Graphical abstract
Keywords: Barcoding, Chiroptera, Ecology, Evolution, Trypanosomes
Highlights
-
•
We observed the first evidence of TcBat in Colombia.
-
•
Trypanosoma cruzi, Trypanosoma cruzi marinkellei, Trypanosoma dionisii, Trypanosoma rangeli were the most frequent.
-
•
The most frequent DTUs in bats were TcI, TcII, TcIII, TcIV and TcBat.
-
•
Bats play a relevant role in the evolution of T. cruzi.
Abstract
Bats (Chiroptera) are the only mammals naturally able to fly. Due to this characteristic they play a relevant ecological role in the niches they inhabit. These mammals spread infectious diseases from enzootic to domestic foci. Rabbies, SARS, fungi, ebola and trypanosomes are the most common pathogens these animals may host. We conducted intensive sampling of bats from the phyllostomidae, vespertilionidae and emballonuridae families in six localities from Casanare department in eastern Colombia. Blood-EDTA samples were obtained and subsequently submitted to analyses of mitochondrial and nuclear genetic markers in order to conduct barcoding analyses to discriminate trypanosome species. The findings according to the congruence of the three molecular markers suggest the occurrence of Trypanosoma cruzi cruzi (51%), T. c. marinkellei (9%), T. dionisii (13%), T. rangeli (21%), T. evansi (4%) and T. theileri (2%) among 107 positive bat specimens. Regarding the T. cruzi DTUs, we observed the presence of TcI (60%), TcII (15%), TcIII (7%), TcIV (7%) and TcBAT (11%) being the first evidence to our concern of the foreseen genotype TcBAT in Colombia. These results allowed us to propose reliable hypotheses regarding the ecology and biology of the bats circulating in the area including the enigmatic question whether TcBAT should be considered a novel DTU. The epidemiological and evolutionary implications of these findings are herein discussed.
1. Introduction
Bats are considered the only mammals naturally capable of maintaining a stable flight. These organisms correspond to approximately 20% of known mammal species classified to date and divided into two suborders (megachiroptera and microchiroptera) (Teeling et al., 2005). Most bats are considered insectivores, the rest are frugivores, fish-eaters and blood eaters (vampire bats). They play a relevant role in flower pollination and fruit seed dispersal (Hodgkison et al., 2003). Bats are important components of neo-tropical communities as they occupy a large variety of trophic niches and are often the most species-rich and abundant mammals in the area (Dietz and Kalko, 2006, Giannini and Kalko, 2004). Neo-tropical bats such as Carollia, Myotis, Artibeus and Desmodus play a relevant role in neo-tropical ecosystems as they disperse seeds, pollinate flowers and control insect populations (Bernard, 2001, Kunz et al., 2007). The ecology of these mammals highlight their relevance as reservoirs of infectious diseases and zoonotic pathogens due to their high ability of mobility, broad distribution and social behaviour (communal roosting and fission–fusion social structure). Among these pathogens emerge rabies, severe acute respiratory syndrome (SARS), henipavirus, possibly ebola and the trypanosomes (Guyatt et al., 2003, Li et al., 2005, Halpin et al., 2000, Hamilton et al., 2012a, Hamilton et al., 2012b).
Trypanosomes (genus Trypanosoma) are usually transmitted by arthropods or leech vectors. Among the Trypanosoma genus exists four clear clades depending on the host that the parasite infects (aquatic clade, mammalian clade, terrestrial clade and avian clade); within the mammalian clade emerges the Trypanosoma cruzi clade containing T. c. cruzi, T. c. marinkellei, T. dionisii, T. conorhinii, T. rangeli and other mammalian trypanosomes. This clade is of extreme interest in terms of public health; Chagas disease caused by T. c. cruzi represents an important human pathology in the Americas and the heterogeneity displayed by this taxon is exhibited in at least six discrete typing units (DTUs) widely distributed in humans, insect vectors and reservoirs named from T. c. cruzi I to T. c. cruzi VI (TcI–TcVI) with the emergence of one new genotype strictly associated to bat species (Myotis and Noctilio) in Brazil and Panama (Zingales et al., 2009, Zingales et al., 2012, Marcili et al., 2009a, Pinto et al., 2012). This might suggest that host-fitting is the main mechanism of trypanosomes evolution (Hamilton et al., 2012b). Nevertheless, many authors have suggested that the T. cruzi clade is the sister clade of a recently discovered bat-trypanosome species from Africa named T. erneyi supporting what is now named the bat-seeding hypothesis in the diversification of T. cruzi clade (Lima et al., 2012, Hamilton et al., 2012b).
Different studies have attempted to elucidate the prevalence of Trypanosomes in bats observing the occurrence of 3–9% in surveys conducted in South America (Marinkelle, 1976, Dias et al., 1942, Deane, 1961). In neo-tropical bats the most common trypanosome species detected are T. cruzi cruzi, T. c. marinkellei, T. dionisii, T. rangeli and T. conorhinii (Baker et al., 1978, García et al., 2012, Marcili et al., 2009a, Marcili et al., 2009b). In Colombia exists one single survey where T. cruzi-like infections were detected in a considerable number of bat specimens (Marinkelle, 1976). Despite this effort, no conclusive results associating the biology, ecology and evolution of trypanosomes and bats have emerged. In this context, we undertook intensive sampling of neo-tropical bats from an endemic area of Chagas disease in Colombia with a high rate of T. cruzi transmission in order to unravel the distribution of bat trypanosomes in the area with the aim of obtaining reliable information about the transmission dynamics of these protozoans with special focus on elucidating the relevance of this subpatent infections in the biology, evolution and ecology of neo-tropical bats.
2. Materials and methods
2.1. Study area and specimen identification
Bat specimens were captured in six localities across the Casanare department in Eastern Colombia (Fig. 1 ). A total of 175 specimens were captured in Nunchia (3 bats), San Luis de Palenque (45 bats), Yopal (13 bats), Agua Azul (22 bats), Maní (21 bats) and Tamara (71 bats) using nets and procedures permitted by the Ministerio del Medio Ambiente in Colombia obtaining appropiate geographical coordinates of capture (Table S1). Blood samples (200 μL) were collected in EDTA and transferred to Guanidine-EDTA buffer for complete lysis of the blood and preservation of DNA fragments. Bats were identified to species level using DNA barcoding previously developed for neo-tropical bats (Clare et al., 2011).
Fig. 1.
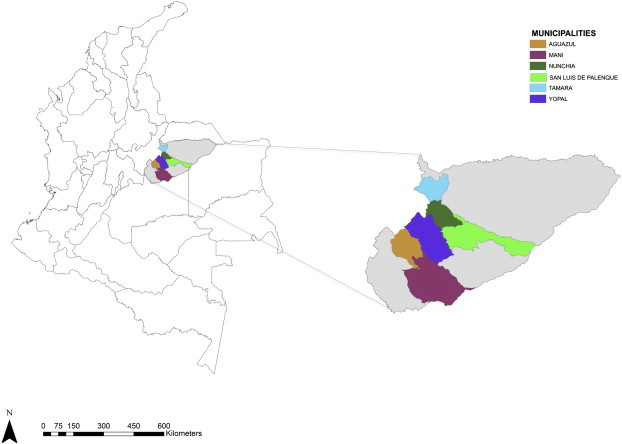
Geographical distribution of the six provinces where bat species were sampled in Eastern Colombia (Nunchía, Yopal, Maní, San Luis de Palenque, Tamará and Agua Azul).
2.2. Molecular identification of trypanosomes species
The blood samples in guanidine buffer (GEB) were submitted for DNA extraction using the Qiamp miniprep kit following the manufactureŕs instructions (Qiagen, Barcelona, Spain). The DNA aliquots were submitted to amplification based on a barcoding approach using three genomic regions; we employed the partial region of cytochrome b gene (cytb), the region V7 of the SSU rDNA 18S gene (SSU rDNA) fragment and gGAPDH as previously established for the discrimination of Trypanosomes species (Marcili et al., 2009a, García et al., 2012). The amplification was accomplished in a final volume of 20 μL using 1× Buffer (Corpogen, COL), 5 mM MgCl2, 1 μM of primers, 10 μM of dNTPs, 0.5U of Taq Tucan (Corpogen, COL) and 20 ng of DNA. The mix was submitted to 29 cycles of amplification, the amplicons were visualised in 2% agarose gels stained with red gel. The PCR products were cleaned up by isopropanol precipitation and sequenced by the dideoxy-terminal method in an automated capillary sequencer (AB3730, Applied Biosystems, UK). The resulting sequences were edited in MEGA 5.0 (Tamura et al., 2011) and aligned using ClustalW 1.8 (Thompson et al., 1994). All edited sequences were deposited in GenBank and assigned accession numbers (KC951574–KC951627). For the identification of Trypanosomes species, the sequences obtained were compared with reference sequences from T. c. cruzi (from the six DTUs and the TcBAT genotype), T. c. marinkellei, T. dionisii, T. conorhinii, T. evansi, T. theileri and T. rangeli retrieved from GenBank.
2.3. Phylogenetic reconstruction
The final set of sequences from each gene fragment was evaluated in ModelTest 3.7 where the most appropriate evolutionary model was selected based on the AIC (Akaike Information Criterion). A maximum composite likelihood (MCL) analysis using a Tamura-3 parameter model and the Neighbour-Joining algorithm was run in RAxML 7.2.5 on the CIPRES project (Cyberinfrastructure for Phylogenetic Research) portal 2.0 servers. Trees were constructed for individual gene fragments in order to unravel the evolutionary drivers among the species detected. To evaluate the robustness of the nodes in the resulting phylogenetic trees, 1000 bootstrap replicates were performed. The final trees were rooted with T. brucei.
3. Results
3.1. Bats species diversity
Neo-tropical bats were captured in six municipalities across the department of Casanare in Colombia where Chagas disease is highly endemic. The results of morphological features and DNA barcoding allowed us to discriminate the bats species diversity as follows: In Nunchia municipality 100% of the bats species sampled was from the species Carollia perspicillata. In Agua azul municipality, four distinct species were sampled observing C. perspicillata (54%), Myotis oxyotus (9%), Desmodus rotundus (23%) and Artibeus planirostris (14%). In Maní municipality we observed the ocurrence of C. perspicillata (43%), M. oxyotus (14%), D. rotundus (10%) and Artibeus planirostris (33%). In San Luis de Palenque municipality we detected C. perspicillata (42%), M. oxyotus (20%), D. rotundus (13%), Rhynchonycteris naso (11%), A. planirostris (8%) and A. fuliginosus (6%). In Tamara municipality we observed C. perspicillata (41%), M. oxyotus (10%), D. rotundus (13%), R. naso (11%), A. planirostris (4%) and A. fuliginosus (21%). Lastly, in Yopal we obtained C. perspicillata (85%) and M. oxyotus (15%) (Fig. 2 ).
Fig. 2.
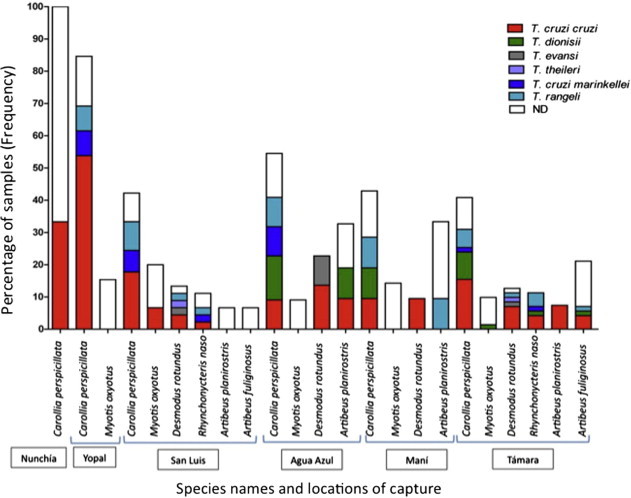
Frequency distribution plot of the trypanosome species detected among the 175 bats sampled according to the geographical area.
3.2. Prevalence of trypanosomes infection
After observing the distribution of bat species we employed the barcoding approach based on three independent molecular markers in different regions of the trypanosome genome. There had to be a consensus among the three molecular makers for the accurate discrimination of trypanosome species. In all the cases the three markers showed congruence. A high prevalence of trypanosome infection was detected among the bats sampled finding a rate of infection of 61.2% across the 175 specimens analysed. In the 107 bats positive for trypanosomes infection the ocurrence of trypanosome species from the subgenus schyzotrypanum and megatrypanum being as T. cruzi cruzi (51%), T. c. marinkellei (9%), T. dionisii (13%), T. rangeli (21%), T. evansi (4%) and T. theileri (2%) was observed. The distribution of trypanosome species infecting each bat species and geographical niche is observed in Fig. 2. Regarding the rate of infection by T. c. cruzi DTUs as previously described, from the 55 bats infected by this trypanosome we observed the presence of TcI (60%), TcII (15%), TcIII (7%), TcIV (7%) and TcBAT (11%), likewise the distribution of T. c. cruzi DTUs infecting each bat species and geographical niche is observed in Fig. 3 .
Fig. 3.
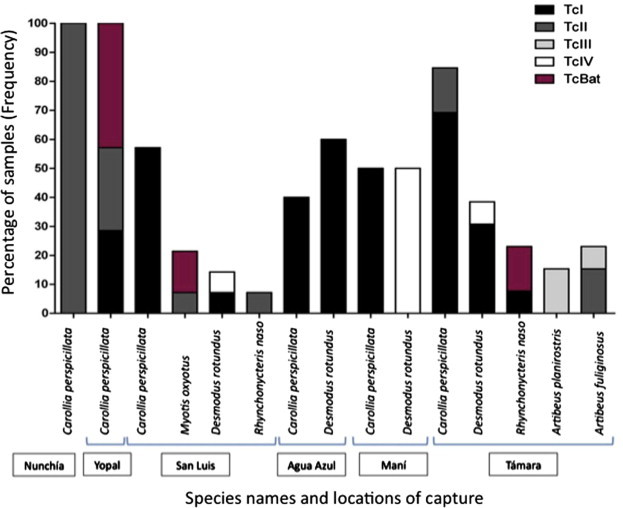
Frequency distribution of the T. c. cruzi genotypes detected among the 175 bats sampled according to the geographical area.
3.3. Phylogenetic relationships of Colombian trypanosomes
Reliable maximum likelihood phylogenetic reconstruction was conducted using the three molecular markers employed (cytb, SSU rDNA and gGAPDH). A total of 8% of the samples showed heterozygous positions in the nuclear gGAPDH loci. Therefore, these sequences were removed from the phylogenetic reconstruction in order to avoid uncertain interpretations regarding the topologies. The topologies of the three markers were congruent. We also employed sequences retrieved from Genbank to detect the genetic relatedness among reference sequences and the sequences obtained from Colombian trypanosomes. The topology of the ML tree corroborated the high affinity of TcBAT to TcI and the status of emergent genotype of TcBAT in the natural history of T. c. cruzi taxon (Fig. 4 ). No outliers were detected on the whole dataset.
Fig. 4.

Maximum Composite Likelihood phylogenetic reconstruction of cytb gene sequences demonstrating the relatedness among the trypanosome species detected and the T. c. cruzi genotypes observed.
4. Discussion
A tailored diversity of trypanosome species from bats was sampled in a relevant area of Casanare department in Colombia. The molecular approach herein reported allowed us to observe a positivity of 61% of infection by trypanosomes from the schyzotrypanum and megatrypanum subgenus. This is a high and not unexpected rate of infection since in similar analyses performed in Panamian bats a rate of infection of 11.6% by T. c. cruzi was detected (Pinto et al., 2012). Also, different surveys in Brazilian bats have shown positivity for trypanosomes of 80% (D’Alessandro et al., 1971, Marinkelle, 1972). In some reports using direct microscopic examination the prevalences range from 2.4% to 9% in Colombian and Brazilian bats (Marinkelle, 1976, Dias et al., 1942, Deane, 1961, Marinkelle and Duarte, 1968). These findings reinforce previous statements showing the important role that bat species play in the transmission of these parasites causing a high risk of pathogen transmission in those areas where bat-human interaction is frequent.
The distribution of the trypanosome species among the bats is strongly linked with the biology and the ecology of these mammals. We sampled bat species that belonged to three distinct families (Phyllostomidae, Vespertilionidae and Emballonuridae). We observed six distinct species after the completion of the survey (Fig. 2). C. perspicillata was the most frequent species in all six provinces surveyed and showed the highest percentage of infection (T. c. cruzi, T. c. marinkellei, T. rangeli and T. dionisii). On the other hand, bats from the genus Myotis, Rhynchonycteris and Artibeus were detected and infected with T. c. cruzi, T. c. marinkellei, T. rangeli and T. dionisii in low proportions. After an exhaustive revision in the literature of the trypanosome species that infect Carollia, Myotis, Rhynchonycteris and Artibeus we observed congruence in the patterns of infection with the ones observed among our dataset (Marinkelle, 1972, García et al., 2012). Moreover, it is necessary to consider the ecology of these bats. These species usually feed on insects, fruits or pollen (Cloutier and Thomas, 1992). It is common that these bats get infected when they ingest insects infected with these trypanosomes. In this sense, triatomine insects are the main hosts of T. c. cruzi, T. c. marinkellei and T. rangeli. Many authors have suggested that Cavernicola pilosa a triatomine bug that can be found on the caves where the bats live is the principal responsible for bat infection (Dias et al., 1942, Marinkelle, 1966, Oliveira et al., 2008). Albeit, C. pilosa has never been found infected with T. rangeli or with T. dionisii.
These bat species usually live on palms of the genus Attalea and Elaeis that are highly infested by Rhodnius prolixus which has been found naturally infected with T. c. cruzi and T. rangeli (Vallejo et al., 2009). This premise also needs to consider the insect vector of T. dionisii where cimids have been detected in the region surveyed. This implies that insectivory life is the best explanation for these bats to be naturally infected. Nevertheless, stercorarian or salivarian and congenital transmission routes cannot be excluded since these bats do inhabit in the canopy of the palms where triatomine bugs may transmit T. rangeli, but further studies are required to ensure this hypothesis. Additionally, recent studies in Venezuelan bats have demonstrated that in Molossus colossus; T. c. cruzi may be transmitted to the offspring suggesting a high risk of kinetoplastid transmission (Añez et al., 2009).
One interesting finding is the ocurrence of T. evansi and T. theileri in merely one bat species. Desmodus rotundus was found infected with T. c. cruzi, T. rangeli, T. theileri and T. evansi; we find it relevant to highlight this bat species since it is the unique hematophagous species sampled and collected in the dwellings of houses. In terms of ecology, the route of infection of this bat species could be double; Bats may acquire T. evansi and T. theileri by blood ingestion from cattle and/or vectorial transmission (Family Tabaniidae). To our concern this is the first report of T. theileri in Colombian bats and is not unexpected since different surveys conducted in the area have demonstrated the presence of these trypanosomes in cattle (Wells et al., 1982, Cassalett et al., 2011). Otherwise, infection by T. rangeli and T. c. cruzi has also been reported in bovines but in a very low proportion (Fujita et al., 1994). The question raises whether the merely unique mechanism for acquisition of infection is by blood ingestion of cattle or if maybe while this bat species inhabit the palms, caves or others, some triatomines may transmit T. c. cruzi/T. rangeli by stercorarian/salivarian routes. Lastly, it is interesting to consider that T. c. cruzi was the most frequent trypanosome species in D. rotundus which implies a tremendous risk of parasite transmission even though knowing per se the high antropophilic behaviour this bat species display.
T. c. cruzi is the agent of Chagas disease, a tropical pathology that affects more than 10 million people in Latin-America (Rassi et al., 2012). Mammals from 100 different orders have been described as potential reservoirs of T. c. cruzi suggesting that this zoonosis cannot be eradicated (Jansen and Roque, 2010). Neo-tropical bats play a relevant role as reservoirs of this parasite as demonstrated by plenty of studies across the continent (Añez et al., 2009, Marcili et al., 2009a, Herrera et al., 2011, García et al., 2012, Marinkelle, 1976). We observed that the most frequent hemoflagellate in all the bats sampled was T. c. cruzi, which corroborates the important epidemiological role of this mammal (Fig. 2). Likewise, we conducted an accurate discrimination of T. c. cruzi DTUs observing the predominance of TcI with the foreseen presence of TcII, TcIII, TcIV and the first report of TcBAT in Colombia (Fig. 3). This is in accordance with the predominance of TcI in humans, reservoirs and insect vectors from the region (Ramírez et al., 2012). This information is of paramount relevance in terms of ecology and phylodynamics of these DTUs; TcI, TcIII and TcIV are mainly associated to the sylvatic cycle of transmission with sporadic reports in domestic foci. Additionally, studies in Brazil have demonstrated that bats are mainly infected by TcI, TcIII and TcIV (Marcili et al., 2009b, Lisboa et al., 2008, Zingales et al., 2012). The interesting findings overcome in the detection of TcII that has been associated to the domestic cycle of transmission with sporadic reports in sylvatic reservoirs (Yeo et al., 2005, Zingales et al., 2012). Despite of the ecological niche that TcII occupies, this DTU has been extensively reported in Brazilian bats from the species Carollia and Noctilio (Phyllostomatidae) (Marcili et al., 2009a, Lisboa et al., 2008). Nevertheless, an emergent genotype previously named as TcBAT was detected in our dataset; in this sense the infection by this genotype was observed in Carollia, Myotis and Rhynchonycteris bats, which is in accordance with previous reports (Marcili et al., 2009a). This suggests that TcBAT is not only encountered in Brazil and Panama (Pinto et al., 2012). Also, the ability of this genotype to infect mammalian cells in vitro strongly suggest the importance of conducting studies to unravel the genetic properties that this genotype displays and whether this genotype should be assigned as a novel DTU (Maeda et al., 2011).
The orthodox overlook of T. c. cruzi DTUs in bat species suggests the importance of these reservoirs in the natural cycle of the parasite. Some authors suggest that marsupials were the first hosts of the T. cruzi taxon but the recent advances in the evolution of bats have allowed some authors to propose the ‘bat-seeding’ hypothesis suggesting that the fact that bat fossils were discovered about 55 mya might imply that the first reservoirs of T. c. cruzi could have been bats; also supported by the speciation processes of trypanosomes and lastly explained by the migration of these mammals (Hamilton et al., 2012a, Hamilton et al., 2012b, Teeling et al., 2005). As herein demonstrated, bats play a paramount role in the dispersion not only of T. c. cruzi DTUs. These mammals may disperse many trypanosome species and the host-fitting features could determine the emergence of new cryptic species, as is the case of TcBAT genotype. The relatively recent arrival of bats to South America and their mobility by flight, suggests that T. cruzi like trypanosomes of bats are derived from either T. cruzi I or T. cruzi II (Z2)/Z3 by habitat sharing or, for insectivorous bats, by consumption of infected triatomine bugs (Gaunt and Miles, 2000). This premise needs to be fully studied and corroborated with the aim of unravelling the natural history of T. c. cruzi.
In summary we conducted, to our knowledge, the first molecular descriptive study of bat trypanosomes in Colombia with direct detection of the parasites from EDTA-blood. The distribution of the trypanosomes identified implies that an intensive and active transmission of parasites is occurring in the region and especially among bat populations. This represents important epidemiological implications with special focus on Chagas disease since this area is endemic for this pathology in Colombia. Also, foreseen trypanosome species were detected with novel reports of T. theileri and TcBAT. Likewise, evolutionary trends could be ruled out observing that T. erneyi was corroborated as the plausible sister clade of T. cruzi and bats as the first and natural reservoirs of Chagas disease in the Americas. The TcBAT genotype needs to be screened in the entire American continent where Chagas disease is endemic to obtain conclusive premises regarding the ecology and distribution of this genotype. Lastly, we suggest conducting new studies in order to unravel the population genetics of TcBAT, schyzotrypanum and megatrypanum subgenus in order to obtain robust and reliable information about the biology of trypanosomes in Colombia as suggested by C.J. Marinkelle in the last decades.
Acknowledgments
This work is dedicated to the memory of Prof. Cornellis Johannes Marinkelle founder of CIMPAT and pioneer in the study of tropical medicine and bat trypanosomes whom died on 18th January of 2012. Financial support was provided by the Project Chagas EpiNet from The European Union Seventh Framework Programme, Contract No. 223034.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2013.06.022.
Appendix A. Supplementary data
Information about the geographical origin of the bats species analysed.
References
- Añez N., Crisante G., Soriano P.J. Trypanosoma cruzi congenital transmission in wild bats. Acta Trop. 2009;109:78–80. doi: 10.1016/j.actatropica.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Baker M.M., Godfrey D.G., Barrett T.V. Biochemical characterization of some species of Trypanosoma, schizotrypanum from bats, Microchiroptera. Am. J. Trop. Med. Hyg. 1978;27:483–491. doi: 10.4269/ajtmh.1978.27.483. [DOI] [PubMed] [Google Scholar]
- Bernard E. Species list of bats, mammalia, chiroptera of santaram area, pará State. Brazil Rev. Bras. Zoolog. 2001;18:455–463. [Google Scholar]
- Cassalett V.J., Parra J.L., Baldrich R.M. Diagnóstico y caracterización molecular de infecciones naturales por Trypanosoma spp. en bovinos de la orinoquía colombiana. Corpoica Ciencia Tecnología Agropecuaria. 2011;12:86–91. [Google Scholar]
- Clare E.L., Lim B.K., Fenton M.B., Hebert P.D.N. Neo-tropical bats: estimating species diversity with DNA barcodes. PLoS One. 2011;6:e22648. doi: 10.1371/journal.pone.0022648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier D., Thomas D.W. Carollia perspicillata. Mammalian Species. 1992;417:1–9. [Google Scholar]
- D’Alessandro A., Barreto B., Duarte R.C.A. Distribution of triatominae-transmitted trypanosomiasis in Colombia and new records of the bugs and infections. J. Med. Entomol. 1971;8:159–172. doi: 10.1093/jmedent/8.2.159. [DOI] [PubMed] [Google Scholar]
- Deane L. Tripanossomídeos de mamíferos da Regiao Amazonica I. Alguns flagelados encontrados no sangue de mamíferos silvestres do Estado do Pará. Rev. Inst. Med. Trop. Sao Paulo. 1961;3:5. [Google Scholar]
- Dias E.M.G., Costa D., Damasceno R., Azevedo M. Investigacoes sobre esquizotripanose de morcegos no Estado do Pará. Encontro do barbeiro “Carvenicola pilosa” como transmisor. Rev. Bras. Biol. 1942;2:03–110. [Google Scholar]
- Dietz M., Kalko E. Seasonal changes in daily torpor patterns of free-ranging female and male Daubenton’s bats, Myotis daubentonii. J. Com. Phys. 2006;176:223–231. doi: 10.1007/s00360-005-0043-x. [DOI] [PubMed] [Google Scholar]
- Fujita O., Sanabria L., Inchaustti A., De Arias A.R., Tomizawa Y., Oku Y. Animal reservoirs for Trypanosoma cruzi infection in an endemic area in Paraguay. J. Vet. Med. Sci. 1994;56:305–308. doi: 10.1292/jvms.56.305. [DOI] [PubMed] [Google Scholar]
- García L., Ortiz S., Osorio G., Torrico M.C., Torrico F., Solari A. Phylogenetic analysis of bolivian bat trypanosomes of the subgenus schizotrypanum based on cytochrome b sequence and minicircle analyses. PLoS One. 2012;7:e36578. doi: 10.1371/journal.pone.0036578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt M., Miles M. The ecotopes and evolution of triatomine bugs, triatominae and their associated trypanosomes. Mem. Inst. Oswaldo Cruz. 2000;95:557–565. doi: 10.1590/s0074-02762000000400019. [DOI] [PubMed] [Google Scholar]
- Giannini N.P., Kalko E.K.V. Trophic structure in a large assemblage of phyllostomid bats in Panama. Oikos. 2004;105:209–220. [Google Scholar]
- Guyatt K.J., Twin J., Davis P., Holmes E.C., Smith G.A., Smith I.L., Mackenzie J.S., Young P.L. A molecular epidemiological study of Australian bat lyssavirus. J. Gen. Virol. 2003;84:485–496. doi: 10.1099/vir.0.18652-0. [DOI] [PubMed] [Google Scholar]
- Halpin K., Young P.L., Field H.E., Mackenzie J.S. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 2000;81:1927–1932. doi: 10.1099/0022-1317-81-8-1927. [DOI] [PubMed] [Google Scholar]
- Hamilton P.B., Cruickshank C., Stevens J.R., Teixeira M.M.G., Mathews F. Parasites reveal movement of bats between the new and old worlds. Mol. Phyl. Evol. 2012;63:521–526. doi: 10.1016/j.ympev.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton P.B., Teixeira M.M.G., Stevens J.R. The evolution of Trypanosoma cruzi: the bat-seeding hypothesis. Trends Parasitol. 2012;28:136–141. doi: 10.1016/j.pt.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Herrera H.M., Rocha F.L., Lisboa C.V., Rademaker V., Mouro G.M., Jansen A.M. Food web connections and the transmission cycles of Trypanosoma cruzi and Trypanosoma evansi, kinetoplastida, trypanosomatidae in the pantanal region. Brazil Trans. R Soc. Trop. Med. Hyg. 2011;105:380–387. doi: 10.1016/j.trstmh.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Hodgkison R., Balding S.T., Zubaid A., Kunz T.H. Fruit bats, chiroptera: pteropodidae as seed dispersers and pollinators in a lowland Malaysian rain forest. Biotropica. 2003;35:491–502. [Google Scholar]
- Jansen A.M., Roque A.L.R. 11 – Domestic and wild mammalian reservoirs. In: Jenny T., Michel T., editors. American Trypanosomiasis. Elsevier; London: 2010. pp. 249–276. [Google Scholar]
- Kunz T.H., Arnett E.B., Cooper B.M., Erickson W.P., Larkin R.P., Mabee T., Morrison M.L., Strickland M.D., Szewczak J.M. Assessing impacts of wind-energy development on nocturnally active birds and bats: a guidance document. J. Wild. Manage. 2007;71:2449–2486. [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of sars-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Lima L., Maia da Silva F., Neves L., Attias M., Takata C.S.A., Campaner M., de Souza W., Hamilton P.B., Teixeira M.M.G. Evolutionary insights from bat trypanosomes: morphological, developmental and phylogenetic evidence of a new species, Trypanosoma Schizotrypanum erneyi sp. nov., in African bats closely related to trypanosoma, schizotrypanum cruzi and allied species. Protist. 2012;163:856–872. doi: 10.1016/j.protis.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Lisboa C.V., Pinho A.P., Herrera H.M., Gerhardt M., Cupolillo E., Jansen A.M. Trypanosoma cruzi, kinetoplastida, trypanosomatidae genotypes in neo-tropical bats in Brazil. Vet. Parasitol. 2008;156:314–318. doi: 10.1016/j.vetpar.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Maeda F.Y., Alves R.M., Cortez C., Lima F.M., Yoshida N. Characterization of the infective properties of a new genetic group of Trypanosoma cruzi associated with bats. Acta Trop. 2011;120:231–237. doi: 10.1016/j.actatropica.2011.09.001. [DOI] [PubMed] [Google Scholar]
- Marcili A., Lima L., Cavazzana M.J., Junqueira A.C.V., Veludo H.H., Maia da Silva F., Campaner M., Paiva F., Nunes V.L.B., Teixeira M.M.G. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitology. 2009;136:641–655. doi: 10.1017/S0031182009005861. [DOI] [PubMed] [Google Scholar]
- Marcili A., Lima L., Valente V.C., Valente S.A., Batista J.S., Junqueira A.C.V., Souza A.I., da Rosa J.A., Campaner M., Lewis M.D., Llewellyn M.S., Miles M.A., Teixeira M.M.G. Comparative phylogeography of Trypanosoma cruzi TCIIc: new hosts, association with terrestrial ecotopes, and spatial clustering. Infect. Genet. Evol. 2009;9:1265–1274. doi: 10.1016/j.meegid.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Marinkelle C.J. Observations on human, monkey and bat trypanosomes and their vectors in Colombia. Trans. R Soc. Trop. Med. Hyg. 1966;60:109–116. [Google Scholar]
- Marinkelle C.J., Duarte R. Trypanosoma pifanoin. sp. from Colombian bats. J. Euk. Microbiol. 1968;15:621–627. doi: 10.1111/j.1550-7408.1968.tb02182.x. [DOI] [PubMed] [Google Scholar]
- Marinkelle C.J. Colombian triatominae and their infestation with trypanosomatid flagellates. Mitt. Inst. Colombo-Alemán Invest. Qentif Colombia. 1972;6:13–29. [Google Scholar]
- Marinkelle C.J. Biology of the trypanosomes of bats. In: Lumsden W.H.R., Evans D.A., editors. Vol. 1. Academic Press; London: 1976. pp. 175–216. (Biology of the Kinetoplastida). [Google Scholar]
- Oliveira R.L.F., Carneiro M.A., Diotaiuti L. Ecology of Cavernicola pilosa Barber, 1937, hemiptera: reduviidae: triatominae in the boa esperanca cave, Tocantins, Brazil. Econtropica. 2008;14:63–68. [Google Scholar]
- Pinto C.M., Kalko E.K.V., Cottontail I., Wellinghausen N., Cottontail V.M. TcBat a bat-exclusive lineage of Trypanosoma cruzi in the Panama Canal Zone, with comments on its classification and the use of the 18S rRNA gene for lineage identification. Infect. Genet. Evol. 2012;12:1328–1332. doi: 10.1016/j.meegid.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Ramírez J.D., Guhl F., Messenger L., Lewis M., Montilla M., Cucunubá Z., Miles M., Llewellyn M. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol. Ecol. 2012;21:4216–4226. doi: 10.1111/j.1365-294X.2012.05699.x. [DOI] [PubMed] [Google Scholar]
- Rassi A., Jr., Rassi A., Marcondes de Rezende J. American trypanosomiasis, chagas disease. Infect. Dis. Clin. N. Am. 2012;26:275–291. doi: 10.1016/j.idc.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling E.C., Springer M.S., Madsen O., Bates P., O’Brien S.J., Murphy W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580–584. doi: 10.1126/science.1105113. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo G.A., Guhl F., Schaub G.A. Triatominae-Trypanosoma cruzi/T. rangeli: Vector-parasite interactions. Acta Trop. 2009;110:137–147. doi: 10.1016/j.actatropica.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Wells E., Ramirez L., Betancourt A. Trypanosoma vivax in Colombia: interpretation of field results. Trop. Anim. Health Prod. 1982;14:141–150. doi: 10.1007/BF02242144. [DOI] [PubMed] [Google Scholar]
- Yeo M., Acosta N., Llewellyn M., Sánchez H., Adamson S., Miles G.A.J., López E., González N., Patterson J.S., Gaunt M.W., de Arias A.R., Miles M.A. Origins of Chagas disease: didelphis species are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int. J. Parasitol. 2005;35:225–233. doi: 10.1016/j.ijpara.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Zingales B., Andrade S., Briones M., Campbell D., Chiari E., Fernandes O., Guhl F., Lages-Silva E., Macedo A., Machado C., Miles M., Romanha A., Sturm N., Tibayrenc M., Schijman A. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI–TcVI. Mem. Inst. Oswaldo Cruz. 2009;104:1051–1054. doi: 10.1590/s0074-02762009000700021. [DOI] [PubMed] [Google Scholar]
- Zingales B., Miles M.A., Campbell D.A., Tibayrenc M., Macedo A.M., Teixeira M.M.G., Schijman A.G., Llewellyn M.S., Lages-Silva E., Machado C.R., Andrade S.G., Sturm N.R. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012;12:240–253. doi: 10.1016/j.meegid.2011.12.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Information about the geographical origin of the bats species analysed.


