Abstract
COVID-19 is a viral respiratory illness caused by a new coronavirus called SARS-CoV-2. The World Health Organization declared the SARS-CoV-2 outbreak a global public health emergency. We performed genetic analyses of eighty-six complete or near-complete genomes of SARS-CoV-2 and revealed many mutations and deletions on coding and non-coding regions. These observations provided evidence of the genetic diversity and rapid evolution of this novel coronavirus.
Keywords: Coronavirus, SARS-CoV-2, Mutations, Genomic diversity
Highlights
-
•
A novel coronavirus is officially known as SARS-CoV-2.
-
•
Eighty-six complete or near-complete genomes of SARS-CoV-2 were analyzed.
-
•
Many mutations and deletions on coding and non-coding regions of SARS-CoV-2 were found.
1. The study
A new coronavirus SARS-CoV-2 is spreading cross the world (Phan, 2020). Since the virus emerged at the seafood wholesale market at the end of last year (Zhu et al., 2019), the number of infected cases has been rising dramatically (Velavan and Meyer, 2020). Human-to-human transmission of SARS-CoV-2 has been confirmed (Nishiura et al., 2020). The virus has been detected in bronchoalveolar-lavage (Zhu et al., 2019), sputum (Lin et al., 2020), saliva (K.K. To et al., 2020), throat (Bastola et al., 2020) and nasopharyngeal swabs (To et al., 2020).
Nucleotide substitution has been proposed to be one of the most important mechanisms of viral evolution in nature (Lauring and Andino, 2010). The rapid spread of SARS-CoV-2 raises intriguing questions such as whether its evolution is driven by mutations. To assess the genetic variation, eighty-six complete or near-complete genomes of SARS-CoV-2 were collected from GISAID [https://www.gisaid.org/]. These SARS-CoV-2 strains were detected in infected patients from China (50), USA (11), Australia (5), Japan (5), France (4), Singapore (3), England (2), Taiwan (2), South Korea (1), Belgium (1), Germany (1), and Vietnam (1). The pair-wise nucleotide sequence alignment was performed by ClustalX2 (Saitou and Nei, 1987), and the sequence of the strain China/WHU01/2020/EPI_ISL_406716 was used as a reference genome.
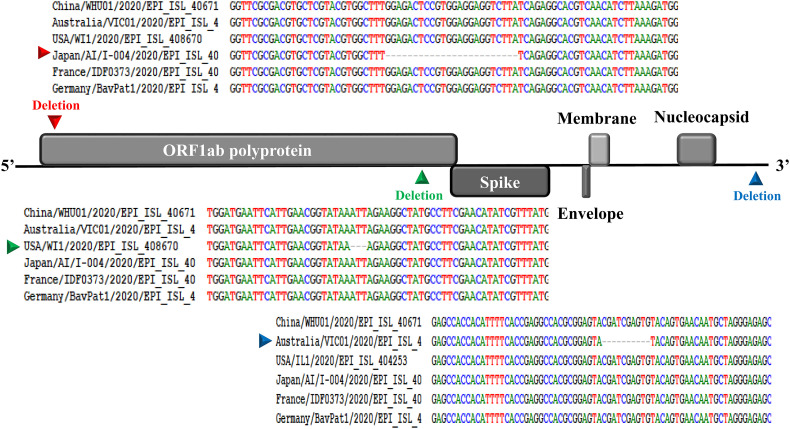
Like other betacoronaviruses, the genome of SARS-CoV-2 has a long ORF1ab polyprotein at the 5′ end, followed by four major structural proteins, including the spike surface glycoprotein, small envelope protein, matrix protein, and nucleocapsid protein (Phan, 2020). Our genetic analysis discovered three deletions in the genomes of SARS-CoV-2 from Japan (Aichi), USA (Wisconsin), and Australia (Victoria) as shown in Fig. 1 . Two deletions (three nucleotides and twenty-four nucleotides) were in the ORF1ab polyprotein, and one deletion (ten nucleotides) was in the 3′ end of the genome.
Fig. 1.
Genomic organization of SARS-CoV-2 and pairwise nucleotide sequence alignment showing deletions in the ORF1ab polyprotein and in the 3′ end of the genome.
It is interesting that our nucleotide sequence alignment also revealed ninety-three mutations over the entire genomes of SARS-CoV-2 (Table 1 ). Forty-two missense mutations were identified in all the major non-structural and structural proteins, except the envelope protein. Twenty-nine missense mutations were in the ORF1ab polyprotein, eight in the spike surface glycoprotein, one in the matrix protein, and four in the nucleocapsid protein. Of note, three mutations (D354, Y364, and F367) located in the spike surface glycoprotein receptor-binding domain. The spike surface glycoprotein plays an essential role in binding to receptors on the host cell and determines host tropism (Fung and Liu, 2019). It is also the major target of neutralizing antibodies (Yu et al., 2020). Mutations in the spike surface glycoprotein might induce its conformational changes, which probably led to the changing antigenicity. To date, a study on localization of amino acids involved in conformational changes of the SARS-CoV-2 spike surface glycoprotein structure is not available. The identification of these amino acids is of significance and should be investigated by further studies.
Table 1.
Mutations found in the entire genome of SARS-CoV-2 strains. The number in the parentheses indicated the location of amino acid in its protein.
| Genomic region | No. nt mutations | Missense mutation | SARV-CoV-2 strain |
|---|---|---|---|
| 5′ UTR | 8 | N/A | |
| ORF1ab polyprotein | 48 | 29 | |
| A (117) → T | USA/CA3/2020/EPI_ISL_408008 USA/CA4/2020/EPI_ISL_408009 |
||
| P (309) → S | France/IDF0515/2020/EPI ISL_408430 | ||
| S (428) → N | USA/CA1/2020/EPI_ISL_406034 | ||
| T (609) → I | USA/CA5/2020/EPI_ISL_408010 | ||
| A (1176) → V | Japan/TY-WK-012/2020/EPI_ISL_408665 | ||
| L (1599) → F | Korea/KCDC03/2020/EPI_ISL_407193 | ||
| I (1607) → V | USA/CA3/2020/EPI_ISL_408008 USA/CA4/2020/EPI_ISL_408009 |
||
| M (2194) → T | Shenzhen/SZTH-004/2020/EPI_ISL_406595 | ||
| L (2235) → I | Wuhan/WH01/2019/EPI_ISL_406798 | ||
| I (2244) → T | Wuhan/IPBCAMS-WH-03/2019/EPI_ISL_403930 | ||
| G (2251) → S | Wuhan/WIV05/2019/EPI_ISL_402128 | ||
| A (2345) → V | Shandong/IVDC-SD-001/2020/EPI_ISL_408482 | ||
| G (2534) → V | Wuhan/IPBCAMS-WH-05/2020/EPI_ISL_403928 | ||
| D (2579) → A | Wuhan/WIV07/2019/EPI_ISL_402130 | ||
| N (2708) → S | Wuhan/IPBCAMS-WH-01/2019/EPI_ISL_402123 | ||
| F (2908) → I | Wuhan/IPBCAMS-WH-01/2019/EPI_ISL_402123 | ||
| T (3058) → I | France/IDF0515/2020/EPI_ISL_408430 | ||
| S (3099) → L | Shenzhen/HKU-SZ-005/2020/EPI_ISL_405839 | ||
| L (3606) → F | Yunnan/IVDC-YN-003/2020/EPI_ISL_408480 Shandong/IVDC-SD-001/2020/EPI_ISL_408482 Chongqing/IVDC-CQ-001/2020/EPI_ISL_408481 Singapore/3/2020/EPI_ISL_407988 France/IDF0515/2020/EPI_ISL_408430 USA/AZ1/2020/EPI_ISL_406223 |
||
| E (3764) → D | Japan/KY-V-029/2020/EPI_ISL_408669 | ||
| N (3833) → K | Wuhan/WH01/2019/EPI_ISL_406798 | ||
| W (5308) → C | Taiwan/2/2020/EPI_ISL_406031 | ||
| T (5579) → I | USA/CA2/2020/EPI_ISL_406036 | ||
| I (6075) → T | England/02/2020/EPI_ISL_407073 England/01/2020/EPI_ISL_407071 |
||
| P (6083) → L | Japan/AI/I-004/2020/EPI_ISL_407084 | ||
| F (6309) → Y | Sichuan/IVDC-SC-001/2020/EPI_ISL_408484 | ||
| E (6565) → D | Shenzhen/SZTH-004/2020/EPI_ISL_406595 | ||
| K (6958) → R | Wuhan/WIV05/2019/EPI_ISL_402128 | ||
| D (7018) → N | Wuhan/WIV02/2019/EPI_ISL_402127 | ||
| Spike polyprotein | 14 | 8 | |
| F (32) → I | Wuhan/HBCDC-HB-01/2019/EPI_ISL_402132 | ||
| H (49) → Y | Guangdong/20SF174/2020/EPI_ISL_406531 Guangdong/20SF040/2020/EPI_ISL_403937 Guangdong/20SF028/2020/EPI_ISL_403936 |
||
| S (247) → R | Australia/VIC01/2020/EPI_ISL_406844 | ||
| N (354) → D | Shenzhen/SZTH-004/2020/EPI_ISL_406595 | ||
| D (364) → Y | Shenzhen/SZTH-004/2020/EPI_ISL_406595 | ||
| V (367) → F | France/IDF0372/2020/EPI_ISL_406596 France/IDF0373/2020/EPI_ISL_406597 |
||
| D (614) → G | Germany/BavPat1/2020/EPI_ISL_406862 | ||
| P (1143) → L | Australia/QLD02/2020/EPI_ISL_407896 | ||
| Intergenic region | 5 | N/A | |
| Envelope protein | 0 | 0 | |
| Matrix protein | 2 | 1 | |
| D (209) → H | Singapore/2/2020/EPI_ISL_407987 | ||
| Intergenic region | 6 | N/A | |
| Nucleocapsid protein | 7 | 4 | |
| T (148) → I | Shenzhen/SZTH-004/2020/EPI_ISL_406595 | ||
| S (194) → L | Shenzhen/SZTH-003/2020/EPI_ISL_406594 Foshan/20SF207/2020/EPI_ISL_406534 USA/CA3/2020/EPI_ISL_408008 USA/CA4/2020/EPI_ISL_408009 |
||
| S (202) → N | Australia/QLD02/2020/EPI_ISL_407896 | ||
| P (344) → S | Guangzhou/20SF206/2020/EPI_ISL_406533 | ||
| 3′UTR | 3 | N/A | |
| Complete genome | 93 | 42 |
Acknowledgments
Acknowledgement
We acknowledge support from Division of Clinical Microbiology, University of Pittsburgh Medical Center.
Declaration of Competing Interest
The author declares no competing financial interests.
References
- Bastola A., Sah R., Rodriguez-Morales A.J., Lal B.K., Jha R., Ojha H.C., Shrestha B., Chu D.K.W., Poon L.L.M., Costello A., Morita K., Pandey B.D. The first 2019 novel coronavirus case in Nepal. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30067-0. (pii: S1473-3099(20)300670), (in press), [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- K.K. To, Tsang O.T., Chik-Yan C.Y., Chan K.H., Wu T.C., Chan J.M.C., Leung W.S., Chik T.S., Choi C.Y., Kandamby D.H., Lung D.C., Tam A.R., Poon R.W., Fung A.Y., Hung I.F., Cheng V.C., Chan J.F., Yuen K.Y. Consistent detection of 2019 novel coronavirus in saliva. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa149. (pii: ciaa149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring A.S., Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Gong Z., Xiao Z., Xiong J., Fan B., Liu J. Novel coronavirus pneumonia outbreak in 2019: Computed tomographic findings in two cases. Korean J. Radiol. 2020 doi: 10.3348/kjr.2020.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura H., Linton N.M., Akhmetzhanov A.R. Initial cluster of novel coronavirus (2019-nCoV) infections in Wuhan, China is consistent with substantial human-to-human transmission. J. Clin. Med. 2020;9 doi: 10.3390/jcm9020488. pii: E488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect. Genet. Evol. 2020;79:104211. doi: 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Velavan T.P., Meyer C.G. The Covid-19 epidemic. Tropical Med. Int. Health. 2020 doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Du L., Ojcius D.M., Pan C., Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020 doi: 10.1016/j.micinf.2020.01.003. pii: S1286-4579(20)300253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., Investigating China Novel Coronavirus. Research Team. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019:2020. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]