Abstract
The ability to use double-stranded RNA to inhibit gene expression sequence-specifically (RNA interference, or RNAi) is currently revolutionizing science and medicine alike. Numerous pre-clinical studies are evaluating RNAi as a novel therapeutic modality in the battle against gain-of-function autosomal dominant diseases, cancer, and viral infections. One emerging concern is that RNAi mono-therapies might ultimately fail to control viruses that can escape silencing by mutation and/or RNAi suppression. Thus, sophisticated strategies are being developed that aim to avert viral resistance by combining RNAi effectors with each other or with further gene expression inhibitors. Several reports already validate this new concept of “combinatorial RNAi” (coRNAi) and illustrate its versatility by describing co-expression of RNAi triggers directed against single or multiple, viral or cellular, targets. Other studies document the successful delivery of these triggers with additional RNA- or protein-based silencers. Moreover, vectors have been engineered to blend RNAi-mediated gene inhibition with conventional gene replacement strategies. Collectively, these efforts open up exciting new therapeutic avenues but could also augment the inherent risks of RNAi technology, including immune responses, off-targeting, and oversaturation of endogenous pathways. Here, we critically review all coRNAi strategies and discuss the requirements for their transition into clinical application.
Introduction
Viral infection remains a critical challenge for modern medicine and continues to pose a complex and global health problem. For instance, more than 500 million people worldwide carry at least one type of hepatitis virus (B or C), and many will develop clinically significant hepatic disease.1 Up to 25% of chronic carriers of hepatitis B virus (HBV) are at high risk of eventually dying from infection-related sequelae, such as end-stage cirrhosis and hepatocellular carcinoma, and an even higher percentage of patients chronically infected with hepatitis C virus (HCV) have an equally somber prognosis. Moreover, approximately 39 million people worldwide were living with human immunodeficiency virus (HIV) in 2005, with approximately 4 million new infections and 3 million deaths that year.2 With a case fatality rate of almost 100%, the HIV/acquired immunodeficiency syndrome epidemic imposes one of the most serious burdens of human mortality. Global pandemics caused by newly emerging viral infections, such as Ebola, severe acute respiratory syndrome coronavirus, and avian influenza (H5N1), present further threats to human health.
The reasons for the persistence of human viruses and the emergence of new infectious diseases are complex. Key is the extensive variation and flexibility of viral genomes, resulting from a combination of minimal generation times, notoriously inaccurate reproduction, and intra-host recombination. Viruses thus have a substantial genetic advantage over their human hosts in the evolutionary “molecular arms race.” This particularly applies to RNA viruses such as HCV, whose RNA-dependent RNA polymerase incorporates the extreme number of 10−3 mutations per viral nucleotide per year (or eight per genome, 100-fold higher than for HBV, a DNA virus).1 Even more worrisome is the rate of 0.2 errors and three recombination events per HIV genome per replication cycle, making it one of the fastest evolving of all organisms.3 Coupled with a logarithmic expansion in the infected host, producing up to 1012 new particles each day, this exerts intense pressure on the natural immune system to control the infection. Further shifting the balance of power is the fact that many viruses exist in genetically distinct quasi-species and subtypes and/or have developed “stealth and cunning” mechanisms to out-maneuver or evade the innate and adaptive immune response.4
Unfortunately, our existing treatment options for viral infections are usually ineffective and very limited. For instance, success rates for HCV are at best 50–60%, even using combinations of the most efficient regimens (pegylated interferon-α and ribavarin).1 Moreover, there is no preventive recombinant vaccine for the virus, or for HIV (two vaccines showed no efficacy in recent phase III clinical trials). The latter is perhaps the most frustrating candidate for development of an anti-viral therapy, as single-drug (e.g., azidothymidine) strategies readily result in the evolution of multiply drug-resistant strains. Even a combination of drugs (highly active anti-retroviral therapies) can typically only delay the onset of acquired immunodeficiency syndrome, not rid the body of virus altogether, owing to the low efficiency of the individual components and constant viral evolution. Moreover, highly active anti-retroviral therapy cocktails (targeting multiple HIV enzymes) are usually associated with severe side effects that are apt to cause metabolic disorders and undermine patient compliance.3 For all these reasons, the development of novel, safer, and more efficacious anti-viral therapies has become a worldwide priority.
RNAi-Based Antiviral Therapies
One particularly promising and powerful recent addition to our arsenal of anti-viral weapons is RNA interference, or RNAi. Hailed as “Scientific breakthrough of the year (2002)” by the journal Science 5 and honored with the 2006 Nobel Prize in Physiology or Medicine (Andrew Z. Fire and Craig C. Mello), RNAi is a natural phenomemon of gene silencing by small duplex RNAs. Originally discovered in plants and nematodes,6 RNAi is now known to be conserved through evolution up to humans, although it might serve different purposes in different species. It likely constitutes the main innate anti-viral defense in plants (and worms and flies),7 yet its primary role in mammals and humans might be the processing of micro RNAs (miRNAs), small regulatory non-coding RNAs.8 Hundreds of miRNAs are encoded in the human genome, many being transcribed in a spatio-temporal manner.9 Following expression as long pri-miRNAs, they are processed by the nuclear enzyme Drosha into shorter pre-miRNAs and then transported into the cytoplasm (via Exportin-5). There, the adenosine triphosphate–dependent RNAse III–like Dicer enzyme generates even shorter (approximately 21 nt) double-stranded RNAs, the small interfering RNAs (siRNAs). Their role is to guide the RNA-induced silencing complex to a homologous messenger RNA (mRNA) and induce either its cleavage or translational repression (depending on the siRNA–mRNA sequence complementarity).10
The extreme efficiency and specificity of this process make RNAi highly attractive for anti-viral therapies. A particular benefit over conventional approaches is that RNAi is an innate cellular pathway, requiring only the introduction of a trigger for its activation, which should minimize the side effects. Importantly, the nature of this trigger can vary and be tailored to the viral target and its unique life cycle.11 , 12 One option well suited to preventing or treating acute infection and providing immediate effects is topical application of siRNAs. However, their use might naturally be limited to mucosal tissues or localized and accessible sites of viral infection (e.g., the respiratory and female genital tracts and the eye).11 Notable recently reported siRNA applications include protection of mice against lethal infection with herpes simplex virus-213 and treatment of acute respiratory syncytial virus infection (a leading cause of child death).14 The latter application is currently being evaluated in a phase I clinical trial. Conversely, a gene therapy approach involving delivery of RNAi expression cassettes is more appropriate for treatment of chronic infections such as HBV, HCV, and HIV.1 Typically, the trigger is a miRNA-like sequence (derived from a natural miRNA15 or an artificial short hairpin RNA (shRNA)) under the control of an RNA polymerase II or III promoter. These cassettes are small and thus readily incorporated into any of the established gene therapy vectors, such as lentiviruses and AAVs (adeno-associated viruses). For instance, we have recently used the latter (AAV serotype 8) to express an anti-HBV shRNA in livers of HBV-transgenic mice, resulting in efficient and persistent viral suppression.16 Another general advantage of RNAi as an anti-viral therapy is that triggers with perfect viral sequence complementarity induce target cleavage. In particular, for positive-strand RNA viruses (e.g., HIV, HCV, and severe acute respiratory syndrome coronavirus), this offers the possibility not only to suppress the pathogen but actually to eradicate it from the host.11 , 12 Last but not least, RNAi silencing requires a minimal target of only 19–21 nt, which might be sufficient to co-suppress related viral isolates. Studies by Lee et al.17 and Kumar et al.18 support this idea by documenting concurrent inhibition of mulitple HIV or flavivirus strains with a single siRNA.
Limitations of RNAi Monotherapies
At this point, a large and growing body of work suggests that RNAi can suppress virtually every class of virus, whether it is based on DNA or RNA and whether it carries double- or single-stranded genomes.11 , 12 Unfortunately, a growing number of studies concurrently suggest that RNAi might face the same obstacles as have hampered other mono-therapies in the past, in particular viral escape. Paradoxically, in this regard, the sequence specificity of RNAi turns into a disadvantage: a single nucleotide change in the target can abolish mRNA degradation and provide the viral mutant with a growth advantage under RNAi pressure. This has already been illustrated by a variety of reports. Among the first, Gitlin et al. noted that prolonged incubation of poliovirus-infected, siRNA-treated cells resulted in enrichment of an RNAi-resistant point mutant.19 , 20 Identical findings were documented for HCV when Randall et al. reported loss of siRNA activity against genotype 1b in a mutant virus differing by only 3 nt.21 Wilson and Richardson validated the fact that subtle changes are indeed sufficient to permit viral escape by showing that successive mono-siRNA treatment of HCV replicon cell lines led to multiple point mutations in the HCV genome and loss of siRNA function.22 The same phenomenon was found consistently for HIV and first reported by Boden et al., who isolated a point mutant after constant HIV-1 growth on anti-tat shRNA-expressing T cells.23 HIV's propensity to escape was confirmed by Das et al.24 and by Lee et al.,17 who identified the emergence of nef or gag mutants under RNAi pressure. Interestingly, Westerhout et al. realized that HIV sporadically escapes through mutations that alter the local genome secondary structure, as opposed to single nucleotide changes.25 Regardless of mechanism, the high propensity to rapid escape mutation, combined with the high natural genetic diversity, makes finding a single conserved and efficient viral target sequence almost impossible.
A second problem with RNAi mono-therapies is that the interaction of human viruses with their hosts might be far more complex than originally thought.26 , 27 , 28 Contrary to the initial belief that RNAi is not part of the human innate anti-viral immune response, growing evidence now suggests the existence of virally encoded “suppressors of RNAi silencing” (SRSs). This should not be surprising, as double-stranded RNA is often generated during viral replication, exposing the virus to host RNAi and likely exerting pressure to evolve an anti-RNAi counter-mechanism. It is fascinating, however, to learn how many different approaches human viruses might have taken to developing SRSs (which are frequent among plant and insect viruses7). The first mammalian examples were the influenza NS1 and vaccinia E3L proteins, which might counteract the RNAi pathway via non-specific sequestering of double-stranded RNA, thus competing for Dicer substrates.29 However, their main function is inhibition of protein kinase R, and RNAi suppression has been shown only in non-vertebrate cells thus far. Adenovirus-encoded virus-associated RNAs, short miRNA-like molecules accumulating to 108 copy numbers per infected cell, are another example.30 Recent studies suggest that virus-associated RNAs can out-compete the Exportin-5–mediated translocation of endogenous miRNAs, and in addition competitively suppress Dicer activity.31 , 32 Similarly, it was proposed that the HCV core and the HIV Tat proteins inhibit RNAi via Dicer binding.33 , 34 HIV might also attenuate RNAi via a second mechanism—sequestering of trans-activating response RNA-binding protein (an essential RNA-induced silencing complex component) through its trans-activation response region element.35 On the other hand, it is striking to note that some human viruses hijack the RNAi machinery to carry out their own replication strategies, which might seem counterintuitive.26 , 27 , 28 One remarkable example is again HCV, which subverts a liver-specific miRNA (miR-122) for its gene expression.36 Moreover, other viruses were recently found to encode their own miRNAs, albeit in most cases (e.g., HIV or Epstein–Barr virus) without evidence for functional or genetic significance.34 , 37 , 38 , 39 , 40 , 41 An exception is an SV40-encoded miRNA whose function appears to be inhibition of the SV40 T antigens late in infection, to reduce the susceptibility of the virus to, and activation of, cytotoxic T lymphocytes.42
Combinatorial RNAi as a Novel Strategy
As our knowledge of the interactions between viruses and the RNAi pathway expands, it will continue to shape our therapeutic strategies. Viral exploitation of cellular factors offers a novel means of clinical interference (e.g., sequestration of endogenous or virally encoded miRNAs). Moreover, the putative presence of viral SRSs raises a need to target these suppressors directly, or to block early stages in the viral life cycle to prevent their accumulation. Combined with the need to compensate for natural diversity and rapid evolution, this makes the development of efficient anti-viral therapies a very challenging goal.
The emerging solution to thwart viral evolution and circumvent the related issues is to multiplex RNAi triggers or to combine them with other silencers of gene expression. This novel approach, comparable to HIV highly active anti-retroviral therapy and best described as “combinatorial RNAi” or “coRNAi,” holds significant promise over conventional mono-therapies. First and foremost, application of a silencer cocktail against multiple conserved viral sequences increases the chances of averting escape mutants, considering the evolutionary leap the virus would have to make. A stochastic computational model predicts that strong expression of four individual anti-HIV shRNAs might already suffice to prevent viral escape.43 Second, coRNAi therapies can blend inhibitors of viral and cellular gene expression to maximize efficacy and further minimize the risk of mutational escape. Particularly promising targets are viral cell surface receptors, whose silencing would not only block the earliest step in viral infection but also strand the virus outside the cell and thus prolong the time available for its detection by the immune system. Third, combining RNAi triggers with ribozymes or other inhibitors of gene expression will allow the concurrent blocking of virally encoded SRSs and direct targeting of the viral genome (and/or cellular factors). Such a mix of RNAi and unrelated silencers will also minimize the potential risks associated with high-level mi/shRNA expression in the cell.
Below and in Figure 1 , we comprehensively summarize the various recently reported coRNAi strategies. We focus on vector-mediated gene therapy approaches, as we believe the main coRNAi application will be long-term suppression of persisting human viruses such as HCV and HIV. We also include a few examples for functional genomics and treatment of non-viral disorders. However, it should be noted that initial proofs-of-principle for coRNAi were obtained using siRNAs. Examples include a study by Kronke et al., who used a library of endoribonuclease-prepared siRNAs to block HCV replicons in cell culture.44 Although their approach seemed useful as a way to inhibit multiple viral genotypes and to avert escape, its clinical applicability remained unclear. Similarly, Wilson and Richardson co-electroporated two distinct siRNAs into replicon cells to suppress resistant HCV mutants,22 and other groups used siRNA cocktails to enhance inhibition of HIV, severe acute respiratory syndrome coronavirus, and foot-and-mouth disease virus.45 , 46 , 47
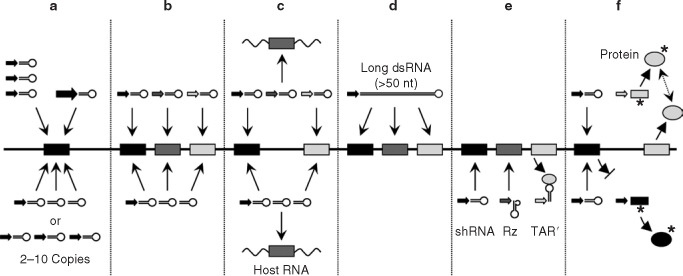
Figure 1.
Strategies for combinatorial RNA interference (coRNAi). Shown is a schematic viral genome (black line) with individual RNAi target sequences (shaded boxes). Depicted are the six different possible scenarios for coRNAi discussed in detail in the text. In brief, in (a), a single region is targeted with multiple hairpin RNAs, expressed either from individual promoters (in separate or a single vector backbone(s)) or as concatemers from one promoter. Another possibility, not mentioned in the text, to increase hairpin RNA levels is the use of an inherently stronger promoter (thicker arrow). Similarly, constructs expressing multiple small hairpin RNAs (shRNAs) from one or separate promoters can be designed simultaneously to target various (b) viral or (c) combinations of viral and cellular genes. (d) A unique means of co-silencing numerous regions on a single target is expression of a long hairpin RNA from one promoter. Recently, coRNAi vectors were refined to co-express short hairpin RNAs together with other nucleic acid–based inhibitors (ribozymes (Rzs) and HIV trans-activation response region (TAR) decoy (e)) or with transdominant ((f), gray circles; the asterisk indicates point mutations)) or therapeutic proteins (f, black circle).
Strategy I: Expression of Hairpin RNA Repeats
Concatemerization of single RNAi triggers (Figure 1a) represents the simplest example of coRNAi and has been reported sporadically to date. Intended to raise intra-cellular sh/miRNA levels, it might be especially useful against viral targets that replicate and spread at high enough rates to out-compete low-level RNAi. It might also be beneficial for limited sites within a single target or for simultaneous inhibition of multiple targets (e.g., viral genotypes) via a conserved site. Likewise, it could balance low vector efficiencies or promoter strengths that would otherwise prevent adequate sh/miRNA expression. However, a better way to increase intra-cellular sh/miRNA levels might be the use of inherently stronger RNAi promoters. Nevertheless, we discuss this strategy here because it essentially proves the feasibility of concatenating hairpin RNAs and illustrates some of the key issues.
Gonzalez et al. used individual U6 promoters to express up to six copies of an shRNA against a target site conserved in all classical HLA class I genes (HLA A, B, and C).48 Construct efficacy was validated through transient transfection of a T-cell line and subsequent measurement of surface HLA ABC antigens. As predicted, raising the shRNA copy number resulted in a progressive (up to 17-fold) RNAi improvement, correlating with increased detectable shRNA expression. The effect was stably maintained for up to 6 months in the absence of interferon (IFN) production and mediated the expected protection against a cytotoxic T-lymphocyte response. Gonzalez et al. were also able to introduce six shRNA copies into primary T cells and knock down HLA A, although the result was not compared with the result of lower shRNA numbers. Interestingly, an increase to 10 shRNA copies further augmented transient RNAi, but the effect was no longer stable. The reasons were not studied but likely included toxicity or genetic instability.
Zhou et al. were among the first to demonstrate the possibility of multiplexing artificial miRNAs.49 Using a modified mIR-30 hairpin to target human superoxide dismutase 1 (SOD1, fused with a luciferase reporter), they expressed two copies (separated by 100 nt) from a single cytomegalovirus promoter. In transiently co-transfected cells and compared with a single hairpin, the tandem vector surprisingly failed to yield higher siRNA levels or better target knockdown. Instead, the authors noted decreased RNAi from the tandem plasmid, for reasons unclear. This study thus strongly suggests the need for a better understanding of the rules for miRNA concatemerization.
Similarly curious results were reported by the same group in transgenic mice.50 Plasmids expressing anti-SOD2 mIR-30 from a ubiquitin C promoter were injected into fertilized eggs, yielding two lines carrying a single or three miRNA copies. The single-copy line had higher siRNA levels, correlating with better SOD knockdown. Crossing both lines yielded bigenic heterozygous mice expressing even higher siRNA levels and showing a near SOD knockout phenotype. This study was the first to describe hairpin RNA multimerization in transgenic mice, but the puzzling lack of correlation between miRNA copy numbers and RNAi efficacy remains to be studied further.
Sun et al. concatenated mIR-30–based anti–S-phase kinase-associated protein 2 or anti-androgen receptor hairpins under a cytomegalovirus promoter.51 In line with the results of Gonzalez et al.,48 an increase in miRNA numbers from one to two resulted in enhanced siRNA levels and RNAi (following lentiviral delivery into 293 cells). However, addition of a third copy yielded only a marginal further (disproportionally smaller) increase. Perplexingly, the efficacy of the single hairpin could also be increased by adding an irrelevant second miRNA to the vector. This suggested a link between miRNA processing and efficacy, rather than a direct effect of copy numbers, but this interesting possibility remains to be investigated.
Recently, Chung et al. tested constructs based on a distinct miRNA, mIR-155, and came to different conclusions.52 Concatemerization of up to eight anti-luciferase miRNAs under a single ubiquitin C promoter resulted in a progressive increase in RNAi. The reasons for the obvious discrepancy with earlier attempts are unclear, but they could relate to the use of two different mIRs (30 versus 155), which may have affected hairpin folding or stability. Moreover, the studies also differed in the positioning of the miRNAs (exons or introns), as well as in the nature and length of the flanking sequences (known to be crucial for miRNA processing). Another remarkable finding in this work was that construct efficacy further increased with inclusion of a second intron or upon removal of the polyadenylation site. Although not confirmed experimentally, an interesting speculation was that these modifications increased nuclear retention of the primary transcripts, thereby facilitating miRNA processing.
Together, these few studies already clearly highlight the feasibility of expressing multiple sh/miRNAs from a single construct, although there exist few therapeutic applications at this point. Vectors employing miRNAs appear to be preferred for expression of multiple hairpins as they allow the use of a single promoter, which offers the potential for spatio-temporal coRNAi control. However, the striking discrepancies among studies exemplify the need for deeper investigation into the rules that govern efficient miRNA processing. Certainly, factors such as hairpin number and positioning, and intervening spacer length and composition, must be tested and optimized individually.
Strategy II: Multimerization of Different Hairpin RNAs
Numerous papers have recently provided proofs-of-concept for the approach of co-expressing multiple sh/miRNAs from a single vector, using non-viral or viral targets (HCV or HIV) as examples (Figure 1b and c). In brief, there were two different goals: (i) to establish coRNAi as a surrogate genetic tool for basic studies, allowing the dissection of overlapping functions of individual factors to biochemical pathways; (ii) more relevant in the context of this review, to elucidate the usefulness of coRNAi for the treatment or prevention of viral infection and escape by co-targeting multiple viral and/or cellular genes.
Yu et al. were among the first to apply a coRNAi approach to the study of gene function, via simultaneous inhibition of multiple endogenous mRNAs.53 They designed two shRNAs (under separate U6 promoters) to target the α- and β-isoforms of glycogen synthase kinase 3, two related enzymes involved in various cellular processes and human disorders. In stably co-transfected cells, coRNAi of both isoforms led to an additive increase in expression of the glycogen synthase kinase 3 target β-catenin, as compared with inhibition of the individual enzymes. This report thus exemplified the usefulness of coRNAi for functional analyses and moreover implied a possible treatment for diseases linked to abnormally high glycogen synthase kinase 3-α/β levels (Alzheimer's or type 2 diabetes).
Jazag et al. provided a similar example for coRNAi-based analyses of complex signal transduction pathways, in which inhibition of individual genes could not account for the whole process.54 Their targets were Smad transcription factors (Smad2, 3, or 4), which mediate the transforming growth factor-β cytostatic response in many cell types. The latter is of clincial interest as its loss contributes to tumorigenesis. Using separate U6-driven shRNAs against the different Smads, the authors established stable cell lines expressing one, two, or all three hairpins. Similar to the results of Gonzalez et al.,48 shRNA expression and Smad knockdown could be maintained for at least 20 passages, likely owing to the small shRNA number and the low expression levels from the integrated plasmids. As hoped, phenotypic analyses of their various cell lines revealed different contributions of all three Smads to parameters such as wound closure and cell migration, providing further insight into the role of Smads in cancer.
The potential of coRNAi for functional gene studies was further substantiated by a series of similar reports in cell cultures or transgenic mice using sh/miRNAs to co-target, e.g., multiple SOD genes,50 , 55 cyclin A and S-phase kinase-associated protein 2,51 and the related kinases B-Raf and c-Raf.52 A particularly remarkable article by Shin et al. reported the use of a tightly regulated tetracycline-inducible coRNAi system.56 In detail, the group engineered lentiviruses conditionally to express two mIR-30 hairpins targeting the heterotrimeric G proteins Gα12 and Gα13. Analyses of reporter gene expression (luciferase fused with a serum response element) allowed them to delineate a specific role of Gα13 of transmitting receptor-mediated serum response element activation. This study is thus another illustration of coRNAi as a powerful experimental platform for analysis of potential redundancy in signaling pathways.
From a clinical standpoint, two of the most interesting targets for therapeutic coRNAi are HCV and HIV. As mentioned before, these two viruses are particularly resistant to therapeutic intervention owing to their extreme natural genetic diversity and potential for mutational escape. Many groups therefore recently began to pursue the strategy of co-targeting viral and cellular genes (encoding host co-factors required for viral uptake, replication, or expression), on the basis that the latter are not under evolutionary pressure and are thus substantially less prone to mutation. A growing number of cellular proteins have already been identified as playing critical roles for these two viruses, and their individual silencing had significant effects on the outcome of infection. For instance, Zhang et al. used adenovirally delivered, U6-driven shRNAs against cellular La, polypyrimidine tract-binding protein, and human vesicle-associated membrane protein-associated protein of 33 kd, all known to interact with HCV.57 Individual inhibition of each gene led to a reduction of HCV amplification in replicon cell lines, suggesting their usefulness for future co-targeting strategies including the viral genome itself. Other promising cellular co-targets are viral (co-)receptors, as their inhibition would actually prevent viral infection and not simply block viral reproduction (when targeting viral genes). However, a fundamental concern with this strategy is that knocking down endogenous genes could create an unacceptable loss-of-function pathology for the cell. Therefore, to maximize patient safety, potential cellular targets must be chosen very carefully and evaluated individually and thoroughly.
This is exemplified by a report by Korf et al., who co-targeted the two cellular HCV co-factors HuR (Hu antigen R, binds to the HCV 3′-untranslated region, resulting in its stabilization) and PSMA7 (proteasome α-subunit 7, modulates HCV-internal ribosome entry site activity), together with the HCV genome (5′- or 3′-untranslated region).58 In transfected HCV replicon cell lines, each of their individual best shRNAs (under separate U6 promoters) caused at least 50% reduction in viral RNA and similar decreases in HCV NS5B protein. Importantly, some combinations of anti-HCV and anti-HuR/PSMA7 shRNAs showed a strong additive effect, illustrated by an increase in viral protein and RNA inhibition of up to approximately 70%. Although not evaluated, this exciting result suggested the potential of this particular coRNAi approach to prevent HCV escape mutants and/or to inhibit multiple genotypes (provided they share the cellular factors). On the other hand, the therapeutic usefulness of the two cellular proteins remained obscure, as both have essential functions in the cell (PSMA7 is a proteasome subunit, and HuR an important regulator of cell proliferation).
Another recent study, by Henry et al., provided the first example for a triple shRNA vector co-targeting the HCV genome (internal ribosome entry site or NS5B) and a host factor (cell surface tetraspanin CD81, binds the HCV envelope protein E2).59 In HCV replicon cells, all individual shRNAs (under separate H1 promoters, delivered by a lentivirus) efficiently reduced replication or expression of their specific target by at least 80%. Similar results (with respect to the individual targets) were obtained for coRNAi vectors expressing two or all three shRNAs simultaneously. Important conclusions were the lack of competition among the individual shRNAs and the absence of non-specific effects from their vectors. However, it must be noted that the H1 promoter is relatively weak and that shRNA levels were not quantified in this study. Moreover, Huh-7 replicon cells have a reduced capacity to produce IFNα, further clouding analysis of potential side effects from their coRNAi vector. Regardless, the finding that HCV inhibition persisted for 17 days in stably transduced cell lines argues against a major adverse effect and provides another proof of the feasibility of establishing long-term coRNAi. It will be exciting to evaluate fully the therapeutic potential of the promising triple vector from this study, in particular the effect of CD81 knockdown on HCV binding or uptake.
A recent unique study by Akashi et al. suggested the feasibility of expressing long (>50 nt) shRNAs in human cells in the absence of an IFN response, allegedly as a result of mismatches in the sense shRNA strand.60 Accordingly, the authors used a plasmid encoding a 51-bp-long, U6-driven shRNA for the efficient co-targeting of the NS5B gene from two distinct HCV strains differing in nine nucleotides. Compared with a conventional 20-mer shRNA, the longer hairpin not only suppressed both isolates but also yielded more rapid knockdown. Although this was not strictly a coRNAi approach, this study is notable because the results implied the generation of multiple different siRNAs from the long precursor (albeit not truly characterized).60 , 61 If confirmed, this strategy could theoretically be exploited to prevent resistance by targeting a long error-prone region in the viral genome (Figure 1d).
Boden et al. were among the first to recognize the need for a coRNAi approach to controlling HIV.23 They engineered an AAV vector to express a single anti-HIV tat shRNA in cultured lymphoma cells and found it suppressed HIV-1 replication for more than 3 weeks. However, its activity was subsequently lost because a highly resistant HIV point mutant emerged within 2 months, prompting the authors to suggest anti-viral coRNAi for future therapies. Similar conclusions were reached by Song et al., who tested a combination of two different siRNAs, targeting the viral p24 or the cellular CCR5 (major HIV-1 co-receptor in macrophages) gene.47 When the siRNAs were co-transfected into monocyte-derived macrophages, they observed a strong synergistic effect and almost complete inhibition of HIV infection, compared with a weaker effect with the individual siRNAs. Similar to Boden et al.,23 the group thus favored a vector-based coRNAi approach, ideally targeting multiple steps in the HIV life cycle. This idea was substantiated by Lee et al.,17 who reported accumulation of gag mutants in HIV-infected CD4+ T cells transduced with an anti-gag shRNA lentivirus. It was also supported by Das et al.,24 who noted the emergence of HIV nef mutants following virus passage on T cells stably expressing a single anti-nef shRNA.
A series of recent papers document the power of co-suppressing cellular HIV co-factors (receptors) to control HIV infection, similar to HCV. Among the first, Anderson et al. inhibited CD4 (primary receptor), CCR5 (co-receptor for monocyte/macrophage-tropic HIV) and CXCR4 (T-cell tropic HIV co-receptor).62 Bi-specific siRNA constructs were engineered to target CXCR4 and either CD4 or CCR5, in vitro transcribed, and transfected into HIV-permissive cells, including peripheral blood mononuclear cells. Virus challenge assays showed a marked protection of the transfected cells from HIV, in particular by the combined CXCR4/CD4 construct. This study was an important proof-of-principle, but the clinical applicability of the bi-specific siRNA is questionable, in particular as it might cause an IFN response.
In a follow-up study, the same group engineered lentiviral vectors to co-express their best shRNAs against CXCR4 and CCR5 (under a U6 or H1 promoter, respectively) in peripheral blood mononuclear cells.32 Similar to their initial study, this resulted in protection against HIV infection in a viral challenge assay. However, it remains to be tested whether such an anti-receptor coRNAi approach will suffice to prevent viral resistance. Moreover, the clinical usefulness of HIV receptors as (sole) therapeutic targets remains to be validated. One concern is that inhibiting a specific receptor may select for viral variants that use a non-targeted, different (co-)receptor, ultimately negating any therapeutic effect.63 Also, CD4 might not be an ideal target, as it is an essential cell surface molecule for immunological function.62 In contrast, CCR5 might be dispensable for life, as there are asymptomatic individuals homozygous for CCR5 mutation.11 , 64 Likewise, CXCR4 mutation did not affect T-cell development and maturation, at least in murine studies. Nevertheless, CXCR4 knockdown is critical in stem cells (a major target for HIV therapies) as this molecule plays a role in cell homing into bone marrow.62
Several other groups have begun to study co-inhibition of HIV genes as a therapeutic modality. Chang et al. used lentiviral vectors to deliver anti-HIV shRNAs to various virus-permissive cell types.65 A critical finding was that a combination of three vectors, directed against highly conserved regions in the viral pol, int, and vpu genes, outperformed the individual shRNAs in terms of suppressing HIV in a stable virus-producer T-cell line. Despite the increased efficacy, the possible formation or prevention of escape mutants was not evaluated in this short-term study.
It was instead addressed in a more comprehensive, very recent study by ter Brake and colleagues.66 In a screen of all HIV-1 subtypes (including the LAI prototype) for highly conserved regions, the authors identified 19 potential targets. Of a battery of 86 shRNAs against these targets, 21 were found to be transiently effective from an H1 promoter. Further studies of three of them, directed against the gag or pol genes, revealed strong individual protection against HIV challenge in stably transduced permissive cells (including peripheral blood mononuclear cells). Intriguingly, all three shRNAs combined in one lentiviral vector conferred near-resistance to viral infection. Most important, the group also studied the emergence of viral resistance in cells expressing only one or two of their most effective shRNAs. As hoped, viral inhibition was more durable in the coRNAi cell line, although eventually (day 22) most cultures were positive, regardless of shRNA copy number. Although they do not show the data, the authors also mentioned an improved vector expressing four different shRNAs (from separate and distinct promoters) and able to further delay viral escape for up to 60 days. The strength of this article is that it is the first (and only to date) to validate a multiple shRNA approach for suppression of HIV escape. It is also particularly noteworthy that the shRNAs were carefully chosen to concurrently target all HIV-1 subtypes, although this was not confirmed experimentally. Important for future use of this particular system will be to investigate the genetic stability of the threefold or fourfold shRNA lentiviral vectors, as well as the potential side effects from these unique constructs.
Similarly intriguing recent work by Nishitsuji et al. showed the feasibility of efficiently co-targeting two other viral regions, the integrase gene (int) and the U3 attachment site (att).67 When transduced via lentiviral vectors into CD4+ T cells, the individual shRNAs showed a more potent inhibitory effect on HIV replication than an anti-tat construct (the same one used by Boden et al.23). Remarkably, at a higher HIV dose where the single shRNAs were no longer able to control the virus, a combination of the int- and att-specific shRNA vectors still gave strong suppression for almost 3 weeks. In line with previous work, the group noted the emergence of resistant point mutants after HIV infection of single shRNA-expressing cells. Subsequently generated shRNAs specific for these mutants could suppress their replication, but, unexpectedly, a combination of wild-type and mutant shRNAs had less effect on preventing viral escape. The authors hypothesized intra-cellular competition of the various shRNA vectors for the same target site, but this idea was not validated. As in the HCV studies by Akashi et al.60 and Watanabe et al.,61 the group also tested a long (50-nt) hairpin RNA covering the target region of their anti-int shRNA. Interestingly, this construct was able to co-suppress both wild-type and mutant HIV strains, but the effect was weak and only transient, for reasons unknown. Nonetheless, these articles together suggest that when the technical problems have been overcome and the safety of long hairpins can be guaranteed, combining multiple short and long RNAs might further increase the power of coRNAi to control viral resistance.
Strategy III: Co-Expression of Small RNAS and Other RNA-Based Inhibitors
Collectively, the studies reviewed above clearly validate the promise of coRNAi to suppress viral infection and escape. However, they also provide evidence for potential setbacks from co-expression of multiple hairpin RNAs in the same cell and from the same vector. The issues include genetic instabilities, promoter or hairpin interference, and toxic side effects (see also Conclusion). In attempts to circumvent these problems, a few groups have begun to combine sh/miRNAs with other silencers of gene expression or with cDNAs encoding therapeutic proteins (Figure 1e and f, and next section).
A noteworthy early proof-of-concept came from Hemmings-Mieszczak et al., who mixed siRNAs with antisense oligonucleotides to inhibit a rat pain-related cation channel (P2X3, an important target in pain research).68 Rationales were the high costs of siRNA mono-therapies in humans and the low efficiency of antisense oligonucleotides. As hoped for, the coRNAi approach produced synergistic effects in terms of P2X3 knockdown in cultured cells, but, interestingly, only when both agents targeted non-homologous regions. The reasons for this competition remain elusive, but the findings are reminiscent of the study by Nishitsuji and colleagues described above.67 It is unclear whether the molecular mechanisms are related or even identical, but these two studies certainly prompt caution in attempts to target a single site with multiple inhibitors.
Jarczak et al. were among the first to suggest a combination of shRNAs with hammerhead ribozymes (Rzs) for HCV treatment.69 The group targeted the highly conserved 5′- and 3′- viral untranslated regions with various U6-driven shRNAs or Rzs (under U6 or tRNAVal promoters). After individual transfection into HCV replicon cell lines, their best candidates inhibited HCV NS5B expression by approximately 30% (Rzs) or 50% (shRNA). Although mixing various Rzs increased overall inhibition marginally, it was most notable that combining the best Rzs and shRNAs gave an approximately 25% additive effect to each shRNA, irrespective of its initial potential (however, combinations of shRNAs alone were not tested). This study is an important proof-of-concept, in particular as it further confirms the need to target different sites in the viral genome for maximum efficacy, in line with Watanabe's work.61 Moreover, the finding that the use of different promoters for Rz and shRNA expression yielded the strongest additive effects substantiates the need to avoid promoter competition.
At this point, relevant anti-HIV coRNAi approaches have been mostly reported by John Rossi's group, which is currently also preparing a clinical trial. In a 2003 pilot study, this group tested the feasibility of combining two different RNA-based HIV inhibitors in one lentiviral vector, although neither was an RNAi trigger. Instead, Li et al.64 used an anti-CCR5 ribozyme (driven by an adenoviral VA1 promoter) together with a trans-activation response region decoy (U6-promoted and embedded in small nucleolar RNA U16, to ensure co-localization with HIV Tat in nucleoli of infected cells). The resulting vector provided a substantially greater survival advantage to HIV-challenged primary T or CD34+ stem cells compared with the individual ribozyme or decoy. An shRNA against rev and tat tested in parallel was shown to reduce HIV p24 substantially in the same cell types, over the same period, but the results were not directly compared and the constructs were not combined.
Construct combination was reported in a follow-up study in which Rossi's group presented a lentiviral vector combining all three inhibitors from their earlier work (Figure 1e).70 This novel construct suppressed HIV replication for up to 4 weeks in a more-than-additive fashion as compared with single or double vectors. In fact, the triple vector was the only construct that remained inhibitory in CD34+ cells for 28 days, even under challenge with a high HIV dose and from only 1–2 integrated vector copies. Importantly, there was no evidence for evolution of escape mutants with the triple vector, although additional validation might be needed in view of the assays used. The group also reported some minor unexpected, and not yet fully understood, findings with their vector. These include an approximately fourfold drop in viral titers under standard production conditions, approximately threefold lower transduction efficacies in CD34+ cells (versus the single shRNA vector), and a approximately 20% loss in Gfp (green fluorescent protein) expression upon long-term culture of triple-transduced cells (compared with an empty vector). Nevertheless, the overall impressive results with this unique and ingenious construct clearly illustrate the power of coRNAi for HIV therapies. Consequently, a slightly modified vector (deleted for a gfp marker gene) will soon be tested in a clinical trial using autologous hematopoietic stem cells from acquired immunodeficiency syndrome/lymphoma patients and bone marrow transplantation.
Strategy IV: Co-Expression of Small RNAS and Proteins
Another inventive anti-HIV coRNAi strategy recently reported by Rossi's group involved combination of an shRNA with a humanized, transdominant negative mutant HIV Rev protein (huRevM10)71 (Figure 1f). Rev is an attractive anti-viral target because it mediates nuclear export of singly spliced and unspliced full-length genomic RNAs in the HIV life cycle. Point mutations in the nuclear export signal provide the M10 Rev variant with the ability to inhibit HIV replication, rendering it a powerful therapeutic, and it is already under clinical evaluation. Unwalla et al. created a lentiviral vector in which an anti-rev shRNA was expressed from an HIV-inducible RNA polymerase II promoter. Termination by a weak polyadenylation signal permitted read-through of the downstream RevM10 coding sequence, thus generating both functional shRNA and protein from the same promoter. In stably transduced and HIV-challenged T cells, the double vector mediated 90% inhibition of HIV p24 protein and a high cell survival rate (although a direct comparison with single vectors was not provided). Again, there was no evidence for viral escape, and, in fact, an HIV point mutant that had previously evolved in the presence of the anti-rev shRNA alone remained approximately 80% inhibited by the double vector. Together, these impressive results illustrate the power of this particular coRNAi approach to prevent HIV resistance, identical to the triple RNA-based construct described above.70
Last but not least, in addition to co-expression of transdominant anti-viral proteins, one can also envision synergistic gene silencing/addition in other therapeutic contexts. One remarkable example by Samakoglu et al. documented a coRNAi–protein approach for treatment of sickle cell anemia.72 A lentiviral vector was engineered to express a recombinant γ-globin gene from a β-globin promoter/enhancer and to carry an intronic shRNA against the human sickle β-globin (β S) mRNA. CD34+ cells from healthy humans or sickle cell anemia patients were transduced with the vector and then differentiated into erythroid cells. Impressively, all cells showed similar levels of γ-globin and normal β-globin, but β S was specifically and more than 70% reduced in the patient cells. These data imply that synergistic globin protein expression and RNAi-mediated β S knockdown holds promise as a stem cell therapy for sickle cell anemia. Generally, together with the Rossi group's lab work, this latest study paves the way for the development of coRNAi strategies to add and delete gene functions concurrently for the treatment of human disorders.
Conclusions and Perspectives
Although this is still a fledgling area, a rapidly growing body of evidence already illustrates the far-reaching potential of coRNAi technology. It is emerging as a powerful modality to battle some of the most notoriously challenging clinical targets (HCV, HIV, and other human viruses), and initial studies also affirm its great potential for treatment of metabolic or blood disorders or cancer. Concurrently, coRNAi is quickly exceeding our expectations for its use in the study of basic processes, such as signaling or transformation.
However, several critical issues associated with this novel approach must be resolved to permit realization of its promise in humans and to progress to clinical trials. The keys to this transition and to paving the way for coRNAi from bench to bedside will be safety, stability, and efficacy. The safety concern is based on a plethora of earlier reports on unexpected and adverse side effects from mono-RNAi treatments, including “off-target” silencing, IFN responses, and translational inhibition.73 Although it is obvious that these risks may increase proportionally with a coRNAi approach, another specific concern is oversaturation of the endogenous RNAi machinery. This might at least result in competitive reduction of the effects of the individual silencers, which could indeed explain some of the findings described above. Yet, in the worst case, overwhelming of individual factors in the RNAi pathway will result in global dysregulation of endogenous miRNA processing. A dramatic possible outcome is illustrated by our own recent study, where persistent high-level shRNA expression in mouse liver from an optimized AAV vector led to perturbation of miRNA biosynthesis.16 This frequently (36 out of 49 constructs, directed against six different targets) resulted in liver toxicity and cellular regeneration, eventually leading to loss of the RNAi effect over several weeks. However, in almost 50% of all cases (23/49), shRNA-associated toxicity was so severe that the animals failed to recover and ultimately died from the treatment.16 Although the detailed mechanism of toxicity is still under investigation, in line with our in vivo data, other groups have consistently reported concentration-dependent cytotoxic effects from shRNA overexpression in cultured cells.74 , 75 , 76 Particularly noteworthy is a recent study by An et al.,74 who compared the toxicity and efficacy of shRNAs against CCR5 when expressed from two different RNA polymerase III promoters, U6 or the transcriptionally weaker H1. Not surprisingly, the group found a clear correlation between shRNA expression levels in cultured primary T cells and cytotoxicity, although the underlying mechanisms were unclear and seemed to vary with the U6-driven constructs.74 Nevertheless, the authors concluded that lower shRNA numbers appear to be advantageous to maintaining the transduced cell population. This is also in line with our very recent observations of substantially increased RNAi persistency in mice treated with AAV vectors expressing shRNAs from H1 or 7SK promoters, as compared with the more robust U6 (D.G. and M.A.K., unpublished results). Unfortunately, no published work has yet defined the limit to the number of exogenous hairpin RNAs that can be effectively incorporated into a (co)RNAi treatment, and it will likely vary with the types of cells and organs.77 Until this knowledge is available, one cannot overemphasize the need to find the most potent target sites and hairpin RNAs, to carefully optimize coRNAi vector designs, and to work at the lowest concentrations possible. It might be especially beneficial to adapt the latest conditional promoter systems for coRNAi purposes, as they will ideally allow for restriction of gene silencing to a defined subpopulation of cells, thereby mimizing the global risks for the organism.78 Some exciting recent examples include promoters that are specifically activated in HCV- or HIV-infected cells79 , 80 or can be epigenetically and reversibly controlled using exogenous drugs or small molecules.81 , 82
The use of these alternative promoters might concurrently help to circumvent the second concern with coRNAi, i.e., genetic instability of the multi-component vectors. Although it is technically feasible to accommodate multiple sh/miRNA cassettes into virtually any present viral vector (including the smallest of all, AAV), there are hardly any data at this point on the likely risks of recombination or deletion caused by sequence similarities or identities among the individual elements. Anecdotal evidence suggests this problem exists and could hamper the approach, as it might, for instance, explain the reported difficulties in implementing more than six identical shRNAs into a single plasmid.48 One strategy to circumvent this problem would be the use of different promoters for each hairpin, including conventional as well as the above-mentioned novel systems. However, the resulting disproportional expression levels (based on promoter strength) might inadvertently obscure the contribution of the individual components. Alternatively, as already demonstrated and perhaps preferred, vectors can be engineered to express multiple miRNA-like hairpins from a single RNA polymerase II promoter. This could simultaneously maximize genetic stability and spatio-temporal control. On the other hand, the discrepant findings available on the efficiency of multi-miRNA vectors clearly indicate that implementation of this strategy requires an improved understanding of the cellular mechanisms that govern processing of hairpin concatemers.
As with any novel therapy, a stringent test for coRNAi strategies will be their evaluation in animal models of innate or acquired genetic disease or viral infection. Importantly, in vivo trials will not only allow us to evaluate the efficacy of the new vectors directly but also provide us with better clues on the physiological role of the putative virally encoded RNAi suppressors. Thus far, the majority of related findings have been obtained in artificial systems, using either robust plasmids for SRS expression (as opposed to perhaps low-level expression from the intact virus) or heterologous read-outs (e.g., using non-vertebrate cells for mammalian factors). It is thus very possible that under physiological conditions, it will become obvious that the effect of viral SRSs has been overestimated. This would agree with the perplexing fact that many of the viruses proposed to encode SRSs actually seem to hijack the endogenous RNAi pathway, as they rely on cellular or viral miRNAs for their own agenda. One example is HCV, and as we are now fortunate to have the first replication-competent wild-type isolate available, it will be possible and exciting to study the seemingly intricate interplay of the virus with the RNAi machinery in a natural scenario.1 The lessons learned will certainly influence the future design of coRNAi vectors with respect to the importance of SRS-specific silencers. Generally, we expect to benefit from these studies because they will help us to optimize manifold coRNAi parameters, such as the balance of viral and cellular targets, possible associated toxicities, or vector-related issues such as dosing and delivery. Finally, these studies might reveal whether coRNAi will overcome a particular side effect of conventional anti-HIV highly active anti-retroviral therapy, i.e., the generation of multi-drug-resistant and highly infectious viral strains.3 It is certainly our hope that the extreme efficacy noted thus far means this adverse effect will not occur with coRNAi-based therapies.
In conclusion, we anticipate with excitement the elucidation of whether coRNAi technology will live up to its promise in clinical studies and ultimately prove to be our winning strategy in the battle against evolving targets. We are optimistic that we have an essential advantage in this race, as RNAi might not be the primary human innate immune defense and viruses may still lag in the development of efficient counter-mechanisms.8 The inherent modularity of coRNAi is another particular benefit, as it allows us to combine RNAi with any other efficient anti-viral therapeutic, including conventional small-molecule or protein drugs. Particularly exciting candidates emerging as potential future RNAi partners are aptamers, RNA oligonucleotides able to bind ligands with high specificity and affinity. In fact, recent work demonstrates that RNA aptamers can be expressed from RNA polymerase III promoters, identical to shRNAs, opening up the possibility of combining them with RNAi triggers in a multi-component vector.83 Moreover, aptamers can be fused with siRNAs to permit targeted RNAi delivery84 or can be incorporated into shRNA loops as a regulatory element.81 Last but not least, by drawing upon our growing knowledge of endogenous RNAi pathways, the improvements in viral vector design, and the refinement of bioinformatical models of viral infection, we will be able further to enhance the efficacy of the approach. Therefore, for the first time in the history of anti-viral therapies, the balance of power could be decisively tipped in our favor.
Acknowledgments
The authors wish to thank Theresa A. Storm and Lora Wang (Stanford University, School of Medicine, Stanford, CA, USA) for their critical reading of the manuscript. This work was supported by NIH grants DK78424 and AI71068.
Footnotes
published online 20 February 2007
References
- 1.Grimm D, Kay MA. Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther. 2006;13:563–575. doi: 10.1038/sj.gt.3302727. [DOI] [PubMed] [Google Scholar]
- 2.UNAIDS . Report on the global AIDS epidemic 2006. UNAIDS. 2006. <http://www.unaids.org>. [Google Scholar]
- 3.Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 4.Wieland SF, Chisari FV. Stealth and cunning: hepatitis B and hepatitis C viruses. J Virol. 2005;79:9369–9380. doi: 10.1128/JVI.79.15.9369-9380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Couzin J. Breakthrough of the year. Small RNAs make big splash. Science. 2002;298:2296–2297. doi: 10.1126/science.298.5602.2296. [DOI] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nat Rev Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 8.Cullen BR. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat Immunol. 2006;7:563–567. doi: 10.1038/ni1352. [DOI] [PubMed] [Google Scholar]
- 9.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 11.Dykxhoorn DM, Lieberman J. Silencing viral infection. PLoS Med. 2006;3:e242. doi: 10.1371/journal.pmed.0030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leonard JN, Schaffer DV. Antiviral RNAi therapy: emerging approaches for hitting a moving target. Gene Ther. 2006;13:532–540. doi: 10.1038/sj.gt.3302645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palliser D, Chowdhury D, Wang QY, Lee SJ, Bronson RT, Knipe DM. An siRNA-based microbicide protects mice from lethal herpes simplex virus 2 infection. Nature. 2006;439:89–94. doi: 10.1038/nature04263. [DOI] [PubMed] [Google Scholar]
- 14.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Y, Cai X, Cullen BR. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol. 2005;392:371–380. doi: 10.1016/S0076-6879(04)92022-8. [DOI] [PubMed] [Google Scholar]
- 16.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 17.Lee SK, Dykxhoorn DM, Kumar P, Ranjbar S, Song E, Maliszewski LE. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood. 2005;106:818–826. doi: 10.1182/blood-2004-10-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Lee SK, Shankar P, Manjunath N. A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med. 2006;3:e96. doi: 10.1371/journal.pmed.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 20.Gitlin L, Stone JK, Andino R. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J Virol. 2005;79:1027–1035. doi: 10.1128/JVI.79.2.1027-1035.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson JA, Richardson CD. Hepatitis C virus replicons escape RNA interference induced by a short interfering RNA directed against the NS5b coding region. J Virol. 2005;79:7050–7058. doi: 10.1128/JVI.79.11.7050-7058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- 27.Schutz S, Sarnow P. Interaction of viruses with the mammalian RNA interference pathway. Virology. 2006;344:151–157. doi: 10.1016/j.virol.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 28.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 29.Li WX, Li H, Lu R, Li F, Dus M, Atkinson P. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA. 2004;101:1350–1355. doi: 10.1073/pnas.0308308100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews MB, Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol. 2004;78:12868–12876. doi: 10.1128/JVI.78.23.12868-12876.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjarvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556–9565. doi: 10.1128/JVI.79.15.9556-9565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Kato N, Jazag A, Dharel N, Otsuka M, Taniguchi H. Hepatitis C virus core protein is a potent inhibitor of RNA silencing-based antiviral response. Gastroenterology. 2006;130:883–892. doi: 10.1053/j.gastro.2005.12.028. [DOI] [PubMed] [Google Scholar]
- 34.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 36.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 37.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 39.Omoto S, Fujii YR. Regulation of human immunodeficiency virus 1 transcription by nef microRNA. J Gen Virol. 2005;86:751–755. doi: 10.1099/vir.0.80449-0. [DOI] [PubMed] [Google Scholar]
- 40.Omoto S, Fujii YR. Cloning and detection of HIV-1-encoded microRNA. Methods Mol Biol. 2006;342:255–265. doi: 10.1385/1-59745-123-1:255. [DOI] [PubMed] [Google Scholar]
- 41.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- 43.Leonard JN, Schaffer DV. Computational design of antiviral RNA interference strategies that resist human immunodeficiency virus escape. J Virol. 2005;79:1645–1654. doi: 10.1128/JVI.79.3.1645-1654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kronke J, Kittler R, Buchholz F, Windisch MP, Pietschmann T, Bartenschlager R. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J Virol. 2004;78:3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Ren L, Zhao X, Hung T, Meng A, Wang J. Inhibition of severe acute respiratory syndrome virus replication by small interfering RNAs in mammalian cells. J Virol. 2004;78:7523–7527. doi: 10.1128/JVI.78.14.7523-7527.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kahana R, Kuznetzova L, Rogel A, Shemesh M, Hai D, Yadin H. Inhibition of foot-and-mouth disease virus replication by small interfering RNA. J Gen Virol. 2004;85:3213–3217. doi: 10.1099/vir.0.80133-0. [DOI] [PubMed] [Google Scholar]
- 47.Song E, Ren L, Zhao X, Hung T, Meng A, Wang J. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J Virol. 2003;77:7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez S, Castanotto D, Li H, Olivares S, Jensen MC, Forman SJ. Amplification of RNAi—targeting HLA mRNAs. Mol Ther. 2005;11:811–818. doi: 10.1016/j.ymthe.2004.12.023. [DOI] [PubMed] [Google Scholar]
- 49.Zhou H, Xia XG, Xu Z. An RNA polymerase II construct synthesizes short-hairpin RNA with a quantitative indicator and mediates highly efficient RNAi. Nucleic Acids Res. 2005;33:e62. doi: 10.1093/nar/gni061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia XG, Zhou H, Samper E, Melov S, Xu Z. Pol II-expressed shRNA knocks down Sod2 gene expression and causes phenotypes of the gene knockout in mice. PLoS Genet. 2006;2:e10. doi: 10.1371/journal.pgen.0020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 52.Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu JY, Taylor J, DeRuiter SL, Vojtek AB, Turner DL. Simultaneous inhibition of GSK3alpha and GSK3beta using hairpin siRNA expression vectors. Mol Ther. 2003;7:228–236. doi: 10.1016/s1525-0016(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 54.Jazag A, Kanai F, Ijichi H, Tateishi K, Ikenoue T, Tanaka Y. Single small-interfering RNA expression vector for silencing multiple transforming growth factor-beta pathway components. Nucleic Acids Res. 2005;33:e131. doi: 10.1093/nar/gni130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia XG, Zhou H, Xu Z. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 56.Shin KJ, Wall EA, Zavzavadjian JR, Santat LA, Liu J, Hwang JI. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Yamada O, Sakamoto T, Yoshida H, Iwai T, Matsushita Y. Down-regulation of viral replication by adenoviral-mediated expression of siRNA against cellular cofactors for hepatitis C virus. Virology. 2004;320:135–143. doi: 10.1016/j.virol.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 58.Korf M, Jarczak D, Beger C, Manns MP, Kruger M. Inhibition of hepatitis C virus translation and subgenomic replication by siRNAs directed against highly conserved HCV sequence and cellular HCV cofactors. J Hepatol. 2005;43:225–234. doi: 10.1016/j.jhep.2005.02.046. [DOI] [PubMed] [Google Scholar]
- 59.Henry SD, van der Wegen P, Metselaar HJ, Tilanus HW, Scholte BJ, van der Laan LJ. Simultaneous targeting of HCV replication and viral binding with a single lentiviral vector containing multiple RNA interference expression cassettes. Mol Ther. 2006;14:485–493. doi: 10.1016/j.ymthe.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 60.Akashi H, Miyagishi M, Yokota T, Watanabe T, Hino T, Nishina K. Escape from the interferon response associated with RNA interference using vectors that encode long modified hairpin-RNA. Mol Biosyst. 2005;1:382–390. doi: 10.1039/b510159j. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe T, Sudoh M, Miyagishi M, Akashi H, Arai M, Inoue K. Intracellular-diced dsRNA has enhanced efficacy for silencing HCV RNA and overcomes variation in the viral genotype. Gene Ther. 2006;13:883–892. doi: 10.1038/sj.gt.3302734. [DOI] [PubMed] [Google Scholar]
- 62.Anderson J, Banerjea A, Akkina R. Bispecific short hairpin siRNA constructs targeted to CD4, CXCR4, and CCR5 confer HIV-1 resistance. Oligonucleotides. 2003;13:303–312. doi: 10.1089/154545703322616989. [DOI] [PubMed] [Google Scholar]
- 63.Lee NS, Rossi JJ. Control of HIV-1 replication by RNA interference. Virus Res. 2004;102:53–58. doi: 10.1016/j.virusres.2004.01.015. [DOI] [PubMed] [Google Scholar]
- 64.Li MJ, Bauer G, Michienzi A, Yee JK, Lee NS, Kim J. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther. 2003;8:196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- 65.Chang LJ, Liu X, He J. Lentiviral siRNAs targeting multiple highly conserved RNA sequences of human immunodeficiency virus type 1. Gene Ther. 2005;12:1133–1144. doi: 10.1038/sj.gt.3302509. [DOI] [PubMed] [Google Scholar]
- 66.ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA Interference: a Multiple shRNA Approach. Mol Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Nishitsuji H, Kohara M, Kannagi M, Masuda T. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J Virol. 2006;80:7658–7666. doi: 10.1128/JVI.00078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hemmings-Mieszczak M, Dorn G, Natt FJ, Hall J, Wishart WL. Independent combinatorial effect of antisense oligonucleotides and RNAi-mediated specific inhibition of the recombinant rat P2X3 receptor. Nucleic Acids Res. 2003;31:2117–2126. doi: 10.1093/nar/gkg322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jarczak D, Korf M, Beger C, Manns MP, Kruger M. Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons. FEBS J. 2005;272:5910–5922. doi: 10.1111/j.1742-4658.2005.04986.x. [DOI] [PubMed] [Google Scholar]
- 70.Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- 71.Unwalla HJ, Li HT, Bahner I, Li MJ, Kohn D, Rossi JJ. Novel Pol II fusion promoter directs human immunodeficiency virus type 1-inducible coexpression of a short hairpin RNA and protein. J Virol. 2006;80:1863–1873. doi: 10.1128/JVI.80.4.1863-1873.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samakoglu S, Lisowski L, Budak-Alpdogan T, Usachenko Y, Acuto S, Di Marzo R. A genetic strategy to treat sickle cell anemia by coregulating globin transgene expression and RNA interference. Nat Biotechnol. 2006;24:89–94. doi: 10.1038/nbt1176. [DOI] [PubMed] [Google Scholar]
- 73.Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 74.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fish RJ, Kruithof EK. Short-term cytotoxic effects and long-term instability of RNAi delivered using lentiviral vectors. BMC Mol Biol. 2004;5:9. doi: 10.1186/1471-2199-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Persengiev SP, Zhu X, Green MR. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barik S. RNAi in moderation. Nat Biotechnol. 2006;24:796–797. doi: 10.1038/nbt0706-796. [DOI] [PubMed] [Google Scholar]
- 78.Wiznerowicz M, Szulc J, Trono D. Tuning silence: conditional systems for RNA interference. Nat Methods. 2006;3:682–688. doi: 10.1038/nmeth914. [DOI] [PubMed] [Google Scholar]
- 79.Unwalla HJ, Li MJ, Kim JD, Li HT, Ehsani A, Alluin J. Negative feedback inhibition of HIV-1 by TAT-inducible expression of siRNA. Nat Biotechnol. 2004;22:1573–1578. doi: 10.1038/nbt1040. [DOI] [PubMed] [Google Scholar]
- 80.Strayer DS, Feitelson M, Sun B, Matskevich AA. Paradigms for conditional expression of RNA interference molecules for use against viral targets. Methods Enzymol. 2005;392:227–241. doi: 10.1016/S0076-6879(04)92014-9. [DOI] [PubMed] [Google Scholar]
- 81.An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- 83.Mi J, Zhang X, Rabbani ZN, Liu Y, Su Z, Vujaskovic Z. H1 RNA polymerase III promoter-driven expression of an RNA aptamer leads to high-level inhibition of intracellular protein activity. Nucleic Acids Res. 2006;34:3577–3584. doi: 10.1093/nar/gkl482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McNamara JO, II, Andrechek ER, Wang Y, Viles KD, Rempel RE. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]