction was carried out for 40 cycles with denaturation at 94 °C for 1 min, annealing at 52 °C for 2 min and polymerization at 68 °C for 2 min.
To determine the sequence of the 5’ end of the S gene of 19 CCoV-IIa strains, a RT-PCR assay with El-Ins1 (5′-GGATTACTAARGADKGGTAAGTTGC-3′) and S2 (5′-CAACTTCACTTGAAGCAACA-3′) primers was used, as previously described (Decaro et al., 2007).
To determine the sequence of the 5’ end of the S gene of 13 CCoV-IIb strains, a RT-PCR assay with 20179 (5′-GGCTCTATCACATAACTCAGTCCTAG-3′) and 174–268 (5′-CAACATGTAACCTTTGTCTGTGATCTGC-3′) primers was used, as previously described (Decaro et al., 2010).
The nucleotide sequences were determined in both directions by a commercial facility (Beckman Colter Genomics, United Kingdom). Sequence assembling and analysis were carried out using the BioEdit software package (Hall, 1999) and the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) analysis tool. Phylogenetic analysis was conducted using the MEGA4 program (Tamura et al., 2007). Phylogenetic trees, based on the partial amino acid sequences of S and M proteins, were elaborated using neighbour-joining method, supplying a statistical support with bootstrapping over 1000 replicates.
2.4. GenBank accession numbers
The partial sequences of the S gene of CCoV-I strains LA/10/08/I/GR, TH/95/08/I/GR, TH/24/09/I/GR, TH/69/09/I/GR, AT/02/09/I/GR, LA/12/08/I/GR, TH/101/08/I/GR, VO/34/09/I/GR, AT/05/09/I/GR, TH/10/09/I/GR, TH/25/08/I/GR, TH/24/08/I/GR, TH/37/08/I/GR, TH/82/09/I/GR and TH/83/08/I/GR have been registered in GenBank under accession numbers JQ422551, JQ422552, JQ422553, JQ422554, JQ422555, JQ422556, JQ422557, JQ422558, JQ422559, JQ422560, JQ422561, JQ422562, JQ422563, JQ422564, and JQ422565, respectively.
The partial sequences of the S gene of CCoV-IIa strains TR/01/07/I/GR, TR/01/08/IIa/GR, TR/02/07/IIa/GR, TR/59/08/IIa/GR, TR/56/08/IIa/GR, AT/60/09/IIa/GR, AT/61/09/IIa/GR, TH/81/09/IIa/GR, AT/01/09/IIa/GR, AT/03/09/IIa/GR, AT/04/09/IIa/GR, TH/10/09/IIa/GR, TH/15/09/IIa/GR, AT/20/09/IIa/GR, TH/95/08/IIa/GR, TH/97/08/IIa/GR, TH/98/08/IIa/GR, TH/101/08/IIa/GR and ED/99/08/IIa/GR have been registered in GenBank under accession numbers JQ422566, JQ422567, JQ422568, JQ422569, JQ422570, JQ422571, JQ422572, JQ422573, JQ422574, JQ422575, JQ422576, JQ422577, JQ422578, JQ422579, JQ422580, JQ422581, JQ422582, JQ422583, and JQ422584, respectively.
The partial sequences of the S gene of CCoV-IIb strains AT/02/09/IIb/GR, AT/05/09/IIb/GR, TH/17/09/IIb/GR, TH/22/09/IIb/GR, TH/73/08/IIb/GR, TH/24/09/IIb/GR, TH/74/08/IIb/GR, TH/88/08/IIb/GR, TH/89/08/IIb/GR, TH/90/08/IIb/GR, TH/67/09/IIb/GR, TH69/09/IIb/GR and TH/85/09/IIb/GR have been registered in GenBank under accession numbers JQ422585, JQ422586, JQ422587, JQ422588, JQ422589, JQ422590, JQ422591, JQ422592, JQ422593, JQ422594, JQ422595, JQ422596, and JQ422597, respectively.
2.5. Statistical analysis
Hypothesis test to compare proportions was used to determine significant differences between different dog populations. For all tests, a P-value of less than 0.05 was considered to be statistically significant. Statistical analysis was performed using statistical package Statgraphics Plus for Windows version 4.0, Manugistics Inc., Rockville, MD, USA.
3. Results
3.1. CCoV detection and characterization
By means of RT-nPCR and/or real time RT-PCR, 134 of 206 (65.1%) samples were positive for CCoV (Table 1 ). CCoV was detected in all regions of sampling. By means of genotype specific RT-PCR and/or real time RT-PCR, CCoV-I and CCoV-II were detected in 99 and 94 of 134 CCoV positive samples, respectively (Table 2, Table 3 ).
Table 1.
Detection of CCoV RNA in feces by RT-nPCR and by real time RT-PCR assays. The numbers indicate the positive (+) or negative (-) samples.
| RT-nPCR | Real time RT-PCR |
|
|---|---|---|
| + | – | |
| + | 128 | 2 |
| – | 4 | 72 |
Table 2.
Detection of CCoV-I RNA in feces by RT-PCR and by real time RT-PCR assays. The numbers indicate the positive (+) or negative (-) samples.
| RT-PCR (type I) | Real time RT-PCR (type I) |
|
|---|---|---|
| + | – | |
| + | 89 | 2 |
| – | 8 | 35 |
Table 3.
Detection of CCoV-II RNA in feces by RT-PCR and by real time RT-PCR assays. The numbers indicate the positive (+) or negative (-) samples.
| RT-PCR (type II) | Real time RT-PCR (type II) |
|
|---|---|---|
| + | – | |
| + | 80 | 7 |
| – | 7 | 40 |
By RT-PCR subtyping, CCoV-IIa and CCoV-IIb were detected in 72 and 11 of the 94 CCoV-II positive samples, respectively, while in 10 samples, both subtypes were detected. One of the samples (85/08) was not characterized while no TGEV strains were detected.
3.2. CCoV mixed infections
CCoV-I and CCoV-II were detected in 40 (29.9%) and 30 (22.4%) of the 134 positive samples, respectively, while in 63/134 (47%) samples more than one CCoV genotype/subtype was detected, including 3 samples where triple infections were detected (CCoV-I/CCoV-IIa/CCoV-IIb). The frequency of CCoV genotypes and subtypes detection is shown in Table 4 and the frequency of genotype/subtype combinations in mixed infections is shown in Fig. 1 . Characterization of one sample was not possible (85/08), since by S- gene RT-PCR and M-gene real time RT-PCR, it was characterized as CCoV-II and CCoV-I, respectively.
Table 4.
Number of positive samples and frequency of detection of CCoV genotypes and subtypes. For one sample, characterization was not possible.
| Single infection |
Mixed infection |
|||||||
|---|---|---|---|---|---|---|---|---|
| CCoV | CCoV-I | CCoV-IIa | CCoV-IIb | CCoV-I/CCoV-IIa | CCoV-I/CCoV-IIb | CCoV-IIa/CCoV-IIb | CCoV-I/CCoV-IIa/CCoVIIb | |
| Positive samples | 134 | 40 | 28 | 2 | 44 | 9 | 7 | 3 |
| Frequency (%) | 29.9 | 20.9 | 1.5 | 32.8 | 6.7 | 5.2 | 2.2 | |
Fig. 1.
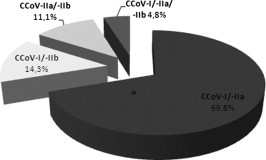
CCoV mixed infections detected (n = 63), by means of RT-PCR and/or real time RT-PCR. Percentages of different genotype/subtype combinations.
3.3. CCoV frequency in different dog populations
CCoV was detected in 99 (77.3%) of 128 dogs, housed in groups (kennels, animal shelters, pet shops) and in 35 (44.9%) of 78 singly housed animals, the difference being statistically significant (p < 0.05).
In addition, regarding CCoV frequency in different age groups, the virus was detected in 67 (73.6%) of 91 dogs younger than 3 months old, and in 67 (58.3%) of 115 dogs at least 3 months of age, the difference between the groups being statistically significant (p < 0.05). The oldest animal tested positive for CCoV was 12 years old.
3.4. Sequencing results and phylogenetic analysis
Sequence analysis of the 15 CCoV-I Greek strains in the 5’ end of the S gene showed a 64.1–99.8% nucleotide (nt) and 60.6–99.6% amino acid (aa) identity to each other. The Greek strains shared the highest genetic relatedness with either one of the two CCoV type I reference strains, Elmo/02 (AY307020) and 23/03 (AY307021). According to the phylogenetic analysis result, the CCoV-I strains segregate with either Elmo/02 (2 strains, 84.9–87.2% nt identity) or 23/03 (74.1–92.8% nt identity), forming two unique clades of CCoV-I viruses. The strains AT/02/09/I/GR and AT/05/09/I/GR, detected in a pet shop outbreak, shared 63% nt identity to each other. All CCoV-I strains formed a separate clade within the same cluster of CCoV-I and FCoV-I viruses, which is separate from that of CCoV-IIa and FCoV-II (Fig. 2 ).
Fig. 2.
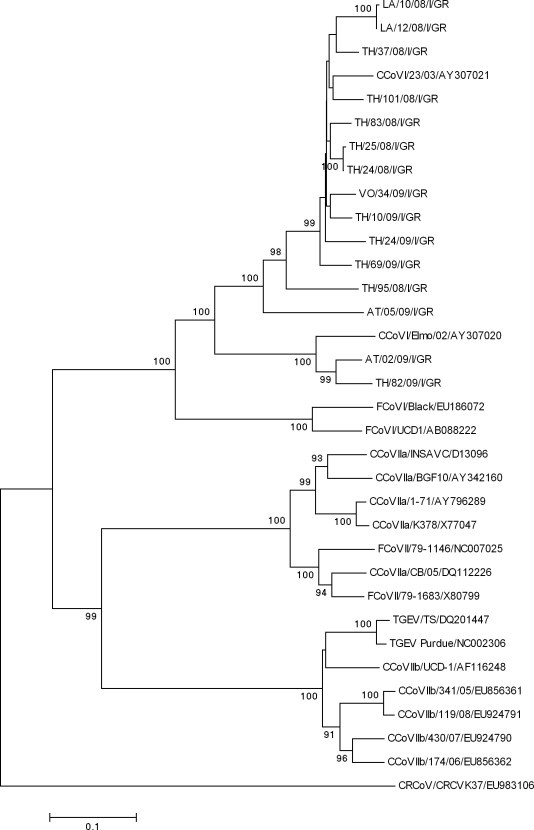
Neighbor-joining tree of the Greek CCoV-I strains, based on the N-terminus of the S protein. The tree is rooted on the Betacoronavirus canine respiratory coronavirus (CRCoV). The numbers represent the percentage of replicate trees based on 1000 bootstrap replicates. The scale bar represents the number of amino acid substitutions per site.
Sequence analysis of the 19 CCoV-IIa Greek strains in the 5’ end of the S gene showed a 79.8–100% nt and 77.7–100% aa identity to each other. By excluding the strain ED/99/08/IIa/GR, the nt and aa identities among Greek CCoV-IIa viruses reached 99.7–100%. By BLAST, it was revealed that ED/99/08/IIa/GR shared the highest identity with the CCoV-IIa reference strain BGF10 (AY342160) (83.7%), while the rest of the strains showed the highest identity to CCoV-IIa reference strain CB/05(DQ112226). The inferred amino acid sequence of ED/99/08/IIa/GR was compared to the analogous sequences available in the online databases. Sequence analysis revealed a highly divergent N-terminal domain of the S protein, 6 amino acids longer with regards to the reference strains, with the insertions being unique to ED/99/08/IIa/GR. The unique residues were found in positions 69–70 (N, I), 147–148 (P, S), and 387–390 (T, L, I, E) of the S protein (Fig. 3 ). According to the phylogenetic analysis result, the CCoV-IIa strains segregated with CB/05, with the exception of ED/99/08/IIa/GR, that formed a unique clade, within the cluster of CCoV-IIa and FCoV-II (Fig. 4 ).
Fig. 3.
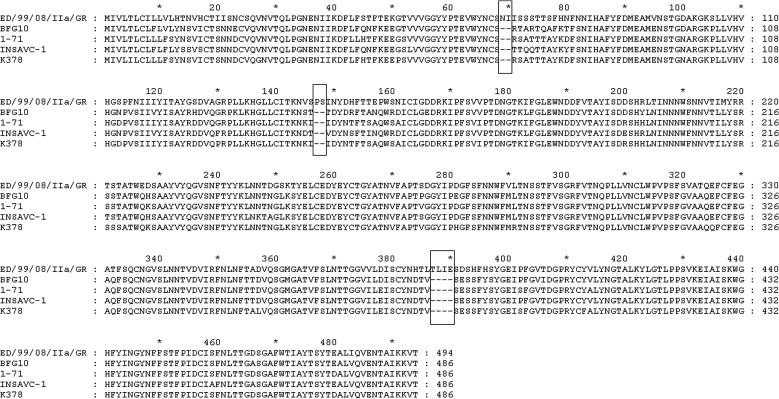
N-terminus of the S protein of the ED/99/08/IIa/GR strain with CCoV-IIa reference strains BGF10, 1-71, INSAVC-1 and K378. The amino acid insertions of ED/99/08/IIa/GR are enclosed in rectangles.
Fig. 4.
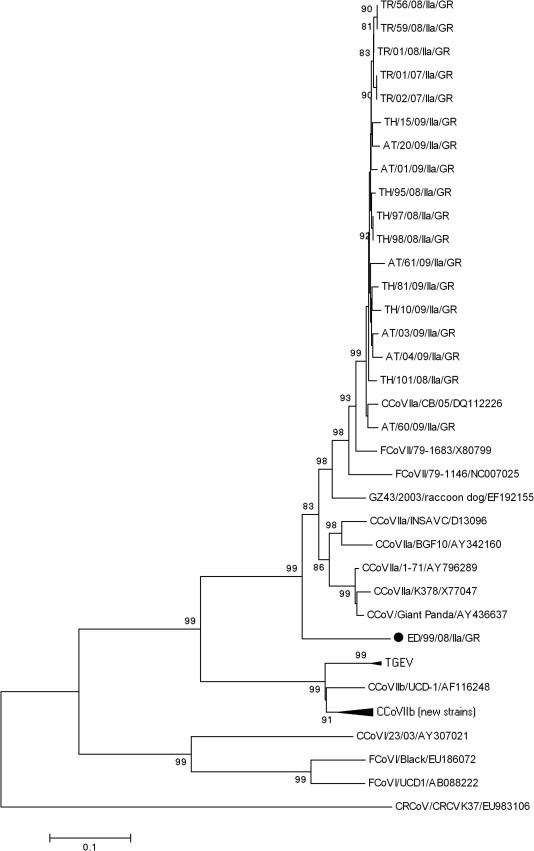
Neighbor-joining tree of the Greek CCoV-IIa strains, based on the N-terminus of the S protein. The tree is rooted on the Betacoronavirus canine respiratory coronavirus (CRCoV). The numbers represent the percentage of replicate trees based on 1000 bootstrap replicates. The scale bar represents the number of amino acid substitutions per site. The divergent strain ED/99/08/IIa/GR is highlighted with a circle.
Sequence analysis of the 13 CCoV-IIb Greek strains in the 5’ end of the S gene showed a 94.1–100% nt identity and 92.3–100% aa identity to each other. The strains shared the highest genetic relatedness with either one of the two CCoV-IIb reference strains, 119/08 (EU924791) and 198/07 (GQ130266). Phylogenetic analysis revealed that the Greek strains were more closely related to CCoV-IIb strains detected in Italy, Romania and Hungary, forming a separate clade from the UK CCoV-IIb strains. In addition, TGEV strains and UCD-1 “old” CCoV-IIb strain formed a separate clade within the same cluster of TGEV and TGEV-like viruses (Fig. 5 ).
Fig. 5.
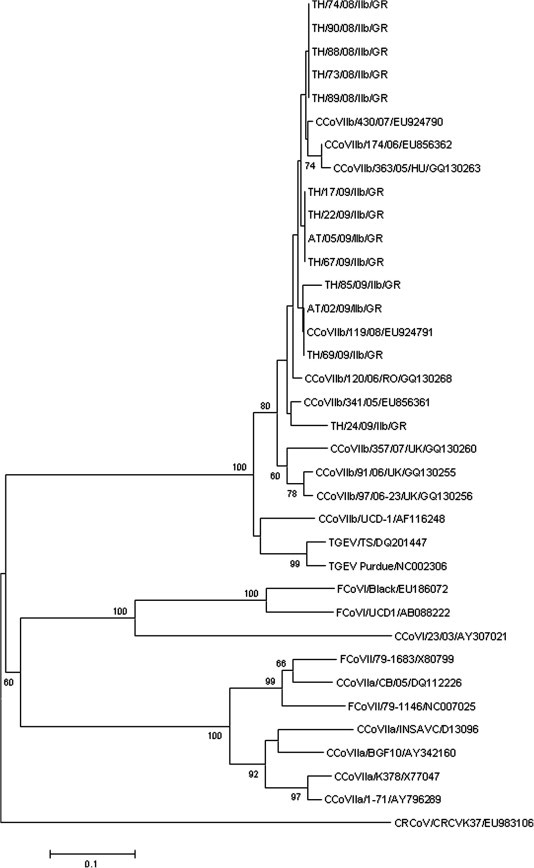
Neighbor-joining tree of the Greek CCoV-IIb strains, based on the N-terminus of the S protein. The tree is rooted on the Betacoronavirus canine respiratory coronavirus (CRCoV). The numbers represent the percentage of replicate trees based on 1000 bootstrap replicates. The scale bar represents the number of amino acid substitutions per site.
4. Discussion
Canine coronavirus is one of the major pathogenic agents of gastroenteritis and diarrhea in dogs. The main purpose of the study was to identify the frequency of CCoV infections in dogs with diarrhea and to characterize the strains that circulate in Greece. By means of molecular techniques, CCoV was detected in 65.1% of the dogs presenting diarrhea. The frequency of CCoV detection was higher than the frequency reported in other countries including Italy, Turkey, England, Japan and Portugal (Bandai et al., 1999, Decaro et al., 2010, Yesilbag et al., 2004), highlighting the significance of the pathogen as a contributing agent to the manifestation of diarrhea in dogs. In addition, positive samples were identified in all regions sampled, suggesting that the virus is frequently detected and it circulates among canine population in Greece.
According to the results of the present study, CCoV was more frequently detected in dogs housed in groups than in singly housed animals. One possible explanation of the difference is the close contact of the animals, the fecal-oral transmission of the disease and the long-term viral shedding. As it has been suggested, dogs may be contagious for a period varying from 2 weeks to several months (Keenan et al., 1976, Pratelli et al., 2001a, Pratelli et al., 2001b), highly contaminating their environment. Likewise, similar differences between singly and group housed dogs have been identified in previous studies, by means of serological assays (Naylor et al., 2001, Rimmelzwaan et al., 1991).
Dogs of all breeds and ages appear to be susceptible to CCoV infection (Appel, 1987, Carmichael and Binn, 1981). In the present study, CCoV was more frequently detected in dogs younger than 3 months old than in dogs of at least 3 months of age. Puppies usually become susceptible after the decline of the maternally derived antibodies, which last for a period of 4–5 weeks (Appel, 1987). In contrast, older animals appear to be less susceptible, due to acquired immunity, occurring as a result of prior exposure to the pathogen. In the present study, the oldest animal tested positive was a 12 year old dog. The results indicate that CCoV is more widespread in puppies, though older animals can also be presented with a coronaviral infection.
Concerning the detection and characterization of CCoV, both RT-PCR and real time RT-PCR were used. It was shown, that there were strains either detected (4/206) or characterized (8/134 CCoV-I, 7/134 CCoV-II) only by real time RT-PCR, as it was expected, since these assays have higher sensitivity compared to the RT-PCR assays (Decaro et al., 2004, Decaro et al., 2005). However, in the present study some of the strains were either detected (2/206) or characterized (2/134 CCoV-I, 7/134 CCoV-II) only by RT-PCR assays, which may be explained by the fact that the assays amplify different parts of the genome. Since coronavirus mutation rates are considered to be high due to the infidelity of RNA-dependent RNA polymerase (Woo et al., 2009), false-negative results are expected, when either primers or probes lack the ability to anneal. It is obvious that the results may be more reliable by using both assays, since no assay has been demonstrated to represent the gold standard for the identification and characterization of CCoV.
Regarding genotype/subtype identification, it was shown that CCoV-I was more frequently detected than “classical” CCoV-IIa or CCoV-IIb viruses. However, in most of the positive samples (47.01%) more than one strain was detected. In addition, in the present study triple CCoV infections (CCoV-I/IIa/IIb), representing almost 5% of the mixed infections, were detected for the first time, since so far only double infections have been identified in all possible combinations (Decaro et al., 2009, Decaro et al., 2010, Erles and Brownlie, 2009). Mixed CCoV infections are usual, although their significance is not completely understood (Pratelli et al., 2004). Preliminary studies have indicated that they may be related to more severe symptoms (Decaro et al., 2005), but the inability to cultivate CCoV-I so far, impedes the elucidation of its pathogenesis and its role in mixed infections.
In the present study, the 5’ end of the S gene was chosen for sequence and phylogenetic analysis of CCoV strains, since mutations that are related to antigenicity, virulence and viral tropism are often observed in this region (Lai and Holmes, 2001, Sanchez et al., 1999). Regarding CCoV-I, the Greek strains showed low genetic similarity to each other and to the prototype CCoV-I strains. Most of the CCoV-I Greek strains shared the highest similarity with 23/03 prototype strain, while only two with Elmo/02, revealing that 23/03-like strains are more frequently detected in Greece. In addition, the two different CCoV-I strains detected in the same pet shop outbreak (AT/02/09/I/GR and AT/05/09/I/GR) indicate that different strains of the same genotype/subtype can be involved in an outbreak.
Regarding CCoV-IIa, the Greek strains showed high genetic similarity to each other and to the prototype CB/05, with the exception of one strain (ED/99/08/IIa/GR). CCoV-IIa strains shared much higher identity to each other and to the prototype strains than CCoV-I strains. Recently, it was suggested that CCoV-I represents a possible parental biotype of CCoV-IIa (Lorusso et al., 2008). If this is the case, the genome of CCoV-IIa is being subjected to the evolutionary pressure of the host immune system for a shorter period of time, with respect to the CCoV-I genome. This fact makes tempting to assume that the higher homogeneity of the CCoV-IIa strains’ genome may be attributed to the shorter circulation time, with respect to strains of type I. Sequence analysis of more CCoV-I strains, coming from different geographic regions may help acquire more information regarding their variability, since so far only few CCoV-I strains have been analyzed.
Of significant interest is the ED/99/08/IIa/GR strain that appears to be divergent, indicating the plasticity of the 5′-end of the CCoV S gene. The receptor-binding domain of coronaviruses is localized within the N-terminus of the S protein and it has been shown that amino acid changes in the region are associated with changes in the tropism and virulence of the virus (Lai and Holmes, 2001, Sanchez et al., 1999). Similar divergent strains have been identified in the past in Australia and in the United Kingdom and they have been associated with severe gastroenteritis outbreaks (Naylor et al., 2002, Sanchez-Morgado et al., 2004). Isolation and experimental infection could give information regarding ED/99/08/IIa/GR mutations and their possible relation to viral pathogenicity.
Regarding CCoV-IIb, it was shown that the newly emerged strains circulate in Greece with higher frequency than previously reported (Decaro et al., 2010). Sequence and phylogenetic analysis of CCoV-IIb strains revealed for the first time that the Greek viruses belong to the newly emerged TGEV-like strains and not to the “old” UCD1 TGEV-like strain. In addition, the Greek strains shared high similarity to each other and were genetically related to strains from Hungary and Romania and distant from UK strains. The relation among strains could be explained based on the trends in commercial trade practices, since Greece, as Italy (Decaro et al., 2010) imports dogs mainly from countries of Eastern Europe.
5. Conclusions
From the results of this study, it can be concluded that CCoV infection is widespread in Greece, especially in young puppies and dogs housed in groups, presenting with diarrhea. Mixed infections prevail among CCoV infections, with the triple (CCoV-I/IIa/IIb) infections being reported for the first time. Sequence analysis of different strains revealed high variability among CCoV-I strains at the 5’ end of the S gene. In addition, the identification of a divergent strain is reported. The circulation of CCoV-I highly variable and TGEV-like emerging strains, as well as the detection of divergent strains, raise significant concerns over the spread of the strains in dogs. To date, vaccination against CCoV is considered a controversial issue, depending heavily upon the population at risk. Further research on the pathogenicity could inform on the possible need for wider cross-reactive prophylactic vaccines.
Acknowledgements
This work was supported partially by a grant from Pfizer Animal Health. Vasileios Ntafis is grateful to Alexander S. Onassis Public Benefit Foundation for doctoral funding. The authors are grateful to Sotirakoglou Kyriaki, Associate Professor in Laboratory of Mathematics and Statistics, Agricultural University of Athens, for her assistance regarding the statistical analysis.
References
- Appel M.J. Canine coronavirus. In: Appel M.J., editor. Virus Infections of Carnivores. Elsevier Science Publishers; The Netherlands: 1987. pp. 115–122. [Google Scholar]
- Appel M.J.G., Cooper B.J., Greisen H., Scott F., Carmichael L.E. Canine viral enteritis. I. Status report on corona- and parvo-like viral enteritides. Cornell Vet. 1979;69:123–133. [PubMed] [Google Scholar]
- Bandai C., Ishiguro S., Masuya N., Hohdatsu T., Mochizuki M. Canine coronavirus infections in Japan: virological and epidemiological aspects. J. Vet. Med. Sci. 1999;61(7):731–736. doi: 10.1292/jvms.61.731. [DOI] [PubMed] [Google Scholar]
- Binn, L.N., Lazar, E.C., Keenan, K.P., Huxsoll, D.L., Marchwicki, R.H., Strano, A.J., 1974. Recovery and characterization of a coronavirus from military dogs with diarrhea. In: Proc.78th Ann. Meeting US Anim. Health Assoc., pp. 359–366. [PubMed]
- Carmichael L.E., Binn L.N. New enteric viruses in the dog. Adv. Vet. Sci. Comp. Med. 1981;25:1–37. [PubMed] [Google Scholar]
- Carstens E.B. Ratification vote on taxonomic proposals to the international committee on taxonomy of viruses. Arch. Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. An update on canine coronaviruses: viral evolution and pathobiology. Vet. Microbiol. 2008;132:221–234. doi: 10.1016/j.vetmic.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Buonavoglia C. Canine coronavirus: not only an enteric pathogen. Vet. Clin. North Am. Small Anim. Pract. 2011;38:799–814. doi: 10.1016/j.cvsm.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Campolo M., Lorusso A., Camero M., Elia G., Martella V., Cordioli P., Enjuanes L., Buonavoglia C. Recombinant canine coronavirus related to transmissible gastroenteritis virus of swine are circulating in dogs. J. Virol. 2009;83(3):1532–1537. doi: 10.1128/JVI.01937-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Mari V., Elia G., Addie D.D., Camero M., Lucente M.S., Martella V., Buonavoglia C. Recombinant canine coronaviruses in dogs. Eur. Emerg. Infect. Dis. 2010;16:41–47. doi: 10.3201/eid1601.090726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Elia G., Campolo M., Desario C., Cirone F., Tempesta M., Buonavoglia C. Molecular characterization of the virulent canine coronavirus CB/05 strain. Virus Res. 2007;125:54–60. doi: 10.1016/j.virusres.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Martella V., Ricci D., Elia G., Desario C., Campolo M., Cavaliere N., Trani L.D., Tempesta M., Buonavoglia C. Genotype-specific fluorogenic RT-PCR assays for the detection and the quantitation of canine coronavirus type I and type II RNA in fecal samples of dogs. J. Virol. Methods. 2005;130:72–78. doi: 10.1016/j.jviromet.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro N., Pratelli A., Campolo M., Elia G., Martella V., Tempesta M., Buonavoglia C. Quantitation of canine coronavirus RNA in the feces of dogs by TaqMan RT-PCR. J. Virol. Methods. 2004;119:145–150. doi: 10.1016/j.jviromet.2004.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjuanes L., Brian D., Cavanagh D., Holmes K., Lai M.M.C., Laude H., Masters P., Rottier P.J.M., Siddell S.G., Spaan W.J.M., Taguchi F., Talbot P. Coronaviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., Carstens E.B., Estes M.K., Lemon S.M., Maniloff J., Mayo M.A., McGeoch D.J., Pringle C.R., Wickner R.B., editors. Virus Taxonomy. Classification and Nomenclature of Viruses. Academic Press; New York: 2000. pp. 835–849. [Google Scholar]
- Erles K., Brownlie J. Sequence analysis of divergent canine coronavirus strains present in UK dog population. Virus Res. 2009;141:21–25. doi: 10.1016/j.virusres.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Keenan K.P., Jervis H.R., Marchwicki R.H., Binn L.N. Intestinal infection of neonatal dogs with canine coronavirus 1–71: studies by virologic, histologic, histochemical and immunofluorescent techniques. Am. J. Vet. Res. 1976;37:247–256. [PubMed] [Google Scholar]
- Lai M.M.C., Holmes K.V. Coronaviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., Griffin D.E., Lamb R.A., Martin M.A., Roizman B., Strais S.E., editors. Fields Virology. fourth ed. Lippincott Williams and Wilkins; Philadelphia: 2001. pp. 1163–1185. [Google Scholar]
- Lorusso A., Decaro N., Schellen P., Rottier P.J., Buonavoglia C., Haijema B.J., de Groot R.J. Gain, preservation and loss of a group 1a coronavirus accessory glycoprotein. J. Virol. 2008;82:10312–10317. doi: 10.1128/JVI.01031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor M.J., Monckton R.P., Lehrbach P.R., Deane E.M. Canine coronavirus in Australian dogs. Aust. Vet. J. 2001;79:116–119. doi: 10.1111/j.1751-0813.2001.tb10718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor M.J., Walia C.S., McOrist S., Lehrbach P.R., Deane E.M., Harrison G.A. Molecular Characterization confirms the presence of a divergent strain of canine coronavirus (UWSMN-1) in Australia. J. Clin. Microbiol. 2002;40:3518–3522. doi: 10.1128/JCM.40.9.3518-3522.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntafis V., Mari V., Danika S., Fragkiadaki E., Buonavoglia C. An outbreak of canine coronavirus in a Greek kennel. J. Vet. Diagn. Invest. 2010;22:320–323. doi: 10.1177/104063871002200231. [DOI] [PubMed] [Google Scholar]
- Pollock R.V.H., Carmichael L.E. Canine viral enteritis, recent developments. Mod. Vet. Pract. 1979;60:375–380. [PubMed] [Google Scholar]
- Pratelli A., Decaro N., Tinelli A., Martella V., Elia G., Tempesta M., Cirone F., Buonavoglia C. Two genotypes of canine coronavirus simultaneously detected in the fecal samples of dogs with diarrhea. J. Clin. Microbiol. 2004;42(4):1797–1799. doi: 10.1128/JCM.42.4.1797-1799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Decaro N., Aliberti A., Buonavoglia D., Tempesta M., Buonavoglia C. Variation of the sequence in the gene encoding for transmembrane protein M of canine coronavirus (CCV) Mol. Cell. Probes. 2001;15:229–233. doi: 10.1006/mcpr.2001.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Martella V., Elia G., Tempesta M., Guarda F., Cappucchio M.T., Carmichael L.E., Buonavoglia C. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. J. Vet. Med. B. 2001;48:385–392. doi: 10.1046/j.1439-0450.2001.00466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Tempesta M., Greco G., Martella V., Buonavoglia C. Development of a nested PCR assay for the detection of canine coronavirus. J. Virol. Methods. 1999;80:11–15. doi: 10.1016/S0166-0934(99)00017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratelli A., Tinelli A., Decaro N., Camero M., Elia G., Gentile A., Buonavoglia C. PCR assay for the detection and the identification of atypical canine coronavirus in dogs. J. Virol. Methods. 2002;106:209–213. doi: 10.1016/S0166-0934(02)00165-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimmelzwaan G.F., Groen J., Egberink H., Borst G.H.A., UytdeHaag F.G.C.M., Osterhaus A.D.M.E. The use of enzyme-linked immunosorbent assay systems for serology and antigen detection in parvovirus, coronavirus and rotavirus infections in dogs in The Netherlands. Vet. Microbiol. 1991;26:25–40. doi: 10.1016/0378-1135(91)90039-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez C.M., Izeta A., Sanchez-Morgado J.M., Alonso A., Sola I., Balasch M., Plana-Duran J., Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 1999;73(9):7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Morgado J.M., Poynter S., Morris T.H. Molecular characterization of a virulent canine coronavirus BGF strain. Virus Res. 2004;104:27–31. doi: 10.1016/j.virusres.2004.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wesley R.D. The S gene of canine coronavirus, strain UCD-1, is more closely related to the S gene of transmissible gastroenteritis virus than to that of feline infectious peritonitis virus. Virus Res. 1999;61:145–152. doi: 10.1016/S0168-1702(99)00032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- Yesilbag K., Yilmaz Z., Torun S., Pratelli A. Canine coronavirus in Turkish dog population. J. Vet. Med. 2004;51:353–355. doi: 10.1111/j.1439-0450.2004.00773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


