Abstract
In 2014, three infectious bronchitis virus (IBV) strains, designated as γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and γCoV/ck/China/I0118/14, were isolated and identified from chickens suspected to be infected with IBV in Guangxi province, China. Based upon data arising from S1 sequence and phylogenetic analyses, the three IBV isolates were genetically different from other known IBV types, which represented a novel genotype (GI-29). Virus cross-neutralization tests, using γCoV/ck/China/I0111/14 as a representative, showed that genotype GI-29 was antigenically different from all other known IBV types, thus representing a novel serotype. Complete genomic analysis showed that GI-29 type viruses were closely related to and might originate from a GX-YL5-like virus by accumulation of substitutions in multiple genes. These GI-29 viral genomes are still evolving and diverging, particularly in the 3′ region, although we cannot rule out the possibility of recombination events occurring. For isolate γCoV/ck/China/I0114/14, we found that recombination events had occurred between nsps 2 and 3 in gene 1 which led to the introduction of a 4/91 gene fragment into the γCoV/ck/China/I0114/14 viral genome. In addition, we found that the GI-29 type γCoV/ck/China/I0111/14 isolate was a nephropathogenic strain and high pathogenic to 1-day-old specific pathogen-free (SPF) chickens although cystic oviducts were not observed in the surviving layer chickens challenged with γCoV/ck/China/I0111/14 isolate.
Keywords: Infectious bronchitis coronavirus, GI-29 genotype, Serotype, Nephropathogenicity
Highlights
-
•
A novel infectious bronchitis virus type, GI-29, has been identified in China.
-
•
Identification of mutations scattered throughout the GI-29 genome.
-
•
The GI-29 type is nephropathogenic to specific pathogen-free chickens.
1. Introduction
Infectious bronchitis virus (IBV) is a gammacoronavirus (family Coronaviridae, order Nidovirales) which causes highly contagious respiratory, reproductive, and urogenital symptoms in chickens (Cavanagh, 2007); consequently, this virus is of economic significance. This virus exists in a wide range of genetically and antigenically distinct types, making the prevention and control of this pathogen very challenging. While it is believed that the natural host of IBV is the chicken, the presence of IBV-like and other avian coronaviruses has been reported in both domestic and wide birds (Cavanagh, 2005, Cavanagh, 2007), making the epidemiology and mechanisms of infection very complex.
The genomic architecture of coronaviruses, such as IBV, is characterized by two large overlapping open reading frames (ORFs), ORF1a and ORF1b, which encode components of the viral replicase and occupy approximately two-thirds of the genome. ORFs encoding the structural proteins, spike (S) glycoprotein, membrane (M) glycoprotein, nucleocapsid (N) phosphoprotein and envelope (E) protein, are located downstream of ORF1a/b and are expressed from 3′ co-terminal sub-genomic mRNAs (Brian and Baric, 2005, Sawicki et al., 2007). The S protein is post-translationally cleaved into S1 and S2 fragments. The S1 subunit of S protein carries the receptor binding site and thus plays an important role in tissue tropism (Belouzard et al., 2012). The S1 protein is also the major inducer of neutralizing and serotype-specific antibodies and thus plays an important role in protective immunity (Wickramasinghe et al., 2011). In addition, S1 is the most variable gene among IBV isolates and such variability may lead to important biological differences between strains; novel serotypic variants could emerge as a result of amino acid changes in spike protein. Generally, the serotypes of IBV differ from each other by 20%–25% at the amino acid level in S1, but may differ by up to 50% (Cavanagh, 2007). In some cases, as little as 2% variation, or 10–15 amino acid changes in the amino acid sequence, can lead to a new IBV serotype because the epitopes eliciting neutralizing antibodies are widely distributed within the spike protein sequence (Cavanagh, 2007, de Wit et al., 2011a, Hodgson et al., 2004). Accordingly, analysis of the S1 gene has been conventionally used to determine viral genetic types (Valastro et al., 2016).
Currently, at least 50 different antigenic and genetic types of IBV that are poorly cross-protective have been discovered and studied by virus-neutralization tests and molecular characterization of the S1 protein gene (de Wit et al., 2011a, de Wit et al., 2011b, Jackwood et al., 2012). Furthermore, new genotypes of IBV and associated variants appear frequently in different parts of the world; these have arisen because of mutations, especially in the S1 subunit, including point mutations, insertions, deletions, and also recombination between different IBV strains (Cavanagh et al., 1992, Jackwood et al., 2012). New genotypes often show antigenic variation and, hence, define new serotypes, and vaccinated poultry may still therefore be infected by serologically distinct strains of IBV. In addition, antigenic variants resulting in a reduced level of cross-protection between different strains represents a major obstacle for the development of an efficacious vaccine.
IBV was recorded in China for the first time in the 1980s (Han et al., 2011). Surveillance studies have reported widespread IBV infections in China since the late 1990s with associated multiple types; LX4 (QX-like) is the predominant type circulating in chicken flocks and first emerged in 1995 (Zhao et al., 2017) and at least five other serotypes (Chen et al., 2017, Gao et al., 2016, Han et al., 2011, Liu et al., 2007) and variants (Han et al., 2016, Liu et al., 2013, Liu et al., 2014, Ma et al., 2012) have been circulating in China over recent years.
In this study, we isolated three IBVs in local chickens from south China (Guangxi province) and investigated genotype, complete genomic characteristics, antigenicity and pathogenicity of each variant in order to understand the origin and evolution of this type of virus.
2. Materials and methods
2.1. Virus isolation
Three IBV viruses, γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and γCoV/ck/China/I0118/14, were isolated in Guangxi province from local yellow feature chickens which showed respiratory signs at 25-, 43- and 53-days-old, respectively. The flock where the γCoV/ck/China/I0118/14 was isolated from was located in Liuzhou, and the other two flocks where the γCoV/ck/China/I0111/14 and γCoV/ck/China/I0114/14 viruses were isolated were located in Guilin. Chickens from the three flocks were spray-vaccinated with IB vaccine H120 when they were 1-day-old followed by a H120 booster vaccination on day 7 of age by eye-drop inoculation. The viruses were isolated from the tracheas of diseased chickens using 9-day-old embryonated specific pathogen-free (SPF) chicken eggs as described previously (Liu and Kong, 2004). Viral titer was determined by titration in 9-day-old embryonated chicken eggs and calculated as previously described (Reed and Muench, 1938) to provide 105.6 of the 50% egg infectious dose (EID50) in 0.1 ml.
In addition, six IBV strains, including ck/CH/LDL/091022 (Liu et al., 2013), ck/CH/LGX/111119 (Chen et al., 2017), ck/CH/LDL/97I (Liu et al., 2007), ck/CH/LDL/140520 (Xu et al., 2016), 4/91 and H120 were used in this study. Strains ck/CH/LDL/091022, ck/CH/LGX/111119, ck/CH/LDL/97I and ck/CH/LDL/140520 represent the four major IBV serotypes circulating in chicken flocks in China. Strains 4/91 and H120 are IB vaccines commonly used in China. All of these viruses were propagated in 9-day-old embryonated SPF eggs and viral titer was determined as mentioned above.
2.2. Chickens and eggs
SPF White Leghorn chickens and fertile eggs from SPF chickens (Harbin Veterinary Research Institute, Harbin, China) were used in this study. Feed and water were free of any antibiotics. All of the experimental procedures were undertaken with the approval of the Harbin Veterinary Research Institute ethical review committee and according to Chinese legislation on the use of animals for experiments, as permitted under project license LHLJ20160321.
2.3. S1 gene sequencing and genotyping of the IBV isolates
A viral RNA extraction kit (TaKaRa Bio Inc., Shiga, Japan) was used to extract total viral RNA from infectious fluids in accordance with the manufacturer's instructions, and S1 genes from the three IBV isolates were amplified using S1Oligo5′ and S1Oligo3′ primers (Adzhar et al., 1997). Amplified PCR products were sequenced directly using sense and antisense primers and sequences were first analyzed with BLASTn (BLASTn: http://blast.ncbi.nlm.nih.gov/Blast.cgi). S1 genes from 96 reference IBV strains were selected and used for phylogenetic analysis with those of our three strains. Phylogenetic analyses were performed using the maximum likelihood method with the general time-reversible nucleotide substitution model and bootstrap tests of 1000 replicates in the MEGA 6 program (Tamura et al., 2013). In addition, pairwise comparison was conducted using S1 genes from 96 references strains and our three isolates according to the results of both BLASTn and phylogenetic analysis.
2.4. Complete genome sequencing and sequence analysis
Our strategy for sequencing the complete genomes of our three IBV isolates was the same as that described previously (Liu et al., 2013). Briefly, twelve overlapped primers were used to amplify the genomes of our three IBV isolates. The extreme 5′ and 3′ termini of the three isolates were determined by rapid amplification of cDNA ends (RACE), using a 3′/5′ RACE kit (Takara Bio Inc.,) according to the manufacturer's instructions. Each of the regions was sequenced at least three times. Nucleotide sequences were edited and assembled, and consensus full-genome sequences were determined and analyzed using the Clustal W method available in Bioedit software package v7.0.3.0. (http://www.mbio.ncsu.edu/bioedit/bioedit) by comparison with a reference, the complete genome of the Beaudette strain published in GenBank. The identities of complete genomic sequences between our three isolates were compared and calculated and then BLASTn was conducted using the complete genomic sequence of isolate γCoV/ck/China/I0111/14. The available complete genomic sequences from 21 reference IBV strains representing different serotypes/genotypes were selected and used for phylogenetic analysis with those of our three strains. Phylogenetic analyses were performed using the maximum likelihood method with the general time-reversible nucleotide substitution model and bootstrap tests of 1000 replicates in the MEGA 6 program (Tamura et al., 2013). Two sequences (GX-YL5 and GX-YL9), which showed the highest identities (97% and 96%, respectively) with the complete genome of our isolate γCoV/ck/China/I0111/14, two other sequences from the vaccine strains (4/91 and H120) which are commonly used in China, and one additional sequence from the predominant IBV serotype (ck/CH/LDL/091022) circulating in China, were selected and included in the alignment using Multiple Alignment with Fast Fourier Transformation (MAFFT) v6 (http://mafft.cbrc.jp/alignment/software/).
To further study the relationship between our three IBV isolates, we created a similarity plot (SimPlot) using a 500 bp window with a 50 bp step with the isolate γCoV/ck/China/I0111/14 as the query strain. Twelve fragments were obtained based upon the results of multiple sequence alignment constructed with MAFFT v6 and SimPlot. To further investigate the genetic relationship between our isolates and the reference strains, and the possibility of recombination events occurring during the origin of our isolates, we constructed maximum likelihood phylogenetic trees based upon the sequences of each fragment with the general time-reversible nucleotide substitution model and bootstrap tests of 1000 replicates in the MEGA 6 program (Tamura et al., 2013).
To further characterize recombinants, the data set was scanned using a Recombination Detection Program (RDP) v2 with the implemented algorithms GENECONV, BootScan, MaxChi, Chimera and SiScan (McKinley et al., 2011). Similarity plot and bootscan analyses were performed using the SimPlot program (Lole et al., 1999) to further identify recombination events and recombination breakpoints. The window width and step size were set to 500 bp and 50 bp, respectively. To confirm recombination breakpoints, pairwise comparisons of the complete genomic sequence of γCoV/ck/China/I0111/14 were performed with those of GX-YL5 and 4/91 strains according to the results of phylogenetic analysis.
The complete genomic sequences of γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and γCoV/ck/China/I0118/14 have been deposited in the GenBank database with the accession numbers KY407557, KY407556 and KY407558, respectively.
2.5. Virus-neutralization testing
Cross virus-neutralization tests were performed using γCoV/ck/China/I0111/14 and six serotypes of IB Viruses (ck/CH/LDL/091022 [LX4 type], ck/CH/LGX/111119 [GI-28 type], ck/CH/LDL/97I [ck/CH/LDL/97I type], 4/91 [793/B type], H120 [Massachusetts type] and ck/CH/LDL/140520 [nrTW I type]), along with serotype-specific antisera, with a protocol described previously (Liu et al., 2013). In brief, two-fold diluted sera were incubated with the same volume (100 μl) of 100 EID50 homologous or heterologous viruses at 37 °C for 1 h, respectively. These virus–serum mixtures were then inoculated into the allantoic cavity of 9-day-old SPF embryonated eggs. Seven days after inoculation, the eggs were examined for typical lesions and the neutralization titer of each serum was determined by the last serum dilution which protected 50% of the embryo and was calculated using the method described by Reed and Muench (1938).
2.6. Infection of SPF chicks
To evaluate the pathogenicity of γCoV/ck/China/I0111/14, twenty 1-day-old SPF chickens were randomly divided into two groups of 10 birds each. The strain γCoV/ck/China/I0111/14 (106.6 EID50/ml) was then given (0.1 ml/bird) by eye-drop inoculation into individual birds in group 1, with the second group serving as negative controls. Clinical signs were observed daily throughout the experimental period. Tissue samples were collected from the trachea, lung, cecal tonsils and kidney of dead chickens and fixed in 10% buffered formalin for histopathology and immunohistochemistry (IHC) which was used for the detection of IBV antigens using a monoclonal antibody 6D10 (Han et al., 2013) directed against nucleoprotein. We collected sera from all birds on day 4, 8, 12, 16 and 20 post challenge (dpc) in order to monitor antibody response against IBV using a commercial enzyme-linked immunosorbent assay kit (IDEXX, Hoofddorp, The Netherlands) in accordance with the manufacturer's instructions. All the birds were humanely killed at 3 months of age. A post-mortem examination was performed. Meanwhile, samples from the kidney were collected in order to detect the presence of IBV by IHC.
3. Results
3.1. A new lineage in genotype I of IBV was identified in South China
Our three strains were genetically related to each other by sequence comparison of S1 and S genes at both nucleotide (97.1% and 98.6%, respectively) and amino acid (95.8% and 97.9%, respectively) levels. BLAST homology searches, using both the S1 and S amino acid sequences of isolate γCoV/ck/China/I0111/14, revealed that GX-YL5 was most similar to γCoV/ck/China/I0111/14, although limited overall sequence identity was observed (89.0% and 93.0%, respectively). These results indicated that our three isolates exhibit unique characteristics, both in S1 and S genes.
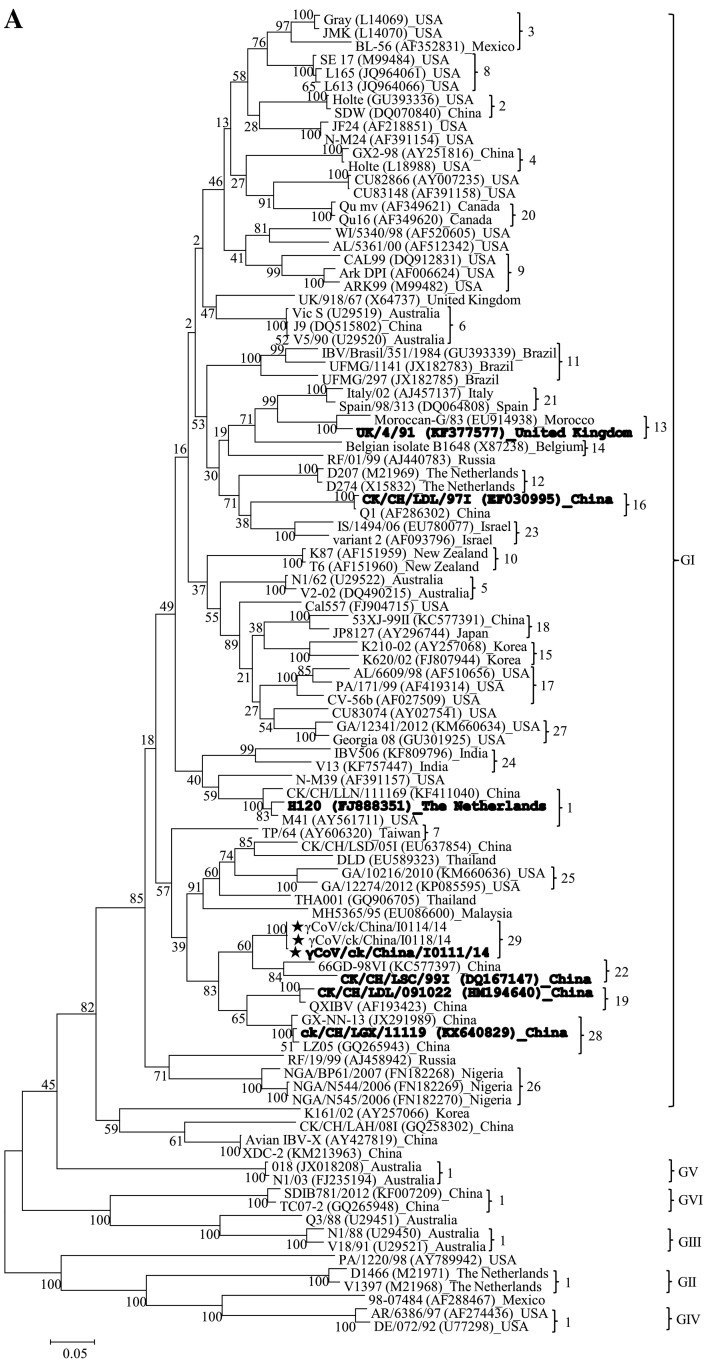
In order to genotype and further analyze the genetic diversity of the three isolates, we constructed phylogenetic trees with 96 IBV reference strains and our three isolates based upon the S1 nucleotide sequences. Results demonstrated that: 1) the 96 reference strains were clustered into six genotypes (GI-1– GI-28; GII– GVI) (Chen et al., 2017, Valastro et al., 2016); 2) Our three isolates were clustered together and clearly set apart in the phylogenetic tree from the 96 reference strains (Fig. 1A) based on the S1 phylogenetic analysis. Hence, we used the IBV classification system proposed by Valastro et al. (2016) and classified our three IBV isolates as a new lineage in genotype I (GI-29). A multiple sequence alignment constructed with MAFFT v6 showed that the mutations appeared to be distributed throughout the S1 genes of our three isolates compared with those of the reference strains; no hot spots or clustering in the location of the mutations were identified (Fig. 2 ).
Fig. 1.
Phylogenetic analysis of our three IBV isolates with other IBV reference strains. Phylogenetic trees, based upon the S1 gene (A) and complete genomic sequences (B), respectively, were constructed using the maximum likelihood method with the general time-reversible nucleotide substitution model and bootstrap tests of 1000 replicates in the MEGA 6 program. Our three IBV isolates in this study are indicated by ★. GenBank accession numbers for the reference IBV strains are shown in parentheses after each strain name. The IBV strains used for cross-neutralization tests are given in bold.
Fig. 2.
Alignment of the complete genome sequences of our three IBV isolates and five other strains was performed using MAFFT. The genome sequence of IBV γCoV/ck/China/I0111/14 was set as the reference sequence. Vertical lines indicate the single nucleotide mutation compared to the reference sequences. The GenBank accession numbers for these genome sequences are same as those in Fig. 1B.
3.2. Genetic characteristics of the GI-29 genome
The complete genomes of our three IBV isolates, γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and γCoV/ck/China/I0118/14, were 27,666, 27,669 and 27,669 nucleotides (nt) in length, respectively, including the 5′- and 3′-UTRs and excluding the poly (A) tail. A three-nucleotide deletion was identified between the sequences of the M and 5a genes in γCoV/ck/China/I0111/14, by comparing with those of isolates γCoV/ck/China/I0114/14 and γCoV/ck/China/I0118/14. The complete genome sequences of the three IBV strains showed a typical IBV gene order of 5′ UTR-1a-1b-S-3a-3b-E-M-5a-5b-N-3′ UTR and shared high genetic identities with each other (> 98.6%); gene lengths are shown in Table 1 .
Table 1.
Gene/Region lengths of our three IBV isolates.
| Virus | 5′ UTR | 1ab | Spike | 3a | 3b | 3c | M | 5a | 5b | N | 3′ UTR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| γCoV/ck/China/I1011/14 | 525 | 19,895 | 3507 | 174 | 189 | 327 | 678 | 198 | 249 | 1230 | 504 |
| γCoV/ck/China/I1014/14 | 525 | 19,895 | 3507 | 174 | 189 | 327 | 678 | 198 | 249 | 1230 | 504 |
| γCoV/ck/China/I1018/14 | 525 | 19,895 | 3507 | 174 | 189 | 327 | 678 | 198 | 249 | 1230 | 504 |
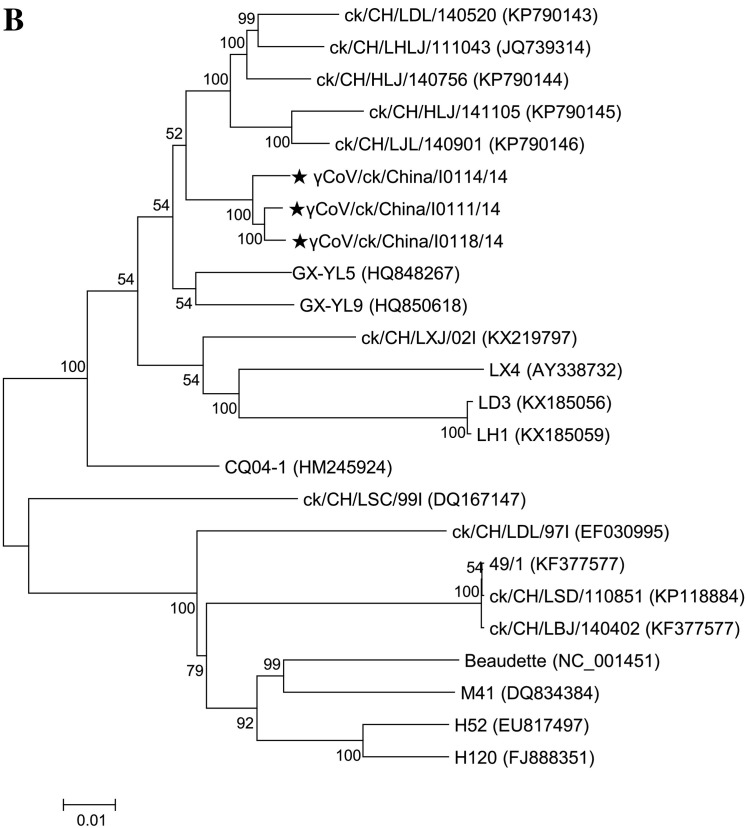
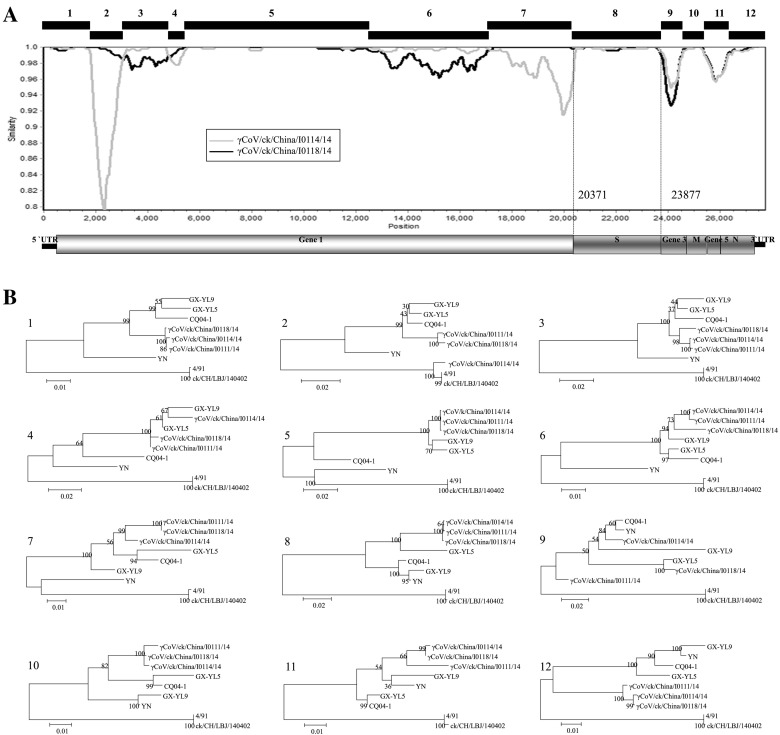
Comparatively, our three isolates were genetically close to two IBV strains, GX-YL5 and GX-YL9 (Fig. 1B), based on the phylogenetic analysis using complete genomic sequences. Multiple sequence alignment data, constructed with MAFFT v6 (Fig. 2) using the complete genomic sequences, confirmed this result and showed that the reference strain GX-YL5 was genetically close to our three isolates. SimPlot analysis also confirmed these results (Fig. 3A). However, topologies from MAFFT v6 and SimPlot analysis showed that our three IBV isolates showed diversity comparing between each other across the genomes. Consequently, 12 gene fragments (Fig. 2A) were manually obtained based on the topologies of SimPlot analysis. Generally, phylogenetic analysis of these fragments confirmed the results from the MAFFT v6 and SimPlot analysis. Phylogenetic trees created from the sequences of fragments 1, 3, 5, 6, 7, 8, 10, 11 and 12 clustered our three isolates into the same genetic sub-groups and were slightly different from the reference strains GX-YL5 and GX-YL9. However, a phylogenetic tree created from fragment 4 clustered our three isolates and reference strains GX-YL5 and GX-YL9 into the same sub-group. Fragment 7, which contained the M gene, showed high genetic diversity among our three isolates and reference strains GX-YL5, GX-YL9, CQ04–1 and YN, although these were different from 4/91 type strains. Interestingly, a sharp change in the topology of the phylogenetic tree constructed using fragment 2 was observed, which clustered isolate γCoV/ck/China/I0114/14 into a group with 4/91 type strains (4/91 and ck/CH/LBJ/140402), suggesting that isolate γCoV/ck/China/I0114/14 might be more likely have undergone two recombination events.
Fig. 3.
Complete genomic analysis of our three IBV isolates and the reference strains. (A) Simplot analysis between our three IBV isolates. The SimPlot was created using a 500 bp window with a 50-bp step with the γCoV/ck/China/I0111/14 isolate as the query strain. Twelve gene fragments were manually obtained based on the topologies of SimPlot analysis. (B) Phylogenetic trees were constructed using the maximum likelihood method with the general time-reversible nucleotide substitution model and bootstrap tests of 1000 replicates in the MEGA 6 program, using the 12 gene fragments (designated as 1–12) based on the above-mentioned results. Three IBV strains isolated in this study and six reference IBV strains were included. The GenBank accession numbers for these genome sequences are as follows: GX-YL9 (HQ850618), GX-YL5 (HQ848267), CQ04-1(HM245924), YN(JF893452), 4/91(KF377577) and ck/CH/LBJ/140402 (KF377577).
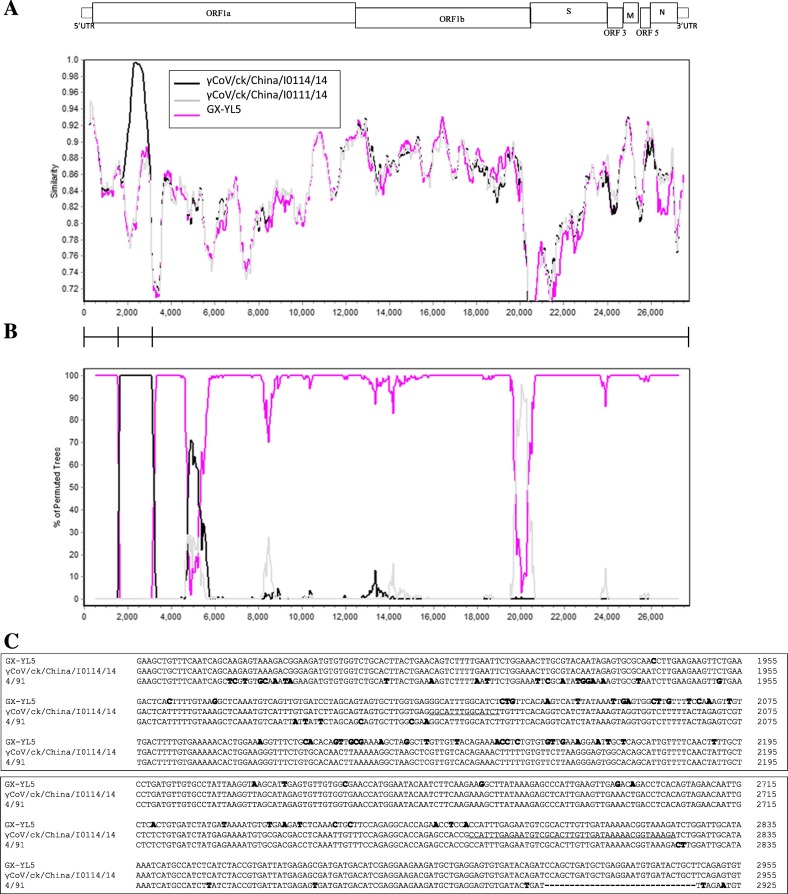
To further examine the relationship of isolate γCoV/ck/China/I0114/14 with the other viruses examined in this study, we conducted RDP analysis, which indicated that the γCoV/ck/China/I0114/14 isolate was a recombinant, with GX-YL5- and 4/91-like viruses as the potential parents (data not shown). Recombination events in γCoV/ck/China/I0114/14 were identified using Simplot, which showed that the γCoV/ck/China/I0114/14 genome is more closely related to the GX-YL5 virus at 5′UTR to the 3′ end of nsp2 in ORF1a, and 5′ end of nsp3 to 3′ UTR. However, the γCoV/ck/China/I0114/14 genome shared a high sequence similarity with the 4/91-like virus at 3′ end of nsp2 to 5′ end of nsp3 in ORF1 (Fig. 4A and B). Pairwise comparisons of the sequences at the 3′ end of nsp2 to the 5′ end of nsp3 from isolate γCoV/ck/China/I0114/14, GX-YL5 and 4/91 strains confirmed this conclusion and revealed two recombination breakpoints located in the nsp2 at nucleotides 2014 to 2027 and nsp3 at 2782 to 2822, respectively (Fig. 4C).
Fig. 4.
SimPlot and sequence analyses to detect recombination and estimate breakpoint within the γCoV/ck/China/I0111/14 genome. (A) Relative gene location and similarity plot. (B) Bootscan analysis showing multiple breakpoints. The 4/91 strain was used as the query sequence. The complete genomic nucleotide sequences used in the analysis were γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and GX-YL5. Both analyses were performed with the F84 distance model, with a window size of 500 bp and step size of 50 bp. (C) Multiple sequence alignment of the predicted breakpoint and flanking sequences among the strains GX-YL5, γCoV/ck/China/I0114/14 and 4/91. Numbers to the right of each alignment show the nucleotide positions in the genome of each virus. Nucleotides differing from those of the γCoV/ck/China/I0114/14 isolate are given in bold.
3.3. Genotype GI-29 represented a unique serotype
To investigate the antigenic serotype of the GI-29 genotype, we performed cross neutralization tests against six major serotypes in China using SPF chicken embryos. Based upon the titers calculated from cross virus-neutralization tests in embryonating eggs, GI-29 was antigenically distinct from not only the vaccine strains but all the circulating serotypes obtained in this study (Table 2 ). This implies that GI-29 was antigenically different from those strains in this study and represents a different serotype.
Table 2.
Results of the virus neutralization tests using γCoV/ck/China/I1011/14 and other IBV strains (serum dilution using a constant amount of virus).
| Virus | Serum |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| 1. γCoV/ck/China/I1011/14 | 7281.4a | 8 | 13 | 32 | 22.6 | 11.3 | 22.6 |
| 2. ck/CH/LGX/111119 | < 2 | 56.9 | – | – | – | – | – |
| 3. ck/CH/LDL/97I | < 2 | – | 90.6 | – | – | – | – |
| 4. 4/91 | 5 | – | – | 786.9 | – | – | – |
| 5. ck/CH/LDL/091022 | 5 | – | – | – | 406.5 | – | – |
| 6. H120 | < 2 | – | – | – | – | 227.5 | – |
| 7. ck/CH/LDL/140520 | 6.4 | – | – | – | – | – | 288 |
–, Not tested.
Reciprocal titer.
3.4. Pathogenicity of GI-29 serotype IBVs
No obvious clinical signs were observed and no birds died during experiments in the control group. However, mild to moderate respiratory signs, such as head shaking and occasionally, open mouth breathing, were observed between 2 and 10 days in chickens challenged with γCoV/ck/China/I0111/14. Of 10 chicks, 4 birds died between 5 and 10 days post-inoculation.
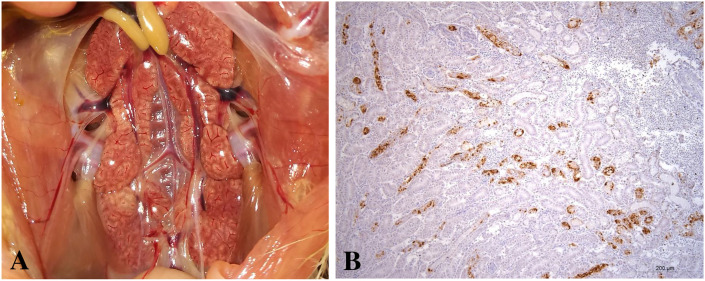
Enlarged kidneys and pale, and urate deposition in the tubules and ureters of the affected kidneys were observed (Fig. 5A), and meanwhile, viral antigens were detected by IHC in the kidneys of dead chickens (Fig. 5B), implicating that the IBV γCoV/ck/China/I0111/14 isolate is a nephropathogenic strain. Hyperemia of the tracheal mucosa was also found in the dead chickens. At 90 days after challenge, no obvious gross lesions were observed in the kidneys of the surviving chickens. Meanwhile, no IHC-positive cells were found in the kidneys of these chickens. In addition, none of the surviving birds exhibited cystic oviducts. IHC-positive cells were observed in the cecal tonsils of dead chickens. No lesions were observed from birds in control group.
Fig. 5.
Renal lesions of the kidney (A) and representation of the type of immunohistochemical (IHC) staining of IBV seen in the kidney of infected chickens (B). Kidneys were swollen, tubules and ureters were distended, and uric acid crystals were present. Obvious IHC IBV positive cells were seen in the kidneys of the chickens following challenge with the IBV isolate γCoV/ck/China/I0111/14.
Some of the birds seroconverted at 8 dpc and all seroconverted at 12 dpc (Table 3 ). No birds showed seroconversion in the control group.
Table 3.
Results of the challenge test using the γCoV/ck/China/I1011/14 strain to infect SPF chickens.
| Group | Morbidity | Mortality | Lesionsa |
IHCb |
Antibody responsec |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nephritis |
Cystic oviducts |
Kidney |
Cystic oviducts |
4 | 8 | 12 | 16 | 20 | 24 | |||||
| Dead | Survived | Survived | Dead | Survived | Survived | |||||||||
| γCoV/ck/China/I1011/14 | 10/10 | 4/10 | 4/4 | 0/6 | 0/5d | 4/4 | 0/6 | 0/5d | 0/10 | 4/7 | 6/6 | 6/6 | 6/6 | 6/6 |
| Negative Control | 0/10 | 0/10 | – | 0/10 | 0/8e | 0/10 | 0/10 | 0/8e | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
Post-mortem examinations were only conducted for chickens that died from a challenge with the γCoV/ck/China/I1011/14 strain, as well as for chickens which survived 90 days post-challenge. All of the dead chickens showed nephritis. The surviving chickens were examined for oviduct abnormalities.
The presence of IBV antigen in the kidneys of dead chickens, and in the kidneys and cystic oviducts of surviving chickens were investigated using IHC.
Antibody responses against IBV were only examined from 4 to 24 days post-challenge.
Five of the 6 surviving chickens were hens in the challenge group.
Eight of the 10 surviving chickens were hens in the negative control group.
4. Discussion
It is generally believed that an S1-based viral classification of IBV might provide data of direct epidemiological relevance and that phylogenetic analysis of the S1 gene is a more appreciable measure of evolutionary history (Valastro et al., 2016). In this study, we isolated three IBVs, γCoV/ck/China/I0111/14, γCoV/ck/China/I0114/14 and γCoV/ck/China/I0118/14, from different regions in Guangxi province, China. As BLAST searches showed that the viruses presented S1 gene amino acid diversity higher than 10% in comparison to other genotypes and variants, we suggest that these isolates are novel IBV variants. It suggested that at least three complete S1 sequences of virus samples collected at least from two different outbreaks should be available for the identification of a novel genotype in the S1-based viral classification system (Valastro et al., 2016). In this study, our three IBV isolates were obtained from different outbreaks. Phylogenetic analysis of their S1 genes showed that they were clustered into a novel lineage in genotype I (GI) and clearly set apart in the phylogenetic tree from the 96 reference strains. Therefore, the three isolates should be classified into a new lineage in genotype I (GI-29) according to the viral classification of IBV suggested by Valastro et al. (2016). This new lineage appears to have the same origin from southern parts of China and is probably related to regional dissemination, although broader epidemiological studies should now be carried out in order to understand the spread of IBV in all Chinese poultry-producing regions.
There was a correlation between the genotype (amino acid sequence identity), serotype and immunotype/protectotype (the level of cross-protection); IBV strains within the same serotype usually share more than 95% amino acid similarity in S1 and belong to the same immunotype (Cavanagh, 2001), whereas different IBV serotypes share less than 85% amino acid similarity (Cavanagh, 2005). However, in some cases, highly similar strains show only limited cross-protection, while a high level of cross-protection may exist for strains with a much lower homology (de Wit et al., 2011a, de Wit et al., 2011b). In this study, the S1 amino acid similarity between GI-29 and the other sequences available in the GenBank database was less than 89% and substitutions were found to be scattered across the S1 gene (Fig. 2), suggesting that the accumulation of mutations in the S1 gene might account for the differing antigenicity of GI-29. Hence, these results indicate that GI-29 comprises not only a novel genotype but a new serotype as well. In addition, these data suggest that GI-29 may be protectotypically different from that of the H120 vaccine strain which might explain why G29-associated outbreaks occurred in H120-vaccination chicken flocks.
Although the GI-29 isolates clustered in the same group based on S1 gene sequences, they also showed different topologies in a multiple sequence alignment constructed with MAFFT v6 and SimPlot analysis using the complete genomic sequences. Based upon these results, 12 gene fragments were obtained and phylogenetically analyzed. Most of the gene fragments of GI-29 correlated with that of the GX-YL5 strain and with each other, although some showed divergence, implicating a common source. It has been speculated that IBV strains substitute large genomic fragments in multiple genes (Lee and Jackwood, 2001). This is probably more likely the case for GI-29 viruses which might originate from a GX-YL5-like virus by accumulation of substitutions in multiple genes which has been evolving and diverging ever since its origin. Alternatively, it is also likely that some gene fragments are exchanged between viruses through recombination. Although we cannot conclude that recombination events have occurred in most parts of the genomes, we can point to the evolutionary trends in the 3′ regions of GI-29 viral genomes as compared to the evolutionary trends in the replicase gene of the reference strains. The trends in the 3′ regions of GI-29 virus genomes, especially in S1 gene, have shown ever increasing diversity, while it appears that the replicase gene is evolving with less variety between GI-29 and reference strains (Fig. 2). This evolutionary trend has also been found in other IBV strains (Mondal and Cardona, 2004). For isolate γCoV/ck/China/I0114/14, we found that recombination events had occurred between nsps 2 and 3 in gene 1 region. The recombination led to the exchange of genetic material between vaccine strain and field isolates, therefore, resulting in the introduction of a 4/91 gene fragment into the virus γCoV/ck/China/I0114/14. A similar phenomenon has also recently been found in Italy and Spain (Moreno et al., 2017).
IBV 793/B-type (4/91) and the Massachusetts-based vaccines are two major antigenic types which have been commonly used in China for a long time. However, the 4/91 vaccine induces only a partial protection against infection of most of the Chinese local IBV strains (Han et al., 2017), such as the GX-YL5-like virus, which allows replication and circulation of both vaccine and field strains in a chicken and produces an environment where co-infection between field (i.e. GX-YL5-like virus) and vaccine (i.e. 793B genotype) strains can enhance the likelihood of recombination. This is likely the case for the emergence of isolate γCoV/ck/China/I0114/14 in this study and also cases found in other countries such as in Italy and Spain (Moreno et al., 2017). In China, only limited numbers of 793/B type viruses have been detected and isolated in chicken flocks (Chen et al., 2015, Han et al., 2017). Furthermore, both the S1 gene and the complete genomic sequence analysis showed that most of the detected 793/B field strains were of the vaccines, or vaccine-derived strains (Han et al., 2017). Meanwhile, recombination events between vaccine strains (i.e. H120 and 4/91), and between field (i.e. GX-YL9-like virus, LX4/QX-like virus) and vaccine (i.e. 4/91) strains have resulted in the emergence of novel serotypes (Han et al., 2017, Liu et al., 2013). Although the spread and effect of these novel strains, such as isolate γCoV/ck/China/I0114/14, on the poultry industry required further evaluation, this study emphasizes the importance of improving vaccination tools in terms of both protection and application (Moreno et al., 2017).
It is believed that all IBV strains initially infect the respiratory tract and causes respiratory signs such as gasping, coughing, sneezing, tracheal rales, and nasal discharge (Cavanagh and Gelb, 2008). Some virus strains may involve nephrosis and nephritis and are referred to as nephropathogenic strains which may not produce significant respiratory lesions or clinical signs. In this study, γCoV/ck/China/I0111/14 did not cause severe respiratory clinical signs; however, its infection produced obvious swollen kidneys with tubules and ureters distended with urates in the dead chickens (Fig. 5A). Furthermore, viral antigens were detected by IHC in the kidneys of all of the dead birds (Fig. 5B), demonstrating that γCoV/ck/China/I0111/14 is a nephropathogenic IBV strain. Infections of layers with many IBV strains in different genotypes/lineages were found to be related to cystic oviducts in layer flocks and involved in egg production problems such as a reduction in egg production and/or a reduction in egg quality. Of the 32 distinct viral lineages classified by Valastro et al. (2016) using all publicly available S1 gene sequences, viruses in 10 lineages were found to be associated with mild to severe reductions in egg production and/or egg quality, and so-called “false layer” syndrome. It was found that no lesions could be observed in the oviducts after infection with the pathogenic 4/91 type (Benyeda et al., 2009, de Wit et al., 2011b), whereas, this genetic type was identified in Canadian outbreaks recently with predominantly respiratory disease and/or egg production problems (Martin et al., 2014), implying that viruses in the same lineage showed different affinity to reproductive tracts and this might be IBV strain specific. In this study, we did not find any obvious lesions which related to cystic oviducts in the five surviving layer chickens challenged with isolate γCoV/ck/China/I0111/14. It is possible that isolate γCoV/ck/China/I0111/14 cannot induce cystic oviducts in layer hens, or alternatively, that the virus can induce cystic oviducts but the small numbers of the chickens experimentally infected in this study were responsible for the differences in virus tropism. However, cystic oviducts were observed in 2 out of the 3, and 4 out of 6 surviving SPF chickens challenged with ck/CH/LGX/111119 (Chen et al., 2017) and ck/CH/LDL/140520 (Gao et al., 2016), respectively, at 1-day-old. This requires further investigation.
The GI-29 type of IBV in this study was isolated from chickens vaccinated twice with H120 vaccine. These viruses are likely able to evade H120 induced immune response based on the results of the current study. However, the use of a combined heterologous vaccination is a common practice and it has been proven to broaden the protection spectrum (Cook et al., 1999). This should be considered for prevention and control of GI-29 type IBVs in China in the future. In addition, it is believed that the wide-spread use of live-attenuated vaccine strains, and the subsequent selective pressure induced by neutralizing antibodies against the spike, may force adaption of the virus to escape immunity, and consequently result in faster evolutionary rates (Jackwood et al., 2012). Hence, active epidemiological surveillance is urgently needed in order to acquire a better understanding of the epidemiology and evolution of GI-29 type viruses, and the updating of vaccines and/or vaccination programs may be required with further experimental studies to control GI-29 type viruses.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgement
This work was supported by grants from the China Agriculture Research System (No. CARS-41-K12), National “Twelfth Five-Year” Plan for Science and Technology Support (2015BAD12B03), and the Provincial Supported Science Foundation of Heilongjiang Province for The National Key Technology R&D Program (GX16B003).
References
- Adzhar A., Gough R.E., Haydon D., Shaw K., Britton P., Cavanagh D. Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian Pathol. 1997;26:625–640. doi: 10.1080/03079459708419239. [DOI] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyeda Z., Mato T., Suveges T., Szabo E., Kardi V., Abonyi-Toth Z., Rusvai M., Palya V. Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol. 2009;38:449–456. doi: 10.1080/03079450903349196. [DOI] [PubMed] [Google Scholar]
- Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 2001;30:109–115. doi: 10.1080/03079450120044506. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34:439–448. doi: 10.1080/03079450500367682. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Gelb J. Infectious bronchitis. In: Saif Y.M., Fadly A.M., Glisson J.R., McDougald L.R., Nolan L.K., Swayne D.E., editors. Diseases of Poultry. 12th edn. Wiley-Blackwell Publishing; Iowa: 2008. pp. 117–135. [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K., Li D., Kant A., Koch G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992;21:33–43. doi: 10.1080/03079459208418816. [DOI] [PubMed] [Google Scholar]
- Chen L., Zhang T., Han Z., Liang S., Xu Y., Xu Q., Chen Y., Zhao Y., Shao Y., Li H., Wang K., Kong X., Liu S. Molecular and antigenic characteristics of Massachusetts genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2015;181:241–251. doi: 10.1016/j.vetmic.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Jiang L., Zhao W., Liu L., Zhao Y., Shao Y., Li H., Han Z., Liu S. Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI-28) in China. Vet. Microbiol. 2017;198:108–115. doi: 10.1016/j.vetmic.2016.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J.K.A., Orbell S.J., Woods M.A., Huggins M.B. Breadth of protection of the respiratory tract provided by different live-attenuated infectioius bronchitis vaccines against challenge with infectious bronchitis viruses of heterologous serotypes. Avian Pathol. 1999;28:477–485. doi: 10.1080/03079459994506. [DOI] [PubMed] [Google Scholar]
- de Wit J.J., Cook J.K., van der Hejden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J., Nieuwenhuisen-van Wilgen J., Hoogkamer A., van de Sande H., Zuidam G.J., Fabri T.H.F. Induction of cystic oviducts and protection against early challenge with infectious bronchitis virus serotype D388 (genotype QX) by maternally derived antibodies and by early vaccination. Avian Pathol. 2011;40:463–471. doi: 10.1080/03079457.2011.599060. [DOI] [PubMed] [Google Scholar]
- Gao M., Wang Q., Zhao W., Chen Y., Zhang T., Han Z., Xu Q., Kong X., Liu S. Serotype, antigenicity, and pathogenicity of a naturally recombinant TW I genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2016;191:1–8. doi: 10.1016/j.vetmic.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Sun C., Yan B., Zhang X., Wang Y., Li C., Zhang Q., Ma Y., Shao Y., Liu Q., Kong X., Liu S. A 15-year analysis of molecular epidemiology of avian infectious bronchitis coronavirus in China. Infect. Genet. Evol. 2011;11:190–200. doi: 10.1016/j.meegid.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhang T., Xu Q., Gao M., Chen Y., Wang Q., Zhao Y., Shao Y., Li H., Kong X., Liu S. Altered pathogenicity of a tl/CH/LDT3/03 genotype infectious bronchitis coronavirus due to natural recombination in the 5′- 17 kb region of the genome. Virus Res. 2013;213:140–148. doi: 10.1016/j.virusres.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhao F., Shao Y., Liu X., Kong X., Song Y., Liu S. Fine level epitope mapping and conservation analysis of two novel linear B-cell epitopes of the avian infectious bronchitis coronavirus nucleocapsid protein. Virus Res. 2016;171:54–64. doi: 10.1016/j.virusres.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Zhao W., Chen Y., Xu Q., Sun J., Zhang T., Zhao Y., Liang S., Gao M., Wang Q., Kong X., Liu S. Genetic, antigenic, and pathogenic characteristics of avian infectious bronchitis viruses genotypically related to 793/B in China. Vet. Microbiol. 2017;203:125–135. doi: 10.1016/j.vetmic.2017.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Casais R., Dove B., Britton P., Cavanagh D. Recombinant infectious bronchitis coronavirus Beaudette with the spike protein gene of the pathogenic M41 strain remains attenuated but induces protective immunity. J. Virol. 2004;78:13804–13811. doi: 10.1128/JVI.78.24.13804-13811.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackwood M.W., Hall D., Handel A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012;12:1305–1311. doi: 10.1016/j.meegid.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.W., Jackwood M.W. Origin and evolution of Georgia 98 (GA98), a new serotype of avian infectious bronchitis virus. Virus Res. 2001;80:33–39. doi: 10.1016/s0168-1702(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Liu S., Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Han Z., Chen J., Liu X., Shao Y., Kong X., Tong G., Rong J. S1 gene sequence heterogeneity of a pathogenic infectious bronchitis virus strain and its embryo-passaged, attenuated derivatives. Avian Pathol. 2007;36:231–234. doi: 10.1080/03079450701338730. [DOI] [PubMed] [Google Scholar]
- Liu X., Ma H., Xu Q., Sun N., Han Z., Sun C., Guo H., Shao Y., Kong X., Liu S. Characterization of a recombinant coronavirus infectious bronchitis virus with distinct S1 subunits of spike and nucleocapsid genes and a 3′ untranslated region. Vet. Microbiol. 2013;162:429–436. doi: 10.1016/j.vetmic.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xu Q., Han Z., Liu X., Li H., Guo H., Sun N., Shao Y., Kong X. Origin and characteristics of the recombinant novel avian infectious bronchitis coronavirus isolate ck/CH/LJL/111054. Infect. Genet. Evol. 2014;23:189–195. doi: 10.1016/j.meegid.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K.S., Bollinger R.C., Paranjape R.S., Gadkari D., Kulkarni S.S., Novak N.G., Ingersoll R., Sheppard H.W., Ray S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Shao Y., Sun C., Han Z., Liu X., Guo H., Liu X., Kong X., Liu S. Genetic diversity of avian infectious bronchitis coronavirus in recent years in China. Avian Dis. 2012;56:15–28. doi: 10.1637/9804-052011-Reg.1. [DOI] [PubMed] [Google Scholar]
- Martin E.A., Brash M.L., Hoyland S.K., Coventry J.M., Sandrock C., Guerin M.T., Ojkic D. Genotyping of infectious bronchitis viruses identified in Canada between 2000 and 2013. Avian Pathol. 2014;43(3):264–268. doi: 10.1080/03079457.2014.916395. [DOI] [PubMed] [Google Scholar]
- McKinley E.T., Jackwood M.W., Hilt D.A., Kissinger J.C., Robertson J.S., Lemke C., Paterson A.H. Attenuated live vaccine usage affects accurate measures of virus diversity and mutation rates in avian coronavirus infectious bronchitis virus. Virus Res. 2011;158:225–234. doi: 10.1016/j.virusres.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S.P., Cardona C.J. Comparison of four regions in the replicase gene of heterologous infectious bronchitis virus strains. Virology. 2004;324:238–248. doi: 10.1016/j.virol.2004.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A., Franzo G., Massi P., Tosi G., Blanco A., Antilles N., Biarnes M., Majó N., Nofrarías M., Dolz R., Lelli D., Sozzi E., Lavazza A., Cecchinato M. A novel variant of the infectious bronchitis virus resulting from recombination events in Italy and Spain. Avian Pathol. 2017;46:28–35. doi: 10.1080/03079457.2016.1200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Sawicki S.G., Sawicki D.L., Siddell S.G. A contemporary view of coronavirus transcription. J. Virol. 2007;81:20–29. doi: 10.1128/JVI.01358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:s2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro V., Holmes E.C., Britton P., Fusaro A., Jackwood M.W., Cattoli G., Monne I. S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect. Genet. Evol. 2016;39:349–364. doi: 10.1016/j.meegid.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe I.N., de Vries R.P., Gröne A., de Haan C.A., Verheije M.H. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J. Virol. 2011;85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Gao M., Xu Q., Xu Y., Zhao Y., Chen Y., Zhang T., Wang Q., Han Z., Li H., Chen L., Liang S., Shao Y., Liu S. Origin and evolution of LX4 genotype infectious bronchitis coronavirus in China. Vet. Microbiol. 2017;198:9–16. doi: 10.1016/j.vetmic.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]