Research highlights
▶ The D–R mathematical model predicts future trends in the spread of human H1N1. ▶ The D–R algorithm may have broader application to other human viral diseases. ▶ D–R modeling will help design surveillance programs for the spread of viral diseases. ▶ The predictive value of the D–R model is superior to the grey prediction model.
Keywords: H1N1, Mathematical model, Grey prediction theory, Infection
Abstract
Incidences of H1N1 viral infections in Mainland China are collected by the Ministry of Health, the People's Republic of China. The number of confirmed cases and the timing of these outbreaks from May 13 to July 22, 2009 were obtained and subjected to a novel mathematical model to simulate the infection profile (time vs number). The model was predicated upon the grey prediction theory which allows assignment of future trends using limited numbers of data points. During the period of our analysis, the number of confirmed H1N1 cases in Mainland China increased from 1 to 1772. The efficiency of our model to simulate these data points was evaluated using Sum of squares of error (SSE), Relative standard error (RSE), Mean absolute deviation (MAD) and Average relative error (ARE). Results from these analyses were compared to similar calculations based upon the grey prediction algorithm. Using our equation, defined herein as equation D–R, results showed that SSE = 6742.00, RSE = 10.69, MAD = 7.07, ARE = 2.47% were all consistent with the D–R algorithm performing well in the estimation of future trends of H1N1 cases in Mainland China. Calculations using the grey theory had no predictive value [ARE for GM(1,1) = −104.63%]. To validate this algorithm, we performed a second analysis using new data obtained from cases reported to the WHO and CDC in the US between April 26 and June 8, 2009. In like manner, the model was equally predictive. The success of the D–R mathematical model suggests that it may have broader application to other viral infections among the human population in China and may be modified for application to other regions of the world.
1. Introduction
Since the World Health Organization (WHO) first identified H1N1 influenza outbreaks in the United States and Mexico in April 2009, the worldwide incidence of this disease has risen dramatically. This infection has had substantial impact on the politics, economies and public health in now endemic regions of the world, so much so that in June 2009, the WHO raised the pandemic alert level to phase 6 (maximum) (http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html). This action prompted many countries to adopt counter-measures such as strengthening prevention and control, perfecting surveillance, and accelerating vaccine development. In Mainland, China, the government and related agencies took rapid and effective measures to curtail the infection. As such, the number of confirmed H1N1 cases in Mainland China increased only moderately. Nevertheless, a better model is needed to predict infection trends of H1N1 and other potential viral infections of humans in order to advance effective prevention and control practices. Precedence exists for such modeling. Several years ago, mathematical models were used to simulate the incidence of Severe Acute Respiratory Syndrome, SARS (Riley et al., 2003, Lipsitch et al., 2003, Dye and Gay, 2003).
Currently there exists established mathematical methods describing the prevalence of diseases over time such as the Time Series Analysis (TSA) which relies on ARMA (Autoregressive Moving Average) and ARIMA (Autoregressive Integrated Moving Average) models extensively (Harris and Sollis, 2003, Pan, 2005, Bell et al., 2004, Campbell et al., 2009). The disadvantage of the TSA algorithm is that the sequence data for TSA must be complete and some unrelated factors need to be excluded. This in turn can result in the lack of qualified data for testing. For this reason, i.e. incomplete data, TSA was considered not be suitable for predicting future trends in H1N1 infections. The grey prediction model overcomes this shortcoming to some extent, because it does not require complete data sequences in order to be effective (Wang et al., 2007, Xiong and Xu, 2005). As such, we used the grey predication model [GM(1,1)] as a basis for constructing the algorithm defined herein. In the case of H1N1, many researchers have analyzed the organism's genetics (McDonald et al., 2007), pathology (Tang and Chong, 2009), evolution and phylogeny (Dunham et al., 2009), mechanism of infection and transmission (Lange et al., 2009), and resistance to drugs (Deyde et al., 2007) to help prevent a pandemic. Here we establish the mathematic model D–R using limited data points, and fit the H1N1 epidemic in Mainland China as a test, then compare this model to the predictive capability of GM(1,1) to show the enhanced predictive value of the new equation. As an additional proof of principle, the validity of the developed model was secondarily demonstrated using data obtained from the US. We trust that such a model will assist in designing effective surveillance strategies for this and other viral diseases in order to determine a trend line for future infections and also determine when and to what extent the infection rate deviates from the norm.
The number of confirmed H1N1 cases in Mainland China is accessible from the official website of the Ministry of Health, the People's Republic of China (http://www.moh.gov.cn/publicfiles//business/htmlfiles/wsb/index.htm). It should be noted that these numbers reflect only confirmed and reported cases. The number of actual cases may be substantially higher. In addition, the number of actual cases and reported cases may differ more in the exponentially increasing period of the epidemic as compared to the start of the epidemic. However, given that these anomalies are partitioned into nearly all public databases of this type and represent an unknown, this factor was not incorporated into the model. The number of the H1N1 cases between May 13 and July 22 was collected and used in this study. During this period, the data on July 8, 10, 12, 14, 16, 18, 19 and 21 were not publicly available. Our model was substantially based upon the grey prediction model then modified using components of the temporal series’ Sliding Average Method (SAM), Weighted Average Method (WAM), ARMA and ARIMA (Harris and Sollis, 2003, Pan, 2005), and statistical regression analysis. The mathematical expression representing increasing tendency of H1N1 cases is given below:
A t−1 + α[K(A t−1 − A t−2) + K(1 − K) (A t−2 − A t−3) + K(1 − K)2(A t−3 − A t−4) +…+K(1−K)(n−1) (A t−n − A t−(n+1))] = F t−β(A t−1 − A t−(n+1))/(n − 1) where A t = the actual value at time t, K = 2/(M + 1) is a fixed value and M = the period of flatness. The following values are also defined: U = γ × F t and L = δ × F t, where F t = predicted value at time t, U = upper limit of the prediction interval, L = lower limit of the prediction interval, and α + β = 1. The values α, β, γ, δ are unfixed values derived from self-adaptation of the model.
The above defined equation for the incidence of H1N1 over time was comprised of two parts; one defines the effect of long-term trends on the data and the other defines the effect of short-term trends on the data. The long-term trend is defined by: (A t−1 − A t−2) + (A t−2 − A t−3) +…+ (A t−n − A t−(n+1))/(n − 1). It describes the average value of the first order difference, and reflects the mean change of the whole series. The short-term trend is K(A t−1 − A t−2) + K(1 − K) (A t−2 − A t−3) + K(1 − K)2(A t−3 − A t−4) +…+ K(1 − K)(n−1) (A t−n − A t−(n−1)) and describes any impact of recent first order differences on the data. The weights given to short-term and long-term trends are designed as α and β, respectively and are derived from self-adaptation of the model. The foundations for the self-adapting model are as follows: (1) if the degree of first order difference and basic data is less than the predefined limit, the values for α and β should decrease and increase, respectively; (2) if the degree of first order difference increases significantly, and the basic data remains less than the predefined limit, the values of α and β will be similar; (3) if the degree of first order difference and basic data increase significantly, the values for α and β will increase and decrease, respectively. Simply put, inasmuch as α is the weight given to the short-term trend and β is the weight assigned to the long-term trend, if the change in short-term data is significant, the long term trend will become less significant requiring an increase in α. If the change in the data support neither long or short term trends, the values of α and β will be congruent. Generally, if the change in the short-term series of numbers is not significant, the long-term trend will have a greater impact on the outcome, thereby increasing the value of β. The mathematical model requires simple procedures to input the original data. The values generated will vary according to changes in the original data and first order differences; however, the process is self-adapting. The γ and δ values are set using the same principles as above.
Information regarding confirmed H1N1 cases during May 17 to July 22, in Mainland China is provided in Table 1 . The fitted curve was derived using this data even though data was collected beginning May 13. This is because the period May 13–16 was used as the minimum dataset upon which to begin building the simulation model which was initiated on May 17. As such it was not included in the table. In this same table, the fitted values of our mathematical model (D–R) and its upper (U) and lower (L) limits representing the trend of H1N1 cases are also summarized along with those calculated using GM(1,1). The degree of accuracy correlating the fitted model to the increasing tendency of H1N1 cases was evaluated from these data using the Sum of squares of error (SSE = Σ(A t − F t)2), Relative standard error (RSE = (SSE/(n − 1))1/2), Mean absolute deviation (MAD = Σ|(A t − F t)|/n) and Average relative error (ARE = (Σ100% × (A t − F t)/A t)/n). Calculation of the fitted curve for equation D–R showed that SSE = 6742.00 relative to that using GM(1,1) where SSE = 1.27 × 108. In our study, RSE is the average value of the quadratic sum of the difference between the actual values and the fitted values. Although the difference between the actual curve and fitted curve shows a gradual increase, the RSE value is only 10.69, indicating that the fitted model is very accurate and far more predictive than that calculated using GM(1,1) where the RSE = 1482.04. The average total absolute value between actual and fitted data is described by MAD. Using equation D–R, we calculated MAD to be 7.07 vs. 901.72 for GM(1,1). In general, MAD and RSE reflect the mean differences between actual and fitted curves. Using large datasets, these differences become insignificant suggesting that the predictive value of the D–R mathematical model is very high. The percent difference between the actual and fitted curves in actual value is defined by ARE. The 2.47% ARE using the D–R equation suggests that the accuracy of the fitted model is approximately 97%; the number (−104.63%) obtained for GM(1,1) is uninformative.
Table 1.
Comparison of actual H1N1 infection values to the values predicted by D–R and GM(1,1).
| Date | At | D–R | U | L | GM(1,1) |
|---|---|---|---|---|---|
| May 17 | 3.00 | 2.28 | 2.16 | 2.39 | 2.79 |
| May 18 | 3.00 | 3.62 | 3.44 | 3.80 | 3.94 |
| May 19 | 4.00 | 3.31 | 3.15 | 3.48 | 4.02 |
| May 20 | 4.00 | 4.62 | 4.39 | 4.85 | 4.95 |
| May 21 | 5.00 | 4.33 | 4.11 | 4.54 | 5.22 |
| May 22 | 5.00 | 5.62 | 5.34 | 5.90 | 6.06 |
| May 23 | 7.00 | 5.33 | 5.07 | 5.60 | 6.41 |
| May 24 | 7.00 | 8.00 | 7.60 | 8.40 | 7.90 |
| May 25 | 11.00 | 7.49 | 7.12 | 7.87 | 8.72 |
| May 26 | 12.00 | 12.82 | 12.18 | 13.47 | 11.43 |
| May 27 | 12.00 | 13.27 | 12.61 | 13.94 | 13.71 |
| May 28 | 13.00 | 12.63 | 11.99 | 13.26 | 15.58 |
| May 29 | 21.00 | 13.76 | 13.08 | 14.45 | 17.32 |
| May 30 | 21.00 | 24.83 | 23.59 | 26.08 | 21.33 |
| May 31 | 26.00 | 22.58 | 21.45 | 23.71 | 25.21 |
| June 1 | 36.00 | 28.91 | 27.47 | 30.36 | 29.70 |
| June 2 | 42.00 | 41.40 | 39.33 | 43.47 | 31.84 |
| June 3 | 51.00 | 46.78 | 44.44 | 49.11 | 34.59 |
| June 4 | 59.00 | 56.80 | 53.96 | 59.64 | 37.21 |
| June 5 | 67.00 | 64.72 | 61.49 | 67.96 | 48.23 |
| June 6 | 70.00 | 72.81 | 69.17 | 76.46 | 70.59 |
| June 7 | 73.00 | 73.77 | 70.08 | 77.46 | 108.57 |
| June 8 | 89.00 | 76.11 | 72.30 | 79.91 | 142.29 |
| June 9 | 100.00 | 99.74 | 94.75 | 104.73 | 168.53 |
| June 10 | 111.00 | 110.43 | 104.91 | 115.96 | 197.29 |
| June 11 | 125.00 | 121.33 | 115.26 | 127.39 | 228.68 |
| June 12 | 141.00 | 137.12 | 130.26 | 143.97 | 262.95 |
| June 13 | 165.00 | 154.84 | 147.10 | 162.59 | 301.10 |
| June 14 | 185.00 | 184.31 | 175.09 | 193.52 | 344.94 |
| June 15 | 226.00 | 203.75 | 193.57 | 213.94 | 395.60 |
| June 16 | 237.00 | 257.26 | 244.40 | 270.12 | 448.46 |
| June 17 | 264.00 | 254.40 | 241.68 | 267.12 | 523.44 |
| June 18 | 297.00 | 286.40 | 272.08 | 300.72 | 608.94 |
| June 19 | 328.00 | 324.76 | 308.53 | 341.00 | 704.10 |
| June 20 | 356.00 | 356.26 | 338.45 | 374.07 | 813.72 |
| June 21 | 414.00 | 382.90 | 363.76 | 402.05 | 938.82 |
| June 22 | 441.00 | 458.48 | 435.56 | 481.41 | 1068.45 |
| June 23 | 490.00 | 472.72 | 449.08 | 496.35 | 1225.40 |
| June 24 | 528.00 | 530.80 | 509.57 | 546.73 | 1395.29 |
| June 25 | 570.00 | 565.32 | 542.70 | 582.28 | 1584.07 |
| June 26 | 618.00 | 608.43 | 584.09 | 626.68 | 1784.71 |
| June 27 | 678.00 | 660.72 | 634.29 | 680.54 | 1994.00 |
| June 28 | 729.00 | 729.33 | 700.15 | 751.21 | 2215.10 |
| June 29 | 766.00 | 777.97 | 746.86 | 801.31 | 2447.58 |
| June 30 | 810.00 | 805.82 | 773.59 | 829.99 | 2679.28 |
| July 1 | 866.00 | 851.00 | 816.96 | 876.53 | 2902.70 |
| July 2 | 915.00 | 914.64 | 878.06 | 942.08 | 3122.86 |
| July 3 | 960.00 | 962.13 | 923.65 | 991.00 | 3334.16 |
| July 4 | 1002.00 | 1004.40 | 964.22 | 1034.53 | 3530.51 |
| July 5 | 1040.00 | 1043.71 | 1022.84 | 1064.59 | 3706.87 |
| July 6 | 1097.00 | 1078.38 | 1056.81 | 1099.95 | 3858.30 |
| July 7 | 1151.00 | 1138.55 | 1115.78 | 1161.32 | 4005.98 |
| July 8 | – | 1194.20 | 1170.31 | 1218.08 | – |
| July 9 | 1223.00 | 1234.74 | 1210.05 | 1259.44 | 4266.10 |
| July 10 | – | 1258.07 | 1232.91 | 1283.23 | – |
| July 11 | 1302.00 | 1293.50 | 1267.63 | 1319.37 | 4442.10 |
| July 12 | – | 1340.92 | 1314.10 | 1367.74 | – |
| July 13 | 1354.00 | 1379.65 | 1338.26 | 1407.24 | 4563.47 |
| July 14 | – | 1383.95 | 1342.43 | 1411.63 | – |
| July 15 | 1444.00 | 1413.19 | 1370.79 | 1441.45 | 4609.30 |
| July 16 | – | 1490.89 | 1446.16 | 1520.71 | – |
| July 17 | 1537.00 | 1536.77 | 1490.67 | 1567.50 | 4664.02 |
| July 18 | – | 1582.27 | 1534.80 | 1613.92 | – |
| July 19 | – | 1626.99 | 1578.18 | 1659.53 | – |
| July 20 | 1668.00 | 1671.06 | 1620.93 | 1704.48 | 4760.80 |
| July 21 | – | 1709.99 | 1658.69 | 1744.18 | – |
| July 22 | 1772.00 | 1751.85 | 1699.29 | 1786.89 | 4834.59 |
This table presents the actual values of H1N1 infections at time t (At) that were used to calculate predicted values using equation D–R, the upper (U) and lower (L) limits, and predicted values using GM(1,1) for the period May 17 to July22. No data was available for the time periods defined by “–”.
As shown in Table 1 and pictorially demonstrated in Fig. 1 , to better display the trend lines and compared datasets, the confirmed H1N1 cases during May 17 to July 22 and those predicted by the model are very similar, with the actual curve appearing higher than the upper limit of the fitted curve at some points. After self-adapting, variation in the fitted curve realigns with the limit values. Self-adaptation is one of the key features of the D–R mathematical model where implementation of the upper and lower limits occurs when the actual values approach or exceed these limits. When the upper limit is exceeded, the infection trend is greater than the norm implying the epidemic is gaining strength. If the actual curve and fitted curve are similar, it suggests there are no significant changes in the epidemic situation. If however, the actual curve approaches the lower limit, it suggests the epidemic is abating. Our results show that the fitted curve derived from the model closely mirrors the actual number of H1N1 cases, indicating the utility of the model. Upon comparing the D–R and GM(1,1) algorithms the data generated using GM(1,1) are initially in line with those using D–R; however, as the dataset increases, substantial deviation occurs in the capabilities of GM(1,1) to mimic the known progression of disease. This loss of predictive value is mirrored in the calculated values for SSE, RSE, MAD and ARE.
Fig. 1.
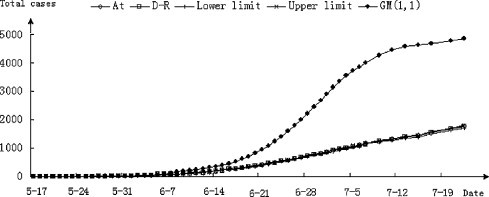
H1N1 infection curves from actual data, and simulated by the D–R and GM(1,1) algorithms. The actual values of H1N1 infections at time t (At) (♢), the predicted values using equation D–R (□), the upper (U) (||) and lower (L) (×) limits, and the values calculated using GM(1,1) (♦) for the period May 17 to July22 were used to create the simulated curves.
To further validate the fitting accuracy and utility of the D–R model, the daily numbers of reported H1N1 cases in the US during the period April 26 to June 8, 2009 were collected from WHO and CDC. The fitted curve was generated using data obtained between April 30 and June 8; the period April 26–29 was used as the minimum dataset upon which to begin building the simulation model and is therefore not presented in Fig. 2 . The initial data were small i.e. on April 26, there were only 20 reported cases; however, by June 8, there were 13,217 reported cases. Beginning June 8, data were provided weekly rather than daily and therefore not used in our analysis. As with the data from Mainland China, fitting was performed by means of the D–R and GM(1,1) algorithms. Calculated values for SSE, RES, MAD and ARE from both the D–R and GM(1,1) models are presented in Table 2 (note; tabulated data points from the WHO and CDC and calculated values for the period April 26 to June 8 used to construct Fig. 2 are available upon request). Values calculated from the D–R model were significantly lower than those from GM(1,1) model. For example, the D–R model ARE value is 2.93% which means that the fitting accuracy reached 97.07% (high precision). In contrast, the accuracy of GM(1,1) was only 39.69%, indicating substantial deviation in the predictive value. As with the data from Mainland China, Fig. 2 demonstrates that the actual curve and D–R fitted curve exhibited similar trends and even overlapped at some time points. In contrast, GM(1,1) model was reasonably predictive during the initial phase of the reporting period similar to that observed with the data from Mainland China; however, beginning May 12, the GM(1,1) curve deviated significantly from the actual curve.
Fig. 2.

H1N1 infection curves from actual data collected from the US, and simulated by the D–R and GM(1,1) algorithms. The actual values of H1N1 infections at time t (At) (♢), the predicted values using equation D–R (□), the upper (U) (||) and lower (L) (×) limits, and the values calculated using GM(1,1) (♦) for the period April 30 to June 8 were used to create the simulated curves.
Table 2.
SSE, RES, MAD and ARE values calculated with D–R and GM(1,1) models using H1N1 data reported from the US.
| SSE | RSE | MAD | ARE | |
|---|---|---|---|---|
| D–R | 7280042.99 | 484.60 | 338.51 | 2.93% |
| GM(1,1) | 923507639.18 | 5458.07 | 4171.35 | −61.31% |
Based on the H1N1 case number reported by the WHO and CDC in the US during the period April 30 to June 8, the SSE, RES, MAD and ARE values were calculated for the D–R and GM(1,1) models.
Others have modeled H1N1 virus transmission under specialized circumstances and in subpopulations of individuals. Fraser et al. (2009) were among the first to present a model of H1N1 transmission based upon data obtained from the outbreak in Mexico. They concluded that transmissibility is substantially higher than that of the seasonal flu, but comparable to previous influenza pandemics with respect to low Basic Reproduction Numbers (R0). Gojovic et al. (2009) generated a simulation model based upon combinatorial uncertainty analysis to project the effects of several strategies to mitigate transmission among Koreans and concluded that if available, massive vaccination would be optimal. Tracht et al. (2010) developed a transmission model predicated on a subpopulation (10%) of individuals that would be willing to correctly use facemasks and concluded a substantial reduction (20%) would ensue. Yet, among these and other extensive studies modeling the effects of mitigation, Coburn et al. (2009) concluded that trying to identify intervention strategies for epidemics that involve recombination of species-specific strains and cross-species transmission, i.e. H1N1 is problematic. The work herein used a subset of data to project future trends and then tested those trend lines against existing data and demonstrated good congruence. This model gives less consideration to micro-environmental factors by establishing upper and lower limits of the prediction intervals and providing self-adapting parameters to account for dominating long or short term effects. As with most models, assimilating trends to regional variation, health care resources and the public health measures to mitigate impact were not evaluated; however, our model is capable of assessing the benefits of intervention strategies.
Taken together, our novel algorithm aligns well with trends observed in the report of H1N1 cases in Mainland China and in the US. This model may not only be used to broadly predict trends in H1N1 cases, but also may be applicable for predicting other epidemics. Although the progression of epidemic diseases is often based upon several hypotheses (Gordis, 2008), for the first time we show that the infection rate of influenza H1N1 can be predicted by a mathematical model that depicts a relationship between the tendency of the data to change, and the fitted and limit values of that data.
Acknowledgements
This work was supported, in part, by the National 973 Plan of China (2010CB534002). We also acknowledge National Natural Science Foundation of China (30700590 and 30972195), Funding supported by Program for New Century Excellent Talents in Heilongjiang Provincial University (1155-NCET-005) and Heilongjiang Provincial Science and Technology Department, China (ZJN0702-01).
References
- Bell M.L., McDermott A., Zeger S.L., Samet J.M., Dominici F. Ozone and short-term mortality in 95 US Urban Communities, 1987–2000. JAMA. 2004;292:2372–2378. doi: 10.1001/jama.292.19.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S.M., Reeves D., Kontopantelis E., Sibbald B., Roland M. Effects of pay for performance on the quality of primary care in England. N. Engl. J. Med. 2009;361:368–378. doi: 10.1056/NEJMsa0807651. [DOI] [PubMed] [Google Scholar]
- Coburn B.J., Wagner B.G., Blower S. Modeling influenza epidemics and pandemics: insights into the future of swine flu (H1N1) BMC Med. 2009;7:30. doi: 10.1186/1741-7015-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyde V.M., Xu X., Bright R.A., Shaw M., Smith C.B., Zhang Y., Shu Y., Gubareva L.V., Cox N.J., Klimov A.I. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 2007;196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- Dunham E.J., Dugan V.G., Kaser E.K., Sarah E., Perkins S.E., Brown L.H., Holmes E.C., Taubenberger J.K. Different evolutionary trajectories of European Avian-Like and Classical Swine H1N1 Influenza A Viruses. J. Virol. 2009;83:5485–5494. doi: 10.1128/JVI.02565-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C., Gay N. Modeling the SARS epidemic. Science. 2003;300:1884–1885. doi: 10.1126/science.1086925. [DOI] [PubMed] [Google Scholar]
- Fraser C., Donnelly C.A., Cauchemez S. Pandemic potential of a strain of influenza A (H1N1): early findings. Science. 2009;324:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojovic M.Z., Sander B., Fisman D., Krahn M.D., Bauch C.T. Modelling mitigation strategies for pandemic (H1N1) CMAJ. 2009;181:673–680. doi: 10.1503/cmaj.091641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordis, L., 2008. Epidemiology: with STUDENT CONSULT Online Access, 4e. Saunders. ISBN: 1416040021/9781416040026.
- Harris R., Sollis R. Jonh Wiley & Sons Ltd; 2003. Applied Time Series Modeling and Forecasting. [Google Scholar]
- Lange E., Kalthoff D., Blohm U., Teifke J.P., Breithaupt A., Maresch C., Starick E., Fereidouni S., Hoffmann B., Mettenleiter T.C., Beer M., Vahlenkamp T.W. Pathogenesis and transmission of the novel swine-origin influenza virus A/H1N1 after experimental infection of pigs. J. Gen. Virol. 2009;90(September(Pt 9)):2119–2123. doi: 10.1099/vir.0.014480-0. Epub2009 Jul 10. [DOI] [PubMed] [Google Scholar]
- Lipsitch M., Cohen T., Cooper B. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald N.J., Smith C.B., Cox N.J. Antigenic drift in the evolution of H1N1 influenza A viruses resulting from deletion of a single amino acid in the haemagglutinin gene. J. Gen. Virol. 2007;88:3209–3213. doi: 10.1099/vir.0.83184-0. [DOI] [PubMed] [Google Scholar]
- Riley S., Fraser C., Donnelly C.A. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science. 2003;300:1961–1966. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- Tang X., Chong K.T. Histopathology and growth kinetics of influenza viruses (H1N1 and H3N2) in the upper and lower airways of guinea pigs H1N1. J. Gen. Virol. 2009;90:386–391. doi: 10.1099/vir.0.007054-0. [DOI] [PubMed] [Google Scholar]
- Tracht S.M., Del Valle S.Y., Hyman J.M. Mathematical modeling of the effectiveness of facemasks in reducing the spread of novel influenza A (H1N1) PLoS One. 2010;5(February (2)):e9018. doi: 10.1371/journal.pone.0009018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.L., Xie M.P., Wang D.L. Application of gray dynamic forecasting model research in urban water use. Res. Soil Water Conserv. 2007;14:430. [Google Scholar]
- World now at the start of 2009 influenza pandemic. Statement to the press by WHO Director-General Dr Margaret Chan. Geneva: World Health Organization; 11 June 2009. Available from: http://www.who.int/mediacentre/news/statements/2009/h1n1_pandemic_phase6_20090611/en/index.html.
- Xiong H.J., Xu H.Z. National Defense Industry Press; 2005. Grey control. ISBN: 7118041440 (in Chinese) [Google Scholar]
- Pan H. University of International Business and Economics Press; Beijing, China: 2005. Time Series Analysis. ISBN:7-81078-567-2 (in Chinese) [Google Scholar]


