Abstract
Rift Valley fever virus (RVFV) is an emergent arthropod-borne zoonotic infectious viral pathogen which causes fatal diseases in the humans and ruminants. Currently, no effective and licensed vaccine is available for the prevention of RVFV infection in endemic as well as in non-endemic regions. So, an immunoinformatics-driven genome-wide screening approach was performed for the identification of overlapping CD8 + and CD4 + T-cell epitopes and also linear B-cell epitopes from the conserved sequences of the nucleocapsid (N) and glycoprotein (G) of RVFV. We identified overlapping 99.39% conserved 1 CD8 + T-cell epitope (MMHPSFAGM) from N protein and 100% conserved 7 epitopes (AVFALAPVV, LAVFALAPV, FALAPVVFA, VFALAPVVF, IAMTVLPAL, FFDWFSGLM, and FLLIYLGRT) from G protein and also identified IL-4 and IFN-γ induced (99.39% conserved) 1 N protein CD4 + T-cell epitope (HMMHPSFAGMVDPSL) and 100% conserved 5 G protein CD4 + T-cell epitopes (LPALAVFALAPVVFA, PALAVFALAPVVFAE, GIAMTVLPALAVFAL, GSWNFFDWFSGLMSW, and FFLLIYLGRTGLSKM). The overlapping CD8 + and CD4 + T-cell epitopes were bound with most conserved HLA-C*12:03 and HLA-DRB1*01:01, respectively with the high binding affinity (kcal/mol). The combined population coverage analysis revealed that the allele frequencies of these epitopes are high in endemic and non-endemic regions. Besides, we found 100% conserved and non-allergenic 2 decamer B-cell epitopes, GVCEVGVQAL and RVFNCIDWVH of G protein had the sequence similarity with the nonamer CD8 + T-cell epitopes, VCEVGVQAL and RVFNCIDWV, respectively. Consequently, these epitopes may be used for the development of epitope-based peptide vaccine against emerging RVFV. However, in vivo and in vitro experiments are required for their efficient use as a vaccine.
Keywords: Rift Valley fever virus, Immunoinformatics, Peptide vaccine, CD8 + T-cell epitope, CD4 + T-cell epitope
Graphical abstract
Highlights
-
•
In total 21 conserved regions were identified from 163 N and 40 G protein of RVFV.
-
•
Immunoinformatics tools were used for CD8 +, CD4 + and B-cell epitopes identification.
-
•
Overlapping 8 CD8 + & 6 IL-4 & IFN-γ induced CD4 + T-cell epitopes were identified.
-
•
High percentage of population coverage was found in endemic & non-endemic regions.
-
•
Docking simulation showed high binding affinity between epitopes and HLA-alleles.
1. Introduction
Rift Valley fever (RVF) is an emergent arthropod-borne zoonotic infectious viral disease caused by the Bunyavirus (genus Phlebovirus) Rift Valley fever virus (RVFV) which is characterized by the hemorrhagic fever, visual impairment, delayed onset encephalitis, neurological disorder, and photophobia in human and mass abortion, hemorrhage, and hepatitis in sheep, cattle, and other ruminants (Hartman, 2017, Linthicum et al., 2016, Rolin et al., 2013) RVFV presents serious threats to global public health and agriculture in parts of Africa, Madagascar, and the Middle East that is meticulously associated with high-rainfall conditions (Davies et al., 1985, Linthicum et al., 2016). RVFV first identified in Rift valley in Kenya in 1930 as a member of the Bunyaviridae family, which is transmitted to the human through infected mosquito vectors as well as through aerosolization, but the initial transmission of the virus to human is thought to be via mucous membrane revelation or inhalation of viral particles during the management of infected animals (Anyangu et al., 2010, Daubney et al., 1931, Hartman, 2017, Ikegami and Makino, 2011).
RVFV is considered as a potential bioterrorism agent by the United States government due to its virulence and the potential for rapid spread (Borio et al., 2002, Mandell and Flick, 2011) and also categorized as a category-A priority pathogen by the National Institute of Allergy and Infectious Diseases (NIAID). The World Organization for Animal Health (OIE) specified the RVFV as a high consequence pathogen (Mandell and Flick, 2010), while the United States Department of Agriculture, Animal and Plant Health Inspection Service (USDA, APHIS) designated the RVFV as the third most dangerous animal threat (Rolin et al., 2013).
The genome of RVFV is a tri-segmented (Large (L), Medium (M) and Small (S)) negative sense, single stranded RNA (ssRNA) encoding seven proteins (Bouloy and Weber, 2010). The L segment encodes a single protein, RNA-dependent RNA polymerase (L protein), while the M segment codes for the precursor for two glycoproteins (Gc and Gn), as well as two major non-structural proteins, the 78-kDa NSm1 and the 14-kDa NSm2. The S segment encodes a structural protein, the nucleocapsid protein (N), as well as a nonstructural protein (NSs) (Ikegami et al., 2005, Won et al., 2006, Won et al., 2007). Both L and N proteins are indispensable for viral RNA replication and transcription, while Gc and Gn protein make up the protein component of the viral envelope as a glycoprotein. The nonstructural proteins, NSs play a key role in the pathogenesis of the diseases and have been recognized as a virulence factor because of its ability to inhibit the host's immune response by thwarting the antiviral interferon (IFN) response (Billecocq et al., 2004, Bouloy et al., 2001, Ikegami, 2009).
At the beginning of the 21st century, a severe RVFV outbreak in the human happened for the first time in Yemen and Saudi Arabia, in which about 1087 suspected cases were reported, including 121 deaths (Balkhy and Memish, 2003). The biggest outbreaks occurred in Kenya, Somalia, and Tanzania including 234, 51, and 109 deaths, respectively in 2006 (Anyamba et al., 2010, Nguku et al., 2010). The severe form of human outbreaks also ensued in Egypt in 2003, Sudan in 2007 and 2010 (Hassan et al., 2011), Madagascar in 2008, South Africa and Namibia from 2009 to 2011 (Métras et al., 2012, Monaco et al., 2013), Mauritania in 2010 and Mauritania and Senegal from 2012 to 2015 (Sow et al., 2014, Sow et al., 2016), and also Niger and Uganda in 2016 with 1220 confirmed human deaths and > 0.5 million estimated human cases (Dar et al., 2013, Hartman, 2017). However, due to the higher fatality rates, RVFV could be a greater threat to the public health, especially in non-endemic regions (Xu et al., 2013). To date, no licensed vaccine or approved therapeutics is available.
Vaccination is considered as the most effective approaches to avert virus infection, especially in the absence of effective treatment drugs. So, continuous outbreak of RVFV in different endemic and non-endemic regions highlight the exigent need for the development safe and effective vaccine. Among the seven RVFV proteins, the recent development focused on the viral nucleocapsid (N) and glycoprotein (G) based subunit vaccine that elicited robust virus neutralizing antibody responses in RVFV infected sheep (Faburay et al., 2014, Faburay et al., 2016, López-Gil et al., 2013, Pichlmair et al., 2010, van Vuren et al., 2007), and therefore, these two viral proteins are deliberated to be the promising target for effective RVFV vaccine design. Besides, several formalin-inactivated whole virus vaccine (Randall et al., 1964, Rusnak et al., 2011), live-attenuated vaccines (Clone 13 and MP-12) (Dungu et al., 2010, Muller et al., 1995), DNA vaccines (Lagerqvist et al., 2009, Spik et al., 2006), Baculovirus expressed protein-based vaccines (Faburay et al., 2014, Schmaljohn et al., 1989), Virus like particles (Mandell et al., 2010, Näslund et al., 2009), and virus-vectored vaccines (Kortekaas et al., 2012, Wallace et al., 2006) were generated and tested on animals for the effectiveness, but these vaccines, except MP-12 did not get license due to the lack of potential safety and efficacy limitation (Ikegami, 2017).
T-cell, especially CD4 + T-cell based cellular immunity is essential for the clearance of RVFV from the peripheral tissues because of its robust IgG and neutralizing antibody responses (Dodd et al., 2013). So, the most conserved target that stimulates both cellular immunity and neutralizing antibodies against RVFV is indispensable for an effective vaccine development. Epitopes are mostly recognized as the antigenic determinants that represent the immunogenic portion of the protein antigen and provoke precise immune responses (Shi et al., 2015, Vivona et al., 2008). Several research findings revealed that epitope-based peptide vaccine could efficiently elicit defensive immune responses against miscellaneous pathogens, such as malaria (Oyarzún and Kobe, 2016), influenza virus (Staneková and Varečková, 2010), hepatitis B virus, hepatitis C virus (Firbas et al., 2006, He et al., 2015, Sominskaya et al., 2010), and HIV (human immunodeficiency virus) (Jin et al., 2009). However, In spite of the availability of the genome sequence in the GenBank database for emerging infectious pathogen, like RVFV, the immunity accompanying with protection is now largely unknown to the researchers. So, the development of a vaccine against this infectious pathogen is more difficult and challenging work. Therefore, immunoinformatics study could be an effective approach for the screening of dominant immunogens from the available genome sequence data for effective vaccine design against emerging infectious and neglected tropical disease (Terry et al., 2015).
In this study, we used immunoinformatics-driven vaccine targets screening approaches for the identification of different epitope vaccine candidates, like CD4 + T-cell, CD8 + T-cell, and B-cell that could elicit defensive humoral and cellular immune responses. The predicted results provided new T-cell and B-cell epitopes as vaccine candidates for the development of RVFV vaccine and indicated that immunoinformatics-driven dominant immunogen identification is a promising approach for accelerating vaccine development against emerging infectious diseases.
2. Materials and methods
A flow chart representing the overall procedures of epitope-based peptide vaccine target identification for RVFV N and G protein has been illustrated in Fig. 1 .
Fig. 1.
Schematic representation of the overall procedures of the vaccine target identification for RVFV nucleocapsid (N) and glycoprotein (G).
2.1. Retrieving protein sequences and identification of conserved sequences
The sequences of N and G protein of different isolates of RVFV were retrieved from the ViPR database (Virus Pathogen Database and Analysis Resource) (http://www.viprbrc.org/), an integrated, exposed Bioinformatics database and powerful analysis resource for several virus families and their respective species (Pickett et al., 2012). The Fasta formatted protein sequences were retrieved from the ViPR database. The retrieved sequences were from different countries of origin, such as Kenya, South Africa, Central African Republic, Tanzania, Madagascar, Mauritania, China, Angola, Uganda, Saudi Arabia, Sudan, Egypt, Somalia, Zambia, Namibia, and Zimbabwe from the year 1944 to 2016.
The retrieved protein sequences were subjected to the CLUSTALW server (http://www.genome.jp/tools-bin/clustalw) (Thompson et al., 2002), for attaining the multiple sequence alignment (MSA). The MSA was performed with default parameters. We selected those conserved regions of N and G protein, which have minimum 15 amino acid residues, as both CD8 + T-cell epitopes (MHC-I binding epitopes) and CD4 + T-cell epitopes (MHC-II binding epitopes) predicted from the conserved sequences. These sequences were primarily selected for the antigenicity and transmembrane helix property prediction.
2.2. Prediction of transmembrane and antigenicity features of the conserved sequences
The antigenicity is the ability of an antigen that is recognized by and interacts with an immunologically specific T-cell receptor or antibody. In this study, the antigenicity of the conserved sequences of N and G protein was anticipated by the VaxiJen v2.0 Server, an alignment independent protective antigen, tumor antigens and subunit vaccines prediction server (Doytchinova and Flower, 2007). The virus was selected as the target organism and the threshold value was set at default parameter.
On the other hand, the TMHMM v2.0 server (http://www.cbs.dtu.dk/services/TMHMM/) was used for the prognosis of transmembrane protein topology property from the antigenic conserved sequences of N and G protein. TMHMM was developed based on the hidden Markov model and it has been esteemed best in an independent evaluation of programs for prediction of transmembrane helices (Krogh et al., 2001, Moller et al., 2001). However, this server was used in order to scrutinize the soluble and membrane protein sequences among all the conserved sequences of N and G protein with both specificity and sensitivity better than 99% (Krogh et al., 2001).
2.3. Prediction of CD8 + T-cell epitopes and their MHC-I binding alleles
The T-cell epitopes are typically peptide fragments which are immunodominant and central to the control and development of precise immune responses, important for epitope-based peptide vaccine design. So, the immunoinformatics tool NetCTL v1.2 was introduced to prophesy nonamers that have the capacity to bind major histocompatibility complex (MHC) class I (HLA alleles) molecules (Larsen et al., 2007). NetCTL server can predict CD8 + T-cell epitopes for 12 supertypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58 and B62) of MHC class I, taking into account Proteasomal C terminal cleavage and MHC class I binding, TAP transport efficiency, whereas MHC class I binding & proteasomal C terminal cleavage and TAP transport efficiency is predicted using artificial neural networks and weight matrix, respectively (Larsen et al., 2007). In this study, the threshold was set at 0.5 for epitope identification which has a sensitivity and specificity of 0.89 and 0.940, respectively. The weight on C terminal cleavage and TAP transport efficiency used as default parameters.
For the identification of frequently occurring MHC class I binding alleles, the epitopes were analyzed by the Stabilized Matrix Base Method (SMM) in the IEDB analysis tool (http://tools.iedb.org/mhci/) (Peters and Sette, 2005). The amino acid length of peptide is 9.0, the IC50 value < 250, and the human as MHC source species was selected as parameters for the identification of MHC-I binding alleles.
2.4. CD4 + T-cell epitopes and their MHC-II binding allele prediction
The MHC class II complex molecule puts forward the peptides to CD4 + T helper lymphocytes (HTLs) that can induce cellular and humoral immunity against the pathogenic micro-organism or foreign antigen. So, the prediction of peptides that bind to MHC class II molecules can play an important role in rational vaccine design (Nielsen et al., 2007). The CD4 + T helper lymphocyte (peptide) and their corresponding MHC class II alleles were predicted through the IEDB MHC II binding tool (http://tools.iedb.org/mhcii/). Among seven peptide prediction method, the novel SMM-align (stabilization matrix alignment) method was used for the direct prediction of peptide and MHC II binding alleles from the conserved sequences of N and G protein of RVFV (Nielsen et al., 2007). The SMM-align predicts peptides and their core sequences binding to 5 Human HLA-DP, 6 HLA-DQ, and 15 HLA-DR alleles. For the identification of CD4 + T-cell epitopes and corresponding HLA class-II alleles with good affinity, the threshold value for IC50 (minimum inhibitory concentration of a biological substance) was taken as < 250 nM in the SMM-align method.
2.5. Immunogenicity, conservancy and allergenicity prediction of the CD8 + and CD4 + T-cell epitopes
Immunogenicity states the capability of an antigen to provoke an immune response against the pathogenic micro-organism aiming at protecting the human or animal being. Immunogenicity is mainly associated with vaccine development and it is considered as the center of the vaccine adeptness (Leroux-Roels et al., 2011). So, the immunogenicity of the predicted CD8 + T-cell epitopes was observed by the MHC I immunogenicity tool of IEDB (http://tools.iedb.org/immunogenicity/) (Calis et al., 2013). Conserved epitope identification is an essential criterion in epitope-based vaccine design due to the broader protection against several strains of the selected species. Due to the greater importance of epitope conservancy, the CD8 + and CD4 + T-cell epitopes were subjected to the IEDB conservation across antigen tool (http://tools.iedb.org/conservancy/) for epitope conservancy prediction (Bui et al., 2007). The allergenicity was anticipated through the AllerHunter web-server (http://tiger.dbs.nus.edu.sg/AllerHunter/). AllerHunter is a Support Vector Machine (SVM) and pairwise sequence similarity based cross-reactive allergen prediction program (Muh et al., 2009).
2.6. Identification of IFN-γ and IL-4 inducer property of the CD4 + T-cell epitopes
The provocation of T-helper cells is crucial for the development of a vaccine or immunotherapy. The CD4 + T-cell epitopes can stimulate the T-helper cells by producing different types of cytokines such as interferon-gamma (IFN-γ) and interleukin-4 (IL-4). As a result, they are important for combating intracellular pathogens, including viruses, bacteria, and parasites. So, we subjected the promiscuous CD4 + T-cell epitopes (peptides) to check, whether predicted epitopes are IFN-γ and IL-4 inducer or non-inducer in nature. In this study, the IFN-γ inducing CD4 + T-cell epitopes was predicted by the IFNepitope tool (http://crdd.osdd.net/raghava/ifnepitope/), an in silico server for predicting and designing interferon-gamma inducing epitopes (Dhanda et al., 2013). The SVM (support vector machine) based method and the IFN-γ versus non IFN-γ model was set as parameters for the prediction of the nature of epitopes. On the other hand, the IL-4 inducer was predicted by the IL4pred tool (http://crdd.osdd.net/raghava/il4pred/), an in silico platform for designing and discovering of interleukin-4 inducing peptides (Dhanda et al., 2013). For the prediction of this nature, the SVM based model was used, whereas the threshold value was set at 0.1 as a parameter of the SVM model.
2.7. Assessment of HLA-alleles distribution
Different HLA alleles are expressed at dramatically diverse frequencies in diverse ethnicities. Consequently, the HLA-alleles distribution among the world population is crucial for a successful peptide-based vaccine development (Bui et al., 2006). In this study, the HLA-alleles distribution of the predicted both CD8 + and CD4 + T-cell epitopes was analyzed by the IEDB population coverage analysis tool (http://tools.iedb.org/population/). This web-server predicts the population coverage based on the MHC binding alleles (Bui et al., 2006).
2.8. Putative linear B-cell epitope prediction from the conserved sequences
B-cell epitope refers to the antigenic region of a protein which can stimulate the B-cell response. The antigenic regions are recognized by the binding sites of immunoglobulin molecules. These B-cell epitopes play an important role in peptide vaccine design, and also in diagnosis of diseases (Saha and Raghava, 2006). Here, the conserved sequences which fulfilled ≥ 0.4 antigenicity score in Vaxijen server and identified as the outer membrane protein in TMHMM server were subjected to the ABCpred server (http://osddlinux.osdd.net/raghava/abcpred) for linear B-cell epitope prediction. ABCpred is a recurrent neural network (RNN) and standard feed-forward (FNN) based B-cell epitope prediction server (Saha and Raghava, 2006). The window length and the threshold value were set at decamer (10) and 0.51, respectively for the epitope prediction. At the same time, the overlapping sequences were also filtered from the list of the epitope.
2.9. Three-dimensional structure design of the overlapping CD8 + and CD4 + T-cell epitopes
A three-dimensional (3D) structure of the both HLA-allele and epitope is required to perform the docking simulation study. So, we predicted the 3D structure of the both CD8 + and CD4 + T-cell epitopes. However, the 3D structure of the HLA-alleles were collected from the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/pdb/). Herein, we designed the 3D structure of the total thirteen CD8 + T-cell epitopes; whereas two epitopes were overlapping with decamer linear B-cell epitopes and the rest of the other eleven epitopes (1 N and 10 G protein) were overlapping with pentadecamer CD4 + T-cell epitopes (peptides). On the other hand, we predicted the 3D structure of the total nine CD4 + T-cell epitopes (1 N and 8 G protein) having CD8 + T-cell epitopes as the core sequence and IFN-γ & IL-4 inducer property as well. We submitted all the epitopes to the PEP-FOLD peptide structure prediction server at the Ressource Parisienne en Bioinformatique Structurale Mobyle Portal for the 3D structure annotation (Shen et al., 2014, Thévenet et al., 2012).
2.10. Molecular docking simulation of the HLA allele-peptide interaction
To know the interaction between binding alleles and predicted epitope, the molecular docking simulation study was performed using the AutoDock tools (Morris et al., 2009) and AutoDock Vina software (Trott and Olson, 2010). So, to identify the interaction between CD8 + T-cell epitopes and the HLA-alleles, the crystal structure of the most conserved HLA-C*12:03 protein molecule named 1EFX was retrieved from the RCSB Protein Data Bank (PDB) as the PDB format (Boyington et al., 2000). However, the predicted crystal structure was in a complex form with protein and an epitope. So, the Discovery Studio (version 16.1.0.15350) was used to simplify the complex structure. After the separation, the protein molecule and the peptide (epitope) were then converted to PDBQT format through the AutoDock tools. For the identification of binding energy at the binding groove of HLA-C*12:03 with an epitope, the space box center was set at 7.890, 30.823, and 76.726 Å in the X, Y and Z axis, respectively. The size was set at 88, 46 and 64 Å in the X, Y and Z dimensions, respectively, and these analyses were done in 0.314 Å spacing parameter. These parameters were set for all the selected CD8 + T-cell epitopes. Finally, the docking simulation was performed by using the AutoDock Vina. Another docking study was performed as a control for the critical evaluation and scientific acceptance of our docking study. The docking method and parameters were set just like as previously mentioned parameters so that we can easily compare between sample and control. In the control section, the docking simulation was completed between human natural killer cell receptor (epitope-GAVDPLLAL) and HLA-C*12:03 protein molecule. All the PDBQT files were converted to the PDB format through the OpenBabel (version 2.4.1) (O'Boyle et al., 2011) and visualized by the PyMOL molecular graphics system (PyMol Viewer).
On the contrary, the docking parameters for HLA-DRB1*01:01 (PDB id: 2FSE) (Rosloniec et al., 2006) and the selected CD4 + T-cell epitopes (peptides) were set at 53.857, 69.017, − 9.549 Å in the X, Y and Z axis, respectively, and the size was set at 66, 78, and 40 Å in the X, Y and Z dimensions, respectively. This docking was completed in 0.481 Å spacing parameter. Herein, the binding interaction was also evaluated by performing a docking between immunodominant determinant of human type II collagen (epitope-AGFKGEQGPKGEPG) and an HLA-DRB1*01:01 allele.
3. Results
3.1. Retrieval of protein sequences and identification of most conserved sequences
A total of 203 sequences of the N protein (163 sequences) and G protein (40 sequences) of different isolates of RVFV were retrieved from the ViPR database. The length of the predicted each N and G protein was 245 AA and 1197 AA, respectively. The online CLUSTALW server created various conserved sequences with varying lengths. In the complete sequences, a total of 5 and 16 conserved sequences were found in the N and G protein of RVFV, respectively. In this study, the conserved sequences generated by the multiple sequence alignment are shown in Table 1 .
Table 1.
Conserved sequences from the nucleocapsid (N) and glycoprotein (G) of RVFV.
| Conserved sequences | Position | VaxiJen score | Transmembrane helix |
|---|---|---|---|
| Nucleocapsid (N) | |||
| MDNYQELAIQFAAQAVDRNEIEQWVREFAYQGFDARRVI ELLKQYGGADWEKDAKKMIVLALTRGNKPRRMMMKMS KEGKATVEALINKYKLKEGNPSRDELTLSRVAAALAG |
1–113 | 0.3589 | Outside |
| TCQALVVLSEWLPVTGTTMDGLS | 115–137 | 0.4542 | Outside |
| PRHMMHPSFAGMVDPSLP | 141–158 | 0.7857 | Outside |
| DYLRAILDAHSLYLLQFSRVINPNLRGRTKEEVAATFTQP MNAAVNSNFISHEKRR |
160–215 | 0.5662 | Outside |
| FLKAFGLVDSNGKPSAAV | 217–234 | 0.5546 | Outside |
| Glycoprotein (G) | |||
| EAVIRVSLSSTREETCFGDSTNPEMIE | 15–41 | 0.8871 | Outside |
| AWDSLREEEMPEELSCSISGIREVKTSSQELYRALKAIIAAD GLNNITCHGKDPEDKISLIKGPPHKKRVGIVRC |
42–117 | 0.2465 | Outside |
| RETMAGIAMTVLPALAVFALAPVVFAED | 128–155 | 0.6132 | Outside, TMhelix inside |
| HLRNRPGKGHNYIDG | 157–171 | − 0.0385 | inside |
| QEDATCKPVTYAGACSSFDVLLEKGKFPLFQSYAHHRTLL EAVHDTIIAKADPPSCDL |
174–234 | 0.3368 | inside |
| CMKEKLVMKTHCPNDYQSAHYLNNDGKMASVKCPPKYE LTEDCNFCR |
239–285 | 0.3555 | inside |
| MTGASLKKGSYPLQDLFCQSSEDDGSKLKTKMKGVCEVG VQALKKCDGQLSTAHEVVPFAVFKNSKKVYLDKLDLKTE ENLLPDSFVCFEHKGQYKGTM |
287–385 | 0.5831 | Outside |
| SGQTKRELKSFDISQCPKIGGHG | 387–409 | 0.8886 | inside |
| TGDAAFCSAYECTAQYANAYCSHANGSGIVQIQVS | 414–448 | 0.2323 | Outside |
| VWKKPLCVGYERVVVKRELSAKPIQRVEPCTTCITKCEPH GLVVRSTGFKISSAVACASGVCVTGSQSPSTEITLKYPGISQSS |
450–533 | 0.5311 | Outside |
| GDIGVHMAHDDQSVSSKIV | 535–553 | 0.6649 | Outside |
| HTALSAFVVVFVFSS | 580–594 | 0.1602 | Outside |
| AIICLAILYKVLKCLKIAPRKVLNPLMWITAFIRWVYKKM VARVADNINQVNREIGWMEGGQL |
596–658 | 0.5367 | Outside |
| NPAPIPRHAPIPRYSTYLMLLLIVSYASACSELIQASSRITTC STEGVNTRCRLS |
662–716 | 0.3969 | Outside, TMhelix inside |
| DQTKFLKIKTVSSELSCREGQSYWTGSFSPKCLSSRRCHLV GECHVNRCLSWRDNETSAEFSFVGESTTMRENKCFEQCG GWGCGCFNVNPSCLFVHTYLQS VRKEALRVFNCIDWVHKLTLEITDFDGSVSTIDLGASSSRF TNWGSVSLSLDAEGISGSNSFSFIESPGKGYAIVDEPFSEIP RQGFLGEIRCNSESSVLSAHESCLRAPNLISYKPMIDQLECT TNLIDPFVVFERGSLPQTRNDKTFAASKGNRGVQAFSKGS VQADLTL |
740–1013 | 0.6272 | Outside |
| FDNFEVDFVGAAVSCDAAFLNLTGCYSCNAGARVCLSITS TGTGTLSAHNKDGSLHIVLPSENGTKDQCQILHFTVPEVEE EFMYSCDGDERPLLVKGTLIAIDPFDDRREAGGESTVVNP KSGSWNFFDWFSGLMSWFGGPLKTILLICLYVALSIGLFFL LIYLGRTGLSKMWLAATKKAS |
1015–1197 | 0.5974 | Outside, TMhelix inside |
3.2. Transmembrane and antigenicity features analysis of the conserved sequences
The transmembrane topology analysis exposed that a total of 5 (out of 5) and 11 (out of 16) conserved sequences of N and G protein, respectively fulfilled the criteria of exomembrane properties (Table 1). As opposed to, 4 conserved sequences (out of 5) of N protein and 9 (out of 16) conserved sequences of G protein follow the antigenicity property of default threshold value, ≥ 0.4, in Vaxijen v2.0 server (Table 1). The Vaxijen score ranged from 0.3589 to 0.7857 in N protein, whereas G protein showed − 0.0385 to 0.8886 score. Finally, the result analysis revealed that 4 of 5 N protein, as well as 8 of 16 G protein conserved sequences, were fulfilled both the antigenicity and exomembrane characteristics (Table 1) and these sequences were used for further analyses. The lengthiest conserved sequence was found in G protein having 273 amino acid residues and antigenicity score of 0.6272.
3.3. Identification of CD8 + T-cell epitopes and their MHC-I binding allele
Initially, a total of 72 and 502 nonamer CD8 + T-cell epitopes were found from the selected conserved sequences of N and G protein of RVFV, respectively. The CD8 + T-cell epitopes were found from all N and G protein sequences, except only conserved sequence “217FLKAFGLVDSNGKPSAAV234” of N protein. After removing the redundant epitopes (1 epitope from each set having highest combined score was selected) the final dataset was fixed at 279 (31 N and 248 G protein). Finally, 146 CD8 + T-cell epitopes (16 N and 130 G protein) were taken under consideration which exhibited both the combined score, ≥ 0.5 (NetCTL v1.2 server), and antigenicity value of ≥ 0.5 (VaxiJen v2.0 server) (Table S1).
Thereafter, the selected 16 N and 130 G protein CD8 + T-cell epitopes were subjected to the SMM based IEDB MHC-I prediction tool for the prophecy of MHC-I binding alleles with the IC50 value < 250 nM. The resulting analysis showed that the CD8 + T-cell epitopes of the N protein were interacted with two to as many as nine MHC-I alleles, whereas the epitopes of G protein exhibited interaction with one to as many as ten alleles in the above-mentioned IC50 value (Table S1). Herein, the immunogenicity of the selected CD8 + T-cell epitopes was also predicted to ensure the best quality of the epitopes. The higher score designates a greater probability of eliciting an effective immune response. Next step analyses were continued with the CD8 + T-cell epitopes that exhibited interaction with ≥ 5 MHC-I alleles, had the positive immunogenicity score, ≥ 0.5 combined scores, and ≥ 0.5 antigenicity score. So, the analyses revealed that among 146 CD8 + T-cell epitopes only 40 epitopes (3 N and 37 G protein) fulfilled the above-mentioned criteria. The epitopes, their position, combined score, antigenicity, and immunogenicity score are presented in Table 2 . Herein, we found G protein epitope 882FTNWGSVSL890 as the highest antigenic with a 2.0756 VaxiJen score, whereas the epitope 134IAMTVLPAL142 was considered as the lowest antigenic for its lowest, 0.5100 score among all the 40 N and G protein CD8 + T-cell epitopes. On the other hand, we got the G protein epitopes, 625TAFIRWVYK633 and 948LRAPNLISY956 as the highest (0.46351) and the lowest (0.00046) immunogenic, respectively for their immunogenicity score in IEDB tool (Table 2). In this study, we identified 34 MHC class I binding alleles (15 HLA-A, 9 HLA-B, and 10 HLA-C alleles) for 37 CD8 + T-cell epitopes of G protein, whereas 9 alleles (1 HLA-A, 3 HLA-B, and 5 HLA-C alleles) were found for the 3 epitopes of N protein. Among them, the N protein epitope 144MMHPSFAGM152 interacting with 7 HLA alleles possessing the maximum number of alleles, while the G protein epitope 882FTNWGSVSL890 interacting with maximum 10 HLA alleles which is followed by 2 epitopes (625TAFIRWVYK633, 1158KTILLICLY1166) with 9 alleles, 2 epitopes (134IAMTVLPAL142, 1142FFDWFSGLM1150) with 8 alleles, and 3 epitopes (144VFALAPVVF152, 1167VALSIGLFF1175, 1169LSIGLFFLL1177) with 7 HLA alleles. Interestingly, the HLA-C*12:03 was identified as the most conserved MHC class I binding allele for both 3 N and 37 G protein epitopes (Table 2).
Table 2.
Predicted potential CD8 + T-cell epitopes of RVFV interacting with different MHC class I HLA-alleles.
| CD8 + T-cell epitopes | Combined score | Antigenicity score | Immunogenicity | MHC-I alleles | Position | Conservancy score |
|---|---|---|---|---|---|---|
| Nucleocapsid (N) | ||||||
| WLPVTGTTM | 0.8867 | 0.9328 | 0.16308 | HLA-C*12:03,HLA-B*15:02,HLA-C*14:02,HLA-C*03:03,HLA-C*07:02 | 125–133 | 99.39% |
| MMHPSFAGM | 1.1149 | 1.7017 | 0.00126 | HLA-B*15:02,HLA-C*14:02,HLA-B*15:01,HLA-C*03:03,HLA-B*35:01,HLA-A*02:06,HLA-C*12:03 | 144–152 | 99.39% |
| SRVINPNLR | 0.6389 | 1.0188 | 0.11864 | HLA-C*12:03,HLA-C*03:03,HLA-C*07:01,HLA-C*14:02,HLA-C*07:02 | 177–185 | 100.00% |
| Glycoprotein (G) | ||||||
| TMAGIAMTV | 1.2078 | 0.8376 | 0.08771 | HLA-A*02:06,HLA-A*02:01,HLA-C*12:03,HLA-A*68:02,HLA-C*03:03,HLA-C*14:02 | 130–138 | 100.00% |
| AVFALAPVV | 1.0052 | 0.7110 | 0.11816 | HLA-C*12:03,HLA-A*02:06,HLA-C*15:02,HLA-A*02:01,HLA-C*14:02 | 143–151 | 100.00% |
| LAVFALAPV | 0.8628 | 0.9445 | 0.1854 | HLA-C*12:03,HLA-C*03:03,HLA-A*02:06,HLA-A*68:02,HLA-C*15:02,HLA-C*14:02 | 142–150 | 100.00% |
| FALAPVVFA | 0.7907 | 1.0606 | 0.16707 | HLA-C*03:03,HLA-C*12:03,HLA-A*02:06,HLA-C*05:01,HLA-A*68:02,HLA-B*15:02 | 145–153 | 100.00% |
| VFALAPVVF | 1.7269 | 1.3111 | 0.08816 | HLA-C*03:03,HLA-A*23:01,HLA-C*14:02,HLA-C*12:03,HLA-A*24:02,HLA-B*15:02,HLA-C*07:02 | 144–152 | 100.00% |
| MAGIAMTVL | 0.82 | 0.7997 | 0.0746 | HLA-C*12:03,HLA-C*03:03,HLA-B*15:02,HLA-C*15:02,HLA-B*35:01 | 131–139 | 100.00% |
| RETMAGIAM | 1.5256 | 0.8666 | 0.04108 | HLA-B*40:01,HLA-C*03:03,HLA-C*12:03,HLA-B*40:02,HLA-B*15:02,HLA-C*07:02 | 128–136 | 100.00% |
| IAMTVLPAL | 1.0016 | 0.5100 | 0.02532 | HLA-C*12:03,HLA-C*03:03,HLA-C*14:02,HLA-A*02:06,HLA-B*15:02,HLA-B*35:01,HLA-A*02:01,HLA-C*15:02 | 134–142 | 100.00% |
| VCEVGVQAL | 0.7653 | 1.1196 | 0.071 | HLA-B*15:02,HLA-C*03:03,HLA-C*14:02,HLA-C*12:03,HLA-C*07:02 | 321–329 | 100.00% |
| RVEPCTTCI | 0.777 | 0.5474 | 0.00664 | HLA-C*05:01,HLA-C*12:03,HLA-C*03:03,HLA-C*15:02,HLA-A*32:01 | 475–483 | 100.00% |
| TAFIRWVYK | 1.382 | 0.6560 | 0.46351 | HLA-C*12:03,HLA-A*68:01,HLA-A*11:01,HLA-C*03:03,HLA-A*31:01,HLA-A*30:01,HLA-A*03:01,HLA-C*07:01,HLA-C*14:02 | 625–633 | 95.00% |
| IICLAILYK | 1.2334 | 1.4732 | 0.1232 | HLA-A*11:01,HLA-C*12:03,HLA-A*03:01,HLA-A*68:01,HLA-C*03:03,HLA-C*14:02 | 597–605 | 10.00% |
| AIICLAILY | 1.2533 | 1.3231 | 0.12082 | HLA-C*12:03,HLA-A*29:02,HLA-C*03:03,HLA-A*30:02,HLA-A*11:01 | 596–604 | 35.00% |
| LSSRRCHLV | 0.6673 | 1.2811 | 0.01885 | HLA-C*12:03,HLA-C*15:02,HLA-C*07:01,HLA-C*03:03,HLA-C*06:02 | 772–780 | 100.00% |
| RVFNCIDWV | 1.0863 | 1.7981 | 0.25241 | HLA-C*12:03,HLA-A*02:06,HLA-A*02:01,HLA-C*07:01,HLA-A*68:02 | 848–856 | 100.00% |
| ETSAEFSFV | 1.1516 | 1.9805 | 0.12215 | HLA-A*68:02,HLA-C*12:03,HLA-C*15:02,HLA-C*03:03,HLA-A*02:06 | 795–803 | 100.00% |
| FVVFERGSL | 1.1708 | 0.7552 | 0.20936 | HLA-C*03:03,HLA-B*15:02,HLA-C*12:03,HLA-C*14:02,HLA-B*35:03,HLA-A*68:02 | 973–981 | 97.50% |
| LRAPNLISY | 1.263 | 0.9951 | 0.00046 | HLA-C*12:03,HLA-C*03:03,HLA-C*07:01,HLA-C*07:02,HLA-C*14:02,HLA-B*14:02 | 948–956 | 35.00% |
| PRQGFLGEI | 0.5175 | 0.9779 | 0.18716 | HLA-C*12:03,HLA-C*07:01,HLA-C*03:03,HLA-C*14:02,HLA-C*06:02 | 924–932 | 97.50% |
| FTNWGSVSL | 1.2356 | 2.0756 | 0.03624 | HLA-B*15:02,HLA-C*03:03,HLA-C*15:02,HLA-B*39:01,HLA-C*12:03,HLA-A*02:06,HLA-A*68:02,HLA-C*14:02,HLA-B*35:03,HLA-C*07:01 | 882–890 | 100.00% |
| FEQCGGWGC | 0.9179 | 0.7089 | 0.17979 | HLA-C*12:03,HLA-C*03:03,HLA-B*15:02,HLA-C*08:02,HLA-B*40:01 | 815–823 | 100.00% |
| KGNRGVQAF | 1.2086 | 0.8800 | 0.04694 | HLA-C*05:01,HLA-C*07:02,HLA-C*12:03,HLA-C*14:02,HLA-B*15:02,HLA-A*32:01 | 994–1002 | 100.00% |
| YSCNAGARV | 0.7606 | 1.0814 | 0.10925 | HLA-C*12:03,HLA-C*15:02,HLA-C*07:01,HLA-C*14:02,HLA-C*06:02 | 1040–1048 | 100.00% |
| EVDFVGAAV | 0.5976 | 0.9494 | 0.25298 | HLA-C*05:01,HLA-C*12:03,HLA-A*68:02,HLA-C*08:02,HLA-C*14:02 | 1019–1027 | 97.50% |
| FFDWFSGLM | 0.5881 | 0.6472 | 0.21048 | HLA-C*12:03,HLA-B*15:02,HLA-C*14:02,HLA-C*08:02,HLA-C*07:01,HLA-B*35:01,HLA-C*04:01,HLA-C*06:02 | 1142–1150 | 100.00% |
| ALSIGLFFL | 1.3051 | 1.2128 | 0.26998 | HLA-A*02:01,HLA-A*02:06,HLA-C*03:03,HLA-B*15:02,HLA-C*12:03 | 1168–1176 | 97.50% |
| GLFFLLIYL | 1.2359 | 0.6126 | 0.24472 | HLA-A*02:01,HLA-C*14:02,HLA-C*12:03,HLA-A*02:06,HLA-B*15:02 | 1172–1180 | 100.00% |
| ILHFTVPEV | 1.2196 | 0.8591 | 0.2541 | HLA-C*14:02,HLA-A*02:01,HLA-C*12:03,HLA-A*02:06,HLA-C*03:03 | 1085–1093 | 100.00% |
| TILLICLYV | 1.1533 | 0.6223 | 0.05257 | HLA-C*12:03,HLA-A*02:06,HLA-C*15:02,HLA-A*02:01,HLA-C*14:02,HLA-A*68:02 | 1159–1167 | 97.50% |
| ILLICLYVA | 0.8389 | 0.5777 | 0.08838 | HLA-A*02:01,HLA-C*12:03,HLA-A*02:06,HLA-C*14:02,HLA-C*03:03 | 1160–1168 | 95.00% |
| FLLIYLGRT | 0.7428 | 1.1077 | 0.17512 | HLA-C*14:02,HLA-A*02:01,HLA-C*12:03,HLA-C*03:03,HLA-A*02:06 | 1175–1183 | 100.00% |
| KMWLAATKK | 1.5878 | 1.0998 | 0.04243 | HLA-A*03:01,HLA-C*14:02,HLA-C*12:03,HLA-A*11:01,HLA-A*31:01 | 1187–1195 | 100.00% |
| LYVALSIGL | 1.134 | 1.8044 | 0.01836 | HLA-C*14:02,HLA-C*03:03,HLA-C*12:03,HLA-B*15:02,HLA-C*07:02 | 1165–1173 | 97.50% |
| KTILLICLY | 1.9084 | 0.7398 | 0.09454 | HLA-C*12:03,HLA-B*58:01,HLA-A*29:02,HLA-A*30:02,HLA-A*01:01,HLA-A*32:01,HLA-C*03:03,HLA-C*14:02,HLA-B*57:01 | 1158–1166 | 97.50% |
| LLICLYVAL | 1.402 | 1.0546 | 0.03237 | HLA-C*03:03,HLA-A*02:01,HLA-A*02:06,HLA-B*15:02,HLA-C*14:02,HLA-C*12:03 | 1161–1169 | 97.50% |
| VALSIGLFF | 1.2362 | 0.9478 | 0.05047 | HLA-C*03:03,HLA-C*12:03,HLA-B*58:01,HLA-B*35:01,HLA-C*05:01,HLA-B*53:01,HLA-C*14:02 | 1167–1175 | 97.50% |
| LSIGLFFLL | 1.0775 | 0.6297 | 0.26902 | HLA-C*03:03,HLA-A*02:06,HLA-C*12:03,HLA-C*15:02,HLA-A*68:02,HLA-B*58:01,HLA-B*15:02 | 1169–1177 | 100.00% |
We further predicted the allergenicity of the 40 epitopes and we found only 7 CD8 + T-cell epitopes, 143AVFALAPVV151, 142LAVFALAPV150, 321VCEVGVQAL329, 475RVEPCTTCI483, 848RVFNCIDWV856, 815FEQCGGWGC823, and 1040YSCNAGARV1048 of G protein which have no allergenicity (non-allergen) to the human, but we did not get any non-allergenic CD8 + T-cell epitopes of N protein. Interestingly, the conservancy of the seven non-allergenic epitopes was 100% among all G protein sequences. However, the rest of the other epitopes were found as the potential allergen to human with 95%–100% conservancy, except 597IICLAILYK605 (10%), 596AIICLAILY604 (35%) and 948LRAPNLISY956 (35%) (Table 2).
3.4. Identification and analysis of CD4 + T-cell epitopes and their MHC-II binding alleles from conserved sequences
Initially 415 pentadecamer peptides (epitopes) (43 N and 372 G protein) were identified from the conserved sequences of the N and G protein (Table S2). All the predicted peptides were subjected to the IEDB MHC-II binding tool and VaxiJen server for the prediction of HLA alleles and antigenicity score, respectively. Thus, we identified 8 N and 54 G protein peptides (total 62 peptides) which fulfilled the criteria, ≥ 0.5 antigenicity score and had ≥ 5 MHC-II alleles and these peptides were considered as potential CD4 + T-cell epitopes (Table S3). Among all the potential epitopes “173LLQFSRVINPNLRGR187” from N protein and “744FLKIKTVSSELSCRE758” from G protein had the highest VaxiJen scores of 1.2501 and 1.3558, respectively. The allergenicity analysis revealed that these two highest antigenic epitopes were non-allergenic to the human. Besides these two epitopes, 5 epitopes of N protein and 14 epitopes of G protein were also non-allergenic to the human with varying antigenicity scores (Table S3). However, a good CD4 + T-cell epitope should interact with as many HLA- alleles as possible. Among all the 8 N protein epitopes, 171LYLLQFSRVINPNLR185 interacting with 10 HLA alleles with an IC50 value < 250 nM. On the contrary, G protein epitope, 1169LSIGLFFLLIYLGRT1183 interacting with 13 HLA alleles possessing the maximum number of HLA-alleles which is followed by the 2 CD4 + T-cell epitopes (1159TILLICLYVALSIGL1173, 1168ALSIGLFFLLIYLGR1182) with 12 alleles, and 6 CD4 + T-cell epitopes (742TKFLKIKTVSSELSC756, 1163ICLYVALSIGLFFLL1177, 1160ILLICLYVALSIGLF1174, 1171IGLFFLLIYLGRTGL1185, 1172GLFFLLIYLGRTGLS1186, and 1157LKTILLICLYVALSI1171) with 11 HLA alleles.
3.5. Identification and analysis of overlapping CD8 + and CD4 + T-cell epitopes
In the CD8 + T-cell epitope prediction section, we found potential 40 CD8 + T-cell epitopes (3 N and 37 G protein). On the other hand, we found 62 CD4 + T-Cell Epitopes (8 N and 54 G protein) in the previous section. Herein, we overlapped the CD8 + T-cell epitopes on the CD4 + T-Cell Epitopes for the identification of overlapping epitopes. The result analysis exhibited that 6 N protein and 34 G protein CD4 + T-Cell Epitopes were overlapped with the CD8 + T-cell epitopes. The overlapped CD4 + T-Cell Epitopes, their respective MHC-II alleles, antigenicity scores, and the positions are shown in Table 3 . Here, 2 CD8 + T-cell epitopes of N protein and 21 CD8 + T-cell epitopes of G protein were found as the core sequences of the 6 N and 34 G protein CD4 + T-Cell Epitopes, respectively. The CD8 + T-cell epitopes were found as the core sequence of the 1 to as many as 7 CD4 + T-Cell Epitopes of G protein, whereas it was found as 2 to as many as 4 for N protein. The CD8 + T-cell epitopes, 1168ALSIGLFFL1176, 1172GLFFLLIYL1180, 1165LYVALSIGL1173, 1161LLICLYVAL1169, 1167VALSIGLFF1175, and 1169LSIGLFFLL1177 were overlapped on the maximum 7 CD4 + T-Cell Epitopes (Table S4).
Table 3.
Predicted potential overlapping CD4 + T-cell epitopes of RVFV interacting with different MHC class II HLA-alleles.
| Overlapping CD4 + T-Cell Epitopes | MHC-II alleles | Antigenicity | Position |
|---|---|---|---|
| Nucleocapsid (N) | |||
| HMMHPSFAGMVDPSL | HLA-DRB1*0101,HLA-DRB1*0901,HLA-DRB1*0405,HLA-DQA1*0501/DQB1*0301,HLA-DRB1*0401,HLA-DRB1*0701 | 1.0025 | 143–157 |
| MMHPSFAGMVDPSLP | HLA-DRB1*01:01,HLA-DRB1*04:05,HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*04:01,HLA-DRB1*09:01 | 0.9451 | 144–158 |
| LYLLQFSRVINPNLR | HLA-DRB1*01:01,HLA-DRB1*04:05,HLA-DRB1*07:01, HLA-DRB1*04:04,HLA DRB1*04:01,HLA-DRB5*01:01, HLA-DRB1*15:01,HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*11:01,HLA-DRB1*09:01 | 0.6108 | 171–185 |
| YLLQFSRVINPNLRG | HLA-DRB1*01:01,HLA-DRB1*04:05,HLA-DRB1*07:01,HLA-DRB1*04:04,HLA-DRB1*04:01,HLA-DRB5*01:01,HLA-DRB1*15:01,HLA-DRB1*09:01,HLA-DRB1*11:01 | 0.6286 | 172–186 |
| LLQFSRVINPNLRGR | HLA-DRB1*01:01,HLA-DRB1*04:05,HLA-DRB1*07:01,HLA-DRB1*04:04,HLA-DRB1*04:01,HLA-DRB5*01:01,HLA-DRB1*09:01 | 1.2501 | 173–187 |
| LQFSRVINPNLRGRT | HLA-DRB1*04:05,HLA-DRB1*01:01,HLA-DRB1*07:01,HLA-DRB1*04:04,HLA-DRB1*04:01,HLA-DRB5*01:01,HLA-DRB1*09:01 | 0.8316 | 174–188 |
| Glycoprotein (G) | |||
| TMAGIAMTVLPALAV | HLA-DRB1*01:01,HLA-DRB1*04:04,HLA-DQA1*05:01/DQB1*03:01,HLA-DQA1*01:02/DQB1*06:02,HLA-DRB1*09:01,HLA-DRB1*07:01,HLA-DRB1*11:01 | 0.7438 | 130–144 |
| LPALAVFALAPVVFA | HLA-DRB1*01:01,HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*15:01,HLA-DRB1*09:01,HLA-DRB1*04:04 | 0.7636 | 139–153 |
| PALAVFALAPVVFAE | HLA-DRB1*01:01,HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*09:01,HLA-DRB1*15:01,HLA-DRB1*04:04 | 0.7211 | 140–154 |
| VLPALAVFALAPVVF | HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DRB1*12:01,HLA-DRB1*04:04 | 0.6372 | 138–152 |
| MTVLPALAVFALAPV | HLA-DQA1*01:02/DQB1*06:02,HLA-DRB1*01:01,HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*04:04,HLA-DRB1*15:01,HLA-DRB1*12:01,HLA-DRB1*09:01, | 0.6143 | 136–150 |
| MAGIAMTVLPALAVF | HLA-DRB1*01:01,HLA-DRB1*04:04,HLA-DRB1*09:01,HLA-DQA1*01:02/DQB1*06:02,HLA-DRB1*07:01,HLA-DRB1*11:01, HLA-DQA1*05:01/DQB1*03:01 | 0.5647 | 131–145 |
| GIAMTVLPALAVFAL | HLA-DRB1*01:01,HLA-DRB1*04:04,HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*09:01,HLA-DRB1*07:01, HLA-DRB1*11:01,HLA-DQA1*01:02/DQB1*06:02, HLA-DRB1*12:01 | 0.6562 | 133–147 |
| AGIAMTVLPALAVFA | HLA-DRB1*01:01,HLA-DRB1*04:04, HLA-DQA1*01:02/DQB1*06:02, HLA-DRB1*09:01,HLA-DQA1*05:01/DQB1*03:01,HLA-DRB1*07:01,HLA-DRB1*11:01, HLA-DRB1*12:01 | 0.5161 | 132–146 |
| NPLMWITAFIRWVYK | HLA-DRB5*01:01,HLA-DRB1*04:04,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DPA1*01/DPB1*04:01,HLA-DRB1*12:01 | 0.5045 | 619–633 |
| IICLAILYKVLKCLK | HLA-DRB1*11:01,HLA-DRB5*01:01,HLA-DRB1*01:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*15:01 | 0.5904 | 597–611 |
| AIICLAILYKVLKCL | HLA-DRB1*11:01,HLA-DRB5*01:01,HLA-DRB1*01:01,HLADPA1*03:01/DPB1*04:02,HLA-DRB1*15:01 | 0.5698 | 596–610 |
| SGSWNFFDWFSGLMS | HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*04:04,HLA-DRB1*01:01, HLA-DRB1*09:01,HLA-DRB1*07:01, HLA-DRB1*04:05 | 0.7241 | 1137–1151 |
| SWNFFDWFSGLMSWF | HLA-DRB1*01:01,HLA-DRB1*09:01,HLA-DRB1*07:01,HLA-DRB1*04:04, HLA-DRB1*04:05, HLA-DPA1*01:03/DPB1*02:01 | 0.5094 | 1139–1153 |
| GSWNFFDWFSGLMSW | HLA-DRB1*04:04,HLA-DRB1*01:01,HLA-DRB1*09:01,HLA-DRB1*07:01, HLA-DRB1*04:05, HLA-DPA1*01:03/DPB1*02:01 | 0.8838 | 1138–1152 |
| YVALSIGLFFLLIYL | HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*01/DPB1*04:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DQA1*01:01/DQB1*05:01,HLA-DPA1*02:01/DPB1*05:01 | 0.8703 | 1166–1180 |
| VALSIGLFFLLIYLG | HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*01/DPB1*04:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*01:01, HLA-DRB1*15:01,HLA-DQA1*01:01/DQB1*05:01,HLA-DPA1*02:01/DPB1*05:01,HLA-DRB1*11:01 | 0.9838 | 1167–1181 |
| CLYVALSIGLFFLLI | HLA-DPA1*01/DPB1*04:01, HLA-DPA1*02:01/DPB1*01:01, HLA-DPA1*02:01/DPB1*05:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DRB1*01:01, HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*07:01, HLA-DRB1*09:01 | 0.8330 | 1164–1178 |
| LYVALSIGLFFLLIY | HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*01/DPB1*04:01,HLA-DRB1*01:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*07:01,HLA-DPA1*02:01/DPB1*05:01,HLA-DQA1*01:01/DQB1*05:01,HLA-DRB1*15:01 | 0.8518 | 1165–1179 |
| ICLYVALSIGLFFLL | HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DRB1*07:01, HLA-DRB1*15:01,HLA-DRB1*09:01, HLA-DPA1*03:01/DPB1*04:02,HLA-DPA1*01/DPB1*04:01, HLA-DRB5*01:01, HLA-DPA1*02:01/DPB1*05:01,HLA-DRB1*04:05 | 1.0197 | 1163–1177 |
| LICLYVALSIGLFFL | HLA-DRB1*01:01,HLA-DRB1*07:01,HLA-DRB1*09:01,HLA-DRB1*15:01,HLA-DRB5*01:01,HLA-DRB1*04:05,HLA-DRB1*04:04,HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*11:01 | 1.0286 | 1162–1176 |
| ALSIGLFFLLIYLGR | HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*01/DPB1*04:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*01:01, HLA-DPA1*02:01/DPB1*05:01,HLA-DRB5*01:01,HLA-DRB1*11:01,HLA-DRB1*15:01, HLA-DRB1*04:05,HLA-DQA1*01:01/DQB1*05:01,HLA-DRB1*04:04 | 1.0248 | 1168–1182 |
| LSIGLFFLLIYLGRT | HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB5*01:01,HLA-DPA1*01/DPB1*04:01,HLA-DRB1*15:01,HLA-DRB1*11:01,HLA-DRB1*04:04,HLA-DRB1*04:05,HLA-DQA1*01:01/DQB1*05:01,HLA-DRB4*01:01,HLA-DPA1*02:01/DPB1*05:01 | 0.9860 | 1169–1183 |
| IGLFFLLIYLGRTGL | HLA-DRB1*01:01,HLA-DRB1*11:01,HLA-DRB1*15:01, HLA-DPA1*01/DPB1*04:01,HLA-DRB5*01:01, HLA-DRB1*04:04,HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DPA1*01:03/DPB1*02:01, HLA-DRB1*04:05, HLA-DRB4*01:01 | 1.0772 | 1171–1185 |
| GLFFLLIYLGRTGLS | HLA-DPA1*02:01/DPB1*01:01, HLA-DRB1*01:01,HLA-DRB1*11:01,HLA-DRB1*15:01,HLA-DRB5*01:01, HLA-DRB1*04:04,HLA-DPA1*03:01/DPB1*04:02,HLA-DPA1*01:03/DPB1*02:01, HLA-DPA1*01/DPB1*04:01,HLA-DRB1*04:05,HLA-DQA1*05:01/DQB1*03:01 | 1.1893 | 1172–1186 |
| SIGLFFLLIYLGRTG | HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*11:01, HLA-DRB1*04:05, HLA-DRB5*01:01,HLA-DPA1*01:03/DPB1*02:01,HLA-DPA1*01/DPB1*04:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*04:04, | 1.2425 | 1170–1184 |
| TILLICLYVALSIGL | HLA-DRB1*01:01,HLA-DRB1*07:01,HLA-DRB1*04:04,HLA-DRB1*15:01,HLA-DRB1*04:05,HLA-DRB1*09:01,HLA-DRB1*11:01,HLA-DRB1*13:02,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*04:01,HLA-DRB5*01:01,HLA-DRB4*01:01 | 1.2532 | 1159–1173 |
| LKTILLICLYVALSI | HLA-DPA1*03:01/DPB1*04:02, HLA-DRB1*07:01,HLA-DRB1*01:01,HLA-DRB1*04:04,HLA-DRB1*15:01, HLA-DRB1*04:05, HLA-DRB4*01:01,HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*13:02, HLA-DRB1*11:01, HLA-DRB1*04:01, | 0.9089 | 1157–1171 |
| KTILLICLYVALSIG | HLA-DRB1*01:01,HLA-DRB1*07:01,HLA-DRB1*04:04,HLA-DRB1*15:01,HLA-DRB1*04:05,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*04:01,HLA-DRB1*13:02,HLA-DRB1*11:01 | 1.1808 | 1158–1172 |
| PLKTILLICLYVALS | HLA-DRB1*01:01,HLA-DRB1*04:04,HLA-DRB1*15:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*04:05,HLA-DRB4*01:01,HLA-DPA1*02:01/DPB1*01:01,HLA-DRB1*11:01 | 0.7049 | 1156–1170 |
| GPLKTILLICLYVAL | HLA-DPA1*03:01/DPB1*04:02,HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DRB4*01:01, HLA-DRB1*04:04, HLA-DPA1*02:01/DPB1*01:01, | 0.5626 | 1155–1169 |
| ILLICLYVALSIGLF | HLA-DRB1*01:01,HLA-DRB1*07:01,HLA-DRB1*04:04,HLA-DRB1*15:01, HLA-DRB1*04:05,HLA-DRB1*09:01,HLA-DRB1*11:01,HLA-DRB5*01:01,HLA-DRB1*13:02,HLA-DRB1*04:01,HLA-DRB4*01:01 | 0.8879 | 1160–1174 |
| FFLLIYLGRTGLSKM | HLA-DRB1*11:01,HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DRB5*01:01,HLA-DQA1*05:01/DQB1*03:01 | 0.5873 | 1174–1188 |
| LFFLLIYLGRTGLSK | HLA-DRB1*01:01,HLA-DRB1*15:01,HLA-DRB1*11:01,HLA-DRB5*01:01, HLA-DRB1*04:04, HLA-DQA1*05:01/DQB1*03:01,HLA-DPA1*03:01/DPB1*04:02,HLA-DPA1*02:01/DPB1*01:01,HLA-DPA1*01:03/DPB1*02:01, | 0.6358 | 1173–1187 |
| LLICLYVALSIGLFF | HLA-DRB1*01:01,HLA-DRB1*07:01,HLA-DRB1*15:01,HLA-DRB1*09:01,HLA-DRB1*04:04,HLA-DRB1*04:05,HLA-DRB5*01:01,HLA-DRB1*13:02,HLA-DRB1*04:01,HLA-DRB1*11:01 | 1.0899 | 1161–1175 |
3.6. Analysis of the IFN-γ and IL-4 inducer property of the CD4 + T-cell epitopes
The identified potential CD4 + T-cell epitopes were further checked for their immune response using 2 different immunoinformatics tools, IFNepitope and IL4pred. These two tools helped us to know the IFN-γ and IL-4 inducing CD4 + T-cell epitopes. Potential CD4 + T-cell epitopes having both IFN-γ and IL-4 inducing nature were identified. Herein, we found 1 N and 8 G protein CD4 + T-cell epitopes and these epitopes were considered as the most potential CD4 + T-cell epitopes (Table 4 ). However, we further performed the overlapping process and found 11 different CD8 + T-cell epitopes (1 N and 10 G protein) as the core sequence of 9 IFN-γ and IL-4 inducer CD4 + T-cell epitopes (Table 4). The conservancy analysis results revealed that the CD8 + T-cell epitopes, 143AVFALAPVV151, 142LAVFALAPV150, 145FALAPVVFA153, 144VFALAPVVF152, 134IAMTVLPAL142, 1142FFDWFSGLM1150, and 1175FLLIYLGRT1183 had 100.00% conservancy. On the other hand, the CD4 + T-cell epitopes, 139LPALAVFALAPVVFA153, 133GIAMTVLPALAVFAL147, 1138GSWNFFDWFSGLMSW1152, 140PALAVFALAPVVFAE154, and 1174FFLLIYLGRTGLSKM1188 had also 100.00% conservancy among all the 40 complete G protein sequences. On the contrary, we got 99.39% conservancy for both CD8 + T-cell and CD4 + T-cell epitope of N protein among all the 163 complete N protein sequences (Table 4).
Table 4.
Predicted most potential overlapping CD8 + T-cell epitopes and IFN-γ and IL-4 induced CD4 + T-cell epitopes of RVFV.
| CD8 + T-cell epitopes | Conservancy (%) | CD4 + T-cell epitopes | Conservancy (%) | IFN-γ | IL-4 |
|---|---|---|---|---|---|
| Nucleocapsid (N) | |||||
| MMHPSFAGM | 99.39% | HMMHPSFAGMVDPSL | 99.39% | Inducer | Inducer |
| Glycoprotein (G) | |||||
| AVFALAPVV | 100.00% | LPALAVFALAPVVFA | 100.00% | Inducer | Inducer |
| LAVFALAPV | 100.00% | ||||
| FALAPVVFA | 100.00% | ||||
| VFALAPVVF | 100.00% | ||||
| IAMTVLPAL | 100.00% | GIAMTVLPALAVFAL | 100.00% | Inducer | Inducer |
| TAFIRWVYK | 95.00% | NPLMWITAFIRWVYK | 95.00% | Inducer | Inducer |
| IICLAILYK | 10.00% | IICLAILYKVLKCLK | 5.00% | Inducer | Inducer |
| IICLAILYK AIICLAILY |
10.00% 35.00% |
AIICLAILYKVLKCL | 5.00% | Inducer | Inducer |
| FFDWFSGLM | 100.00% | GSWNFFDWFSGLMSW | 100.00% | Inducer | Inducer |
| FALAPVVFA | 100.00% | PALAVFALAPVVFAE | 100.00% | Inducer | Inducer |
| AVFALAPVV | 100.00% | ||||
| VFALAPVVF | 100.00% | ||||
| LAVFALAPV | 100.00% | ||||
| FLLIYLGRT | 100.00% | FFLLIYLGRTGLSKM | 100.00% | Inducer | Inducer |
3.7. Combined MHC-I and MHC-II HLA allele distribution analysis
Different HLA alleles (MHC-I and MHC-II) are exposed at different frequencies in different ethnicities. So, the identification of population coverage or HLA allele distribution is an important factor for an effective vaccine design. Predicted epitopes that bind to different types of MHC-I and MHC-II HLA alleles are considered as the best plausible epitopes if their combined frequency in a population exhibit good coverage by approaching 100% or close to the 100%.
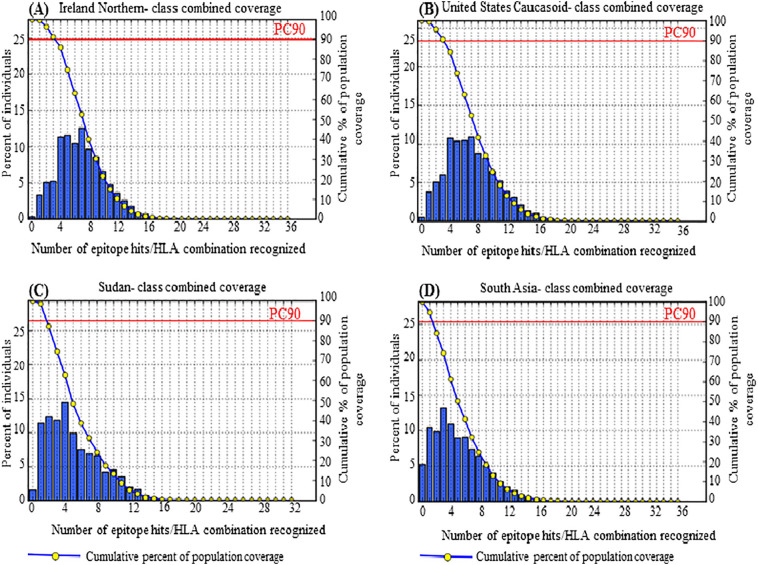
For the G protein, a separate and a combined HLA distribution analysis was performed for 7 CD8 + T-cells and 5 CD4 + T-cells epitopes and their respective HLA alleles. The maximum population coverage was found for the selected CD8 + T-cells and CD4 + T-cells epitopes in 15 different geographic areas of the world (Table 5 ). For CD8 + T-cell epitopes, a good percentage of cumulative population coverage of the 7 epitopes was found in Europe (96.81%), closely followed by North America (95.05%), North Africa (94.17%), and moderately followed by West Africa (89.71%), East Africa (86.45%), South Asia (85.6%), and a poor population coverage was found in South Africa with 1.44% (Table 5). Contrariwise, for CD4 + T-cells epitopes, a reasonable and satisfactory percentage of population coverage of the 5 epitopes was attained in East Asia (69.14%), closely followed by Europe (69.01%), North America (68.07%), and South Asia (63.22%). Herein, the lowest population coverage was also found in South Africa (0.94%) (Table 5). However, a combined HLA distribution analysis using the overlapping CD8 + T-cell epitopes and CD4 + T-cell epitopes showed an excellent population coverage in Europe (99.01%), closely followed by North America (98.42%), East Asia (98.0%), North Africa (97.26%), and moderately followed by West Indies (85.8%). Moreover, maximum coverage, 99.70% was found in the population of Ireland (Northern) and Ireland (Northern) Caucasoid followed by 99.66%, 99.52%, 99.47%, 99.42%, 99.20%, 99.10%, and 98.43% in the population of Mexico Amerindian, England Caucasoid, Germany Caucasoid, United States Caucasoid, France Caucasoid, Italy Caucasoid, and Sudan, respectively. Fig. 2(A), (B), (C), and (D) represent the class combined coverage by the overall population of northern Ireland, United States Caucasoid, Sudan and South Asia, respectively. Though South Africa covered only 0.4%, South Africa (Other) and South Africa (Black) covered 91.28% and 46.34% population, respectively. The population coverage in different countries, area and ethnic groups are shown in Table S5. On the other hand, nonamers CD8 + T-cell epitopes and pentadecamer CD4 + T-cell epitopes derived from N protein showed maximum coverage of 81.25% in the population of United States Polynesian. A medium coverage of 64.04% was found in the population of Mexico Amerindian followed by 61.52%, 59.65%, and 58.65% in the population of Japan Oriental, South Korea, and Finland Caucasoid, respectively (data not shown). Consequently, the population coverage analysis of both N and G protein indicated that the epitopes and the HLA alleles of G protein epitopes are widely distributed throughout the world.
Table 5.
Population coverage rate (%) for the overlapping CD8 + T-cell epitopes and CD4 + T-cell epitopes of the G protein of RVFV.
| Population/area |
Population coverage (%) |
||
|---|---|---|---|
| CD8 + T-cell epitopes | CD4 + T-cell epitopes | Both CD8 + and CD4 + T-cell epitopes | |
| East Asia | 93.52 | 69.14 | 98.0 |
| Northeast Asia | 82.7 | 51.73 | 91.65 |
| South Asia | 85.6 | 63.22 | 94.7 |
| Southeast Asia | 86.07 | 51.25 | 93.21 |
| Southwest Asia | 86.72 | 30.8 | 90.81 |
| Europe | 96.81 | 69.01 | 99.01 |
| East Africa | 86.45 | 51.42 | 93.41 |
| West Africa | 89.71 | 48.28 | 94.68 |
| Central Africa | 90.09 | 39.04 | 93.96 |
| North Africa | 94.17 | 53.0 | 97.26 |
| South Africa | 1.44 | 0.94 | 0.4 |
| West Indies | 67.33 | 56.54 | 85.8 |
| North America | 95.05 | 68.07 | 98.42 |
| South America | 85.49 | 38.24 | 91.04 |
| Oceania | 90.39 | 56.47 | 95.82 |
Fig. 2.
Population coverage by MHC class I and class II restricted epitopes (class combined coverage) predicted from G protein of RVFV. (A) shows maximum coverage by the population of Ireland (Northern) (B) class combined coverage in United States Caucasoid (C) shows population coverage in an endemic country, Sudan (D) represents combined coverage by the overall population of South Asia.
3.8. Putative B-cell epitope prediction and selection
The putative B-cell epitopes from the conserved sequences of N and G protein were predicted according to the criteria set in the ABCpred server. Initially, N protein generated 9 predicted epitopes while G protein generated 69 predicted epitopes from the conserved sequences. Finally, we identified the most probable B-cell epitopes using the criteria, VaxiJen score ≥ 0.5, and non-allergenic nature of the epitopes to the human. So, the results revealed that 1 epitope of N protein and 19 epitopes of G protein maintain both the above-mentioned criteria (Table 6 ). The score ranged from 0.51 to 0.74 in G protein, while the N protein epitope “179VINPNLRGRT188” had 0.82 in ABCpred server. With regard to antigenicity scores attained from VaxiJen server, “179VINPNLRGRT188” and “848RVFNCIDWVH857” sequences derived, respectively, from N and G protein had the highest antigenic scores of 1.9242 and 1.8248, respectively (Table 6). Further analysis showed that the “320GVCEVGVQAL329” and “848RVFNCIDWVH857” decamer epitopes of G protein had the sequence similarity with the nonamer CD8 + T-cell epitopes “321VCEVGVQAL329” and “848RVFNCIDWV856”, respectively (Table 2, Table 6). The conservancy analysis of the B-cell epitopes indicated that all the epitopes had 100.00% conservancy, except the N protein epitope, 179VINPNLRGRT188 (98.77%) and the G protein epitopes, 483ITKCEPHGLV492 (95.0%), 1051SITSTGTGTL1060 (97.50%) and 1087HFTVPEVEEE1096 (97.50%). So, the results revealed that these 100% conserved and non-allergenic B-cell epitopes could be the potential vaccine candidates for epitope based vaccine design.
Table 6.
Putative linear B-cell epitopes from N and G protein of RVFV.
| B-cell epitopes | Position | Abcpred score | Vaxijen score | Conservancy (%) | Allergenicity |
|---|---|---|---|---|---|
| Nucleocapsid (N) | |||||
| VINPNLRGRT | 179–188 | 0.82 | 1.9242 | 98.77 | No |
| Glycoprotein (G) | |||||
| STREETCFGD | 24–33 | 0.66 | 1.2288 | 100.00 | No |
| GASLKKGSYP | 289–298 | 0.74 | 1.3042 | 100.00 | No |
| GVCEVGVQAL | 320–329 | 0.67 | 0.7534 | 100.00 | No |
| DGSKLKTKMK | 310–319 | 0.67 | 1.2747 | 100.00 | No |
| VWKKPLCVGY | 450–459 | 0.65 | 0.9596 | 100.00 | No |
| ITKCEPHGLV | 483–492 | 0.63 | 0.9971 | 95.00 | No |
| RSTGFKISSA | 494–503 | 0.54 | 0.7811 | 100.00 | No |
| SGSNSFSFIE | 898–907 | 0.67 | 0.8627 | 100.00 | No |
| IKTVSSELSC | 747–756 | 0.64 | 0.7460 | 100.00 | No |
| KCFEQCGGWG | 813–822 | 0.64 | 0.5895 | 100.00 | No |
| RVFNCIDWVH | 848–857 | 0.64 | 1.8248 | 100.00 | No |
| FNVNPSCLFV | 826–835 | 0.63 | 1.7464 | 100.00 | No |
| TSAEFSFVGE | 796–805 | 0.59 | 1.5920 | 100.00 | No |
| PKCLSSRRCH | 769–778 | 0.58 | 0.6125 | 100.00 | No |
| SSRRCHLVGE | 773–782 | 0.57 | 1.8137 | 100.00 | No |
| SSELSCREGQ | 751–760 | 0.51 | 1.7370 | 100.00 | No |
| SITSTGTGTL | 1051–1060 | 0.64 | 1.0479 | 97.50 | No |
| HFTVPEVEEE | 1087–1096 | 0.59 | 0.6304 | 97.50 | No |
| GCYSCNAGAR | 1038–1047 | 0.59 | 1.1821 | 100.00 | No |
3.9. Molecular docking simulations of HLA-epitope interaction
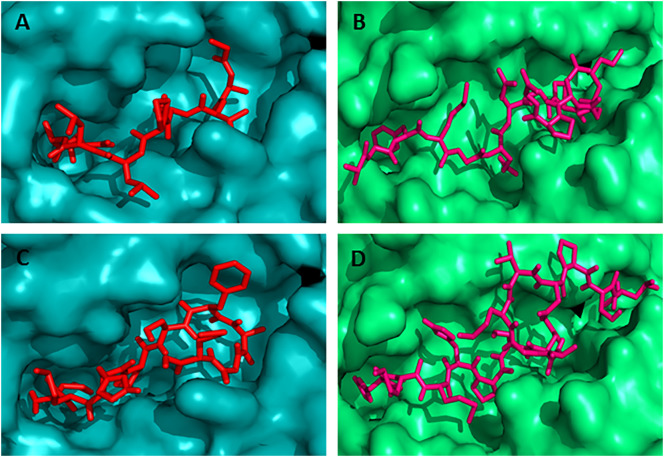
The binding models of the predicted overlapping CD8 + T-cell epitopes and the IFN-γ and IL-4 inducer CD4 + T-cell epitopes with their respective most conserved HLA alleles were generated using AutoDock Vina. The binding affinities (docking score) of the epitopes are presented in the Table 7 . The results showed that the N protein nonamer epitope “MMHPSFAGM” bound to the binding groove of HLA-C*12:03 molecule with the binding energy − 7.5 kcal/mol (Fig. 3(C)), while the control peptide “GAVDPLLAL” bound to the binding grooves of HLA-C*12:03 with the binding affinity of − 8.4 kcal/mol (Fig. 3(A)). The docking simulation of the glycoprotein CD8 + T-cell epitopes showed that the epitopes “AVFALAPVV”, “LAVFALAPV”, “FALAPVVFA”, and “VFALAPVVF” bound to the binding grooves of HLA-C*12:03 with the binding affinity of − 8.8 kcal/mol (Fig. 4(A)), − 9.1 kcal/mol (Fig. 4(B)), − 7.9 kcal/mol (Fig. 4(C)), and − 9.0 kcal/mol (Fig. 4(D)), respectively. The binding energy of the rest of the other 3 CD8 + T-cell epitopes “IAMTVLPAL”, “FFDWFSGLM”, and “FLLIYLGRT” was − 8.5 kcal/mol, − 8.5 kcal/mol, and − 8.7 kcal/mol, respectively (Fig. 5(A), (C) and (E)).
Table 7.
Docking results of the predicted overlapping CD8 + and CD4 + T-cell epitopes of RVFV with respective HLA-alleles.
| CD8 + T-cell epitopes | Docking score (kcal/mol) | No. of H-bond | Allergenicity | CD4 + T-cell epitopes | Docking score (kcal/mol) | No. of H-bond |
Allergenicity | |
|---|---|---|---|---|---|---|---|---|
| Chain C | Chain D | |||||||
| MMHPSFAGM | − 7.5 | 5 | Yes | HMMHPSFAGMVDPSL | − 7.0 | 0 | 3 | Yes |
| AVFALAPVV | − 8.8 | 7 | No | LPALAVFALAPVVFA | − 7.6 | 2 | 5 | Yes |
| LAVFALAPV | − 9.1 | 6 | No | PALAVFALAPVVFAE | − 9.0 | 3 | 3 | Yes |
| FALAPVVFA | − 7.9 | 4 | Yes | |||||
| VFALAPVVF | − 9.0 | 6 | Yes | |||||
| IAMTVLPAL | − 8.5 | 2 | Yes | GIAMTVLPALAVFAL | − 7.6 | 2 | 4 | No |
| FFDWFSGLM | − 8.5 | 6 | Yes | GSWNFFDWFSGLMSW | − 9.0 | 5 | 5 | Yes |
| FLLIYLGRT | − 8.7 | 13 | Yes | FFLLIYLGRTGLSKM | − 6.5 | 6 | 3 | No |
Fig. 3.
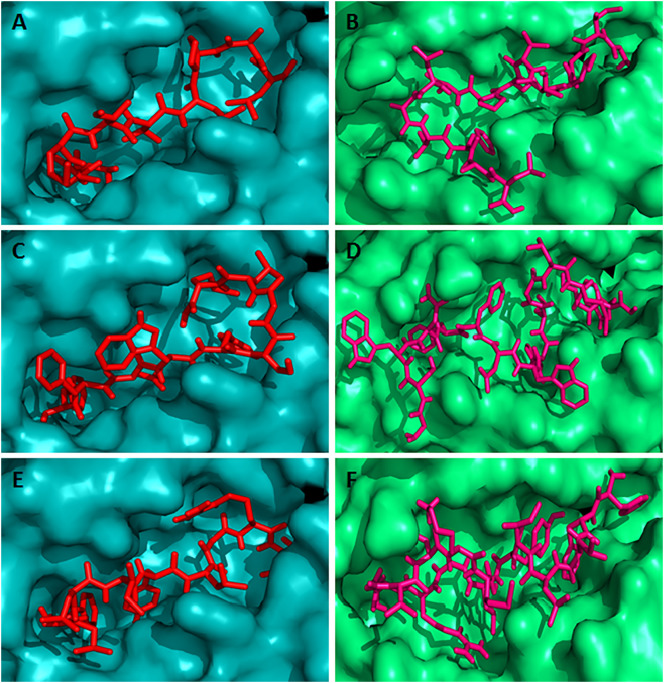
Docking simulation to predict the binding of predicted and control CD8 + and CD4 + T-cell epitopes to MHC class I (HLA-C*12:03) and MHC class II (HLA-DRB1*01:01) molecule, respectively. The colors, teal cyan and lime green indicate the surface structure of HLA-C*12:03 and HLA-DRB1*01:01, respectively. The red and hot pink colors indicate the CD8 + and CD4 + T-cell epitope, respectively (A) binding of control CD8 + T-cell epitope (GAVDPLLAL) to the binding groove of HLA-C*12:03 (affinity: − 8.4 kcal/mol) (B) represents the binding of control CD4 + T-cell epitope (AGFKGEQGPKGEPG) to the binding groove of HLA-DRB1*01:01 (affinity: − 7.3 kcal/mol) (C) shows the binding affinity of N protein CD8 + T-cell epitope “MMHPSFAGM” to the binding groove of HLA-C*12:03 (affinity: − 7.5 kcal/mol) (D) binding of N protein CD4 + T-cell epitope “HMMHPSFAGMVDPSL” to the binding groove of HLA-DRB1*01:01 (affinity: − 7.0 kcal/mol). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Docking simulation to predict the binding of predicted overlapping G protein CD8 + and CD4 + T-cell epitopes to HLA-C*12:03 and HLA-DRB1*01:01 molecules, respectively. The colors, teal cyan and lime green indicate the surface structure of HLA-C*12:03 and HLA-DRB1*01:01, respectively. The red and hot pink colors indicate the CD8 + and CD4 + T-cell epitope, respectively. The binding of CD8 + T-cell epitopes to the binding groove of MHC class I allele HLA-C*12:03 (A) “AVFALAPVV” (affinity: − 8.8 kcal/mol) (B) “LAVFALAPV” (affinity: − 9.1 kcal/mol) (C) “FALAPVVFA” (affinity: − 7.9 kcal/mol) and (D) “VFALAPVVF” (affinity: − 9.0 kcal/mol). On the other hand, the binding of CD4 + T-cell epitopes to the binding groove of MHC class II allele HLA-DRB1*01:01 (E) “LPALAVFALAPVVFA” (affinity: − 7.6 kcal/mol) (F) “PALAVFALAPVVFAE” (affinity: − 9.0 kcal/mol). These 2 CD4 + T-cell epitopes overlapped with the above mentioned 4 CD8 + T-cell epitopes. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 5.
Docking simulation to predict the binding of predicted overlapping G protein CD8 + and CD4 + T-cell epitopes to HLA-C*12:03 and HLA-DRB1*01:01 molecules, respectively. The colors, teal cyan and lime green indicate the surface structure of HLA-C*12:03 and HLA-DRB1*01:01, respectively. The red and hot pink colors indicate the CD8 + and CD4 + T-cell epitope, respectively. (A) CD8 + T-cell epitope “IAMTVLPAL” (affinity: − 8.5 kcal/mol) (B) CD4 + T-cell epitope “GIAMTVLPALAVFAL” (affinity: − 7.6 kcal/mol) (C) CD8 + T-cell epitope “FFDWFSGLM” (affinity: − 8.5 kcal/mol) (D) CD4 + T-cell epitope “GSWNFFDWFSGLMSW” (affinity: − 9.0 kcal/mol) (E) CD8 + T-cell epitope “FLLIYLGRT” (affinity: − 8.7 kcal/mol) and (F) CD4 + T-cell epitope “FFLLIYLGRTGLSKM” (affinity: − 6.5 kcal/mol). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In this study, the HLA allele HLA-DRB1*01:01 was selected as the most conserved allele for the docking simulation study with the CD4 + T-cell epitopes. The binding affinity for the pentadecamer CD4 + T-cell epitopes, “HMMHPSFAGMVDPSL”, “LPALAVFALAPVVFA”, “PALAVFALAPVVFAE”, “GIAMTVLPALAVFAL”, “GSWNFFDWFSGLMSW”, and “FFLLIYLGRTGLSKM” was found − 7.0 kcal/mol (Fig. 3(D)), − 7.6 kcal/mol (Fig. 4(E)), − 9.0 kcal/mol (Fig. 4(F)), − 7.6 kcal/mol (Fig. 5(B)), − 9.0 kcal/mol (Fig. 5(D)), and − 6.5 kcal/mol (Fig. 5(F)), respectively. The binding energy − 7.3 kcal/mol was predicted for the control peptide “AGFKGEQGPKGEPG” in the binding groove of HLA-DRB1*01:01.
4. Discussion
Emerging infectious diseases are typically characterized by very high epidemics mainly caused by the viral pathogens which are accountable for the greatest proportion of the emerging infectious disease threat throughout the world. Over the past thirty-five years, about thirty viral pathogens have emerged due to their human affecting nature. Most of these pathogens are zoonotic and their origins are ominously correlated with ecological, environmental, and socioeconomic factors (Dye, 2014, Nii-Trebi, 2017).Top priority emerging infectious diseases such as Rift Valley fever, Crimean-Congo haemorrhagic fever, Ebola virus disease, Marburg haemorrhagic fever, Middle East respiratory syndrome (MERS), Severe acute respiratory syndrome (SARS), Nipah, and Lassa fever are caused by the viral pathogens which are highly pathogenic to the human (Nii-Trebi, 2017). The immunity associated with protection from these pathogens remains largely unknown to the researchers. So, the vaccine development against these newly emerging infectious diseases is demanding and challenging work (Terry et al., 2015). Therefore, the genome sequence screening of viral pathogens could be the potential step for the identification of vaccine target against the newly emerging infectious diseases.
In this study, we aim to screen and explore the conserved regions of the N and G protein of the RVFV and to find out the CD8 +, CD4 + T-cell epitopes as well as B-cell epitopes and most importantly the overlapping CD8 + and CD4 + T-cell epitopes from the conserved sequences of the proteins by using a strategy namely genome-wide screening of vaccine epitopes. The identified 4 conserved sequences from N protein and 8 conserved sequences of G protein of RVFV based on the antigenicity and exomembrane characteristics are shown in Table 1. We used immunoinformatics approaches for this purpose. The immunoinformatics approaches on the conserved sequences have been used in different viral proteins such as Ebola virus nucleoprotein and glycoprotein (Ali and Islam, 2015, Dash et al., 2017, Dikhit et al., 2015), Zika virus glycoprotein (Alam et al., 2016, Dikhit et al., 2016), Chikungunya virus proteins (Islam et al., 2012, Kori et al., 2015), Nipah virus fusion and glycoprotein (Ali et al., 2015, Sakib et al., 2014), but yet no immunoinformatics approach has been applied to the RVFV protein.
CD8 + T-cells restrict the extent of infection from tissues by identifying and killing infected cells or by the specific antiviral cytokines secretion (Garcia et al., 1999). Therefore, T-cell epitope-based vaccination could be a distinctive process of eliciting a robust immune response against the infectious pathogens, such as viruses (He et al., 2015, Jin et al., 2009, Staneková and Varečková, 2010). According to the Xu et al. (2013) N protein of RVFV is an effective human T-cell immunogen which is able to stimulate comprehensive and potentially protective immunodominant CD8 + T-cell responses. On the other hand, Dodd et al. (2013) demonstrated that, CD4 + T-cells are essential for strong IgG and neutralizing antibody responses that mediate RVFV clearance from peripheral tissues. The researchers also revealed that CD4 + T-cells are responsible for the regulation of immune responses to RVFV infection as well as play a vital role in averting the onset of neurologic disease (Dodd et al., 2013). On the contrary, the researchers Bird and McElroy (2016) reviewed that the contribution and function of CD8 + T-cells and CD4 + T-cells are considerably less known in endorsing and maintaining proper immune control of RVFV infection. However, we screened and identified the promiscuous CD8 + and CD4 + T-cells epitopes (Tables 2 and S3) using in silico approaches for further research in developing epitope-based peptide vaccine in the near future.
A peptide-based vaccine is preferable for its safety, specificity, and absence of the infectious agent (Naylor et al., 2011). However, the small peptide vaccine could be rejected due to the MHC restriction, lack of helper activity, and also weak presentation of antigen by antigen presenting cell (APC), but this rejection may be overcome by using compound peptide vaccines, which include both CD8 + T-cell and CD4 + T-cell epitopes or longer peptides with CD8 + and CD4 + T-cell epitopes (Lohia and Baranwal, 2014, Naylor et al., 2011). The overlapping CD8 + and CD4 + T-cell epitopes have been identified as the vaccine candidates against the cancer Metadherin protein (Dhiman et al., 2016) and H1N1 influenza virus protein (Lohia and Baranwal, 2014, Lohia and Baranwal, 2015). Herein, we identified overlapping CD8 + and CD4 + T-cell epitopes for peptide vaccine design (Table 4). It is noteworthy that the CD8 + T-cell epitopes, 597IICLAILYK605, 596AIICLAILY604, and 625TAFIRWVYK633 were found as the core sequence of the CD4 + T-cell epitopes, but they contained the conservancy score of 10.00%, 35.00%, and 95.00%, respectively. So, these epitopes could not be the vaccine target. Since, both humoral (Th2) and cellular (Th1) mediated immune response produced protective immunity for RVF VLP (Virus like particle) vaccinated animals (Mandell et al., 2010), predicted CD4 + T-cells epitopes were analyzed for Th1 cytokine, IFN-γ and Th2 cytokine, IL-4 production using in silico tools. The experimental study showed that splenocytes from VLP-vaccinated mice secreted IL-4 and IFN-γ which is stimulated by the RVFV-specific antigen, consistent with both humoral and cellular immune responses (Chung et al., 2008, Kortekaas et al., 2012, Mandell et al., 2010). An in silico approach has been used for the identification of IL-4 and IFN-γ induced promiscuous epitopes from Leishmania donovani antigen (Kashyap et al., 2017). Therefore, only those CD4 + T-cell epitopes of G and N protein were selected which were found to be IL-4 and IFN-γ inducers in in silico analysis (Table 4). The results revealed that although the CD4 + T-cell epitopes, 619NPLMWITAFIRWVYK633, 597IICLAILYKVLKCLK611, and 596AIICLAILYKVLKCL610 are IL-4 and IFN-γ inducers, they could not be the potential vaccine candidates due to the low conservancy score of 95.00%, 5.00%, and 5.00%, respectively. In addition, the primary and secondary humoral immunity inducer B-cell epitopes were also predicted and the results demonstrated that two of the G protein B-cell epitopes, “GVCEVGVQAL” and “RVFNCIDWVH” overlapped with the CD8 + T-cell epitopes, “VCEVGVQAL” and “RVFNCIDWV” and all the B-cell epitopes were non-allergenic to the human. The researcher Lal and his coworkers reported that superimposition or overlapping of CD8 + T-cell epitopes with B-cell epitopes would confirm good T-cell response with precise T-cell memory and will be advantageous for the design of the epitope-based vaccine (Lal et al., 2006). So, these epitopes have the potentiality in making an effective vaccine or diagnostic kit.
The severe RVFV outbreak have been recorded in human with high fatality rates for the year 2000 to 2016 in different countries such as Saudi Arabia, Kenya, Somalia, Sudan, South Africa, Namibia, Mauritania, Egypt, Senegal, Niger and Uganda (Balkhy and Memish, 2003, Hartman, 2017, Hassan et al., 2011, Métras et al., 2012, Monaco et al., 2013, Nguku et al., 2010, Sow et al., 2014, Sow et al., 2016). According to the Mansfield et al. (2015) an incursion may occur in Europe due to the geographical range and emergence of RVFV in the northern Egypt and Middle East. So, the higher population coverage in these regions or country is essential to make an effective vaccine against RVFV. In this study, the combined coverage showed that Saudi Arabia, Brazil (Mixed), Uganda, Kenya, and Senegal covered about 98.98%, 98.12%, 90.32%, 89.85%, and 89.00% population, respectively (Table S5). The population coverage for Europe was identified as 99.01% in combined coverage, while the coverage was 98.01% for the World. South Africa covered only 0.4%, but in South African (other) the coverage was found at 91.28% (Table S5). Finally, the results revealed that the epitopes of RVFV G protein would be the effective vaccine target for epitope-based peptide vaccine design. Moreover, molecular docking simulation was performed to explore the binding interaction of the predicted CD8 + and CD4 + T-cell epitopes of the G and N protein. 3D structures of the HLA-C*12:03 and HLA-DRB1*01:01 were available in Protein Data Bank (PDB) with accession id 1EFX and 2FSE, respectively. The docking simulation was performed for those CD8 + and CD4 + T-cell epitopes which were overlapped or superimposed with each other. Herein, the CD8 + T-cell epitopes “AVFALAPVV”, “LAVFALAPV”, “FALAPVVFA”, and “VFALAPVVF” overlapped with both CD4 + T-cell epitopes “LPALAVFALAPVVFA” and “PALAVFALAPVVFAE”, while the rest of the other CD8 + T-cell epitopes, “MMHPSFAGM”, “IAMTVLPAL”, “FFDWFSGLM”, and “FLLIYLGRT” overlapped with “HMMHPSFAGMVDPSL”, “GIAMTVLPALAVFAL”, “GSWNFFDWFSGLMSW”, and “FFLLIYLGRTGLSKM”, respectively and bound with their respective HLA-alleles with significant binding affinity and variable number of hydrogen bonds (Table 7 and Fig. 3, Fig. 4, Fig. 5). So, these epitopes could be the possible vaccine candidates for epitope-based RVFV vaccine design.
5. Conclusion
In this study, overlapping CD8 + and CD4 + T-cell epitopes from RVFV N and G protein were successfully identified by using immunoinformatics-driven genome wide screening approach. The identified epitopes have antigenicity, positive immunogenicity, higher conservancy as well as IL-4 and IFN-γ induced (CD4 + T-cell epitopes) capability. Furthermore, selected CD8 + and CD4 + T-cell epitopes have high binding affinity to their respective HLA-alleles and the allele frequency of these epitopes are high in endemic as well as in many non-endemic regions. Besides, all the predicted 20 B-cell epitopes have also antigenicity, highest conservancy score, and non-allergenic property to the human. Therefore, these epitopes may be used for the development of epitope-based peptide vaccine against emerging RVFV. However, along with this in silico approach, in vivo, and in vitro experiments are required for their efficient use as a vaccine against RVFV.
Acknowledgments
Acknowledgement
The author(s) reply to thanks to the Professor A.K.M Akhtarul Islam, Department of English, Islamic University, Kushtia for editing and checking the spelling and grammatical error of this manuscript.
Conflict of interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2017.10.022.
Appendix A. Supplementary data
Supplementary tables
References
- Alam A., Ali S., Ahamad S., Malik M.Z., Ishrat R. From ZikV genome to vaccine: in silico approach for the epitope-based peptide vaccine against Zika virus envelope glycoprotein. Immunology. 2016;149:386–399. doi: 10.1111/imm.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.T., Islam M.O. A highly conserved GEQYQQLR epitope has been identified in the nucleoprotein of Ebola virus by using an in silico approach. Adv. Bioinforma. 2015 doi: 10.1155/2015/278197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M.T., Morshed M.M., Hassan F. A computational approach for designing a universal epitope-based peptide vaccine against Nipah virus. Interdiscip. Sci. 2015;7:177–185. doi: 10.1007/s12539-015-0023-0. [DOI] [PubMed] [Google Scholar]
- Anyamba A., Linthicum K.J., Small J., Britch S.C., Pak E., de La Rocque S., Formenty P., Hightower A.W., Breiman R.F., Chretien J.-P., Tucker C.J., Schnabel D., Sang R., Haagsma K., Latham M., Lewandowski H.B., Magdi S.O., Mohamed M.A., Nguku P.M., Reynes J.-M., Swanepoel R. Prediction, assessment of the Rift Valley fever activity in east and southern Africa 2006–2008 and possible vector control strategies. Am. J. Trop. Med. Hyg. 2010 doi: 10.4269/ajtmh.2010.09-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anyangu A.S., Hannah Gould L., Sharif S.K., Nguku P.M., Omolo J.O., Mutonga D., Rao C.Y., Lederman E.R., Schnabel D., Paweska J.T., Katz M., Hightower A., Kariuki Njenga M., Feikin D.R., Breiman R.F. Risk factors for severe Rift Valley fever infection in Kenya, 2007. Am. J. Trop. Med. Hyg. 2010 doi: 10.4269/ajtmh.2010.09-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhy H.H., Memish Z.A. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int. J. Antimicrob. Agents. 2003;21:153–157. doi: 10.1016/S0924-8579(02)00295-9. [DOI] [PubMed] [Google Scholar]
- Billecocq A., Spiegel M., Vialat P., Kohl A., Weber F., Bouloy M., Haller O. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 2004 doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird B.H., McElroy A.K. Rift Valley fever virus: unanswered questions. Antivir. Res. 2016;132:274–280. doi: 10.1016/j.antiviral.2016.07.005. [DOI] [PubMed] [Google Scholar]
- Borio L., Inglesby T., Peters C.J., Schmaljohn A.L., Hughes J.M., Jahrling P.B., Ksiazek T., Johnson K.M., Meyerhoff A., O'Toole T., Ascher M.S., Bartlett J., Breman J.G., Eitzen E.M., Hamburg M., Hauer J., Henderson D.A., Johnson R.T., Kwik G., Layton M., Lillibridge S., Nabel G.J., Osterholm M.T., Perl T.M., Russell P., Tonat K. Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- Bouloy M., Weber F. Molecular biology of Rift Valley fever virus. Open Virol. J. 2010 doi: 10.2174/1874357901004010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M., Janzen C., Vialat P., Khun H., Pavlovic J., Huerre M., Haller O. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 2001 doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyington J.C., Motyka S.A., Schuck P., Brooks A.G., Sun P.D. Crystal structure of an NK cell immunoglobulin-like receptor in complex with its class I MHC ligand. Nature. 2000;405:537–543. doi: 10.1038/35014520. [DOI] [PubMed] [Google Scholar]
- Bui H.-H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinforma. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H.-H., Sidney J., Li W., Fusseder N., Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinforma. 2007;8:361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calis J.J.A., Maybeno M., Greenbaum J.A., Weiskopf D., De Silva A.D., Sette A., Keşmir C., Peters B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y.-C., Ho M.-S., Wu J.-C., Chen W.-J., Huang J.-H., Chou S.-T., Hu Y.-C. Immunization with virus-like particles of enterovirus 71 elicits potent immune responses and protects mice against lethal challenge. Vaccine. 2008;26:1855–1862. doi: 10.1016/j.vaccine.2008.01.058. [DOI] [PubMed] [Google Scholar]
- Dar O., McIntyre S., Hogarth S., Heymann D. Rift Valley fever and a new paradigm of research and development for zoonotic disease control. Emerg. Infect. Dis. 2013 doi: 10.3201/eid1902.120941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash R., Das R., Junaid M., Akash M.F.C., Islam A., Hosen S.M.Z. In silico-based vaccine design against Ebola virus glycoprotein. Adv. Appl. Bioinforma. Chem. 2017 doi: 10.2147/AABC.S115859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubney R., Hudson J.R., Garnham P.C. Enzootic hepatitis or rift valley fever. An undescribed virus disease of sheep cattle and man from east africa. J. Pathol. Bacteriol. 1931;34:545–579. doi: 10.1002/path.1700340418. [DOI] [Google Scholar]
- Davies F.G., Linthicum K.J., James A.D. Rainfall and epizootic Rift Valley fever. Bull. World Health Organ. 1985;63(5):941–943. [PMC free article] [PubMed] [Google Scholar]
- Dhanda S.K., Gupta S., Vir P., Raghava G.P.S. Prediction of IL4 inducing peptides. Clin. Dev. Immunol. 2013 doi: 10.1155/2013/263952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhiman G., Lohia N., Jain S., Baranwal M. Metadherin peptides containing CD4(+) and CD8(+) T cell epitopes as a therapeutic vaccine candidate against cancer. Microbiol. Immunol. 2016;60:646–652. doi: 10.1111/1348-0421.12436. [DOI] [PubMed] [Google Scholar]
- Dikhit M.R., Kumar S., Vijaymahantesh Sahoo B.R., Mansuri R., Amit A., Ansari M.Y., Sahoo G.C., Bimal S., Das P. Computational elucidation of potential antigenic CTL epitopes in Ebola virus. Infect. Genet. Evol. 2015;36:369–375. doi: 10.1016/j.meegid.2015.10.012. [DOI] [PubMed] [Google Scholar]
- Dikhit M.R., Ansari M.Y., Vijaymahantesh Kalyani, Mansuri R., Sahoo B.R., Dehury B., Amit A., Topno R.K., Sahoo G.C., Ali V., Bimal S., Das P. Computational prediction and analysis of potential antigenic CTL epitopes in Zika virus: a first step towards vaccine development. Infect. Genet. Evol. 2016;45:187–197. doi: 10.1016/j.meegid.2016.08.037. [DOI] [PubMed] [Google Scholar]
- Dodd K.A., McElroy A.K., Jones M.E.B., Nichol S.T., Spiropoulou C.F. Rift Valley fever virus clearance and protection from neurologic disease are dependent on CD4(+) T cell and virus-specific antibody responses. J. Virol. 2013 doi: 10.1128/JVI.00337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinforma. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dungu B., Louw I., Lubisi A., Hunter P., von Teichman B.F., Bouloy M. Evaluation of the efficacy and safety of the Rift Valley Fever Clone 13 vaccine in sheep. Vaccine. 2010;28:4581–4587. doi: 10.1016/j.vaccine.2010.04.085. [DOI] [PubMed] [Google Scholar]
- Dye C. After 2015: infectious diseases in a new era of health and development. Philos. Trans. R. Soc. B Biol. Sci. 2014 doi: 10.1098/rstb.2013.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faburay B., Lebedev M., McVey D.S., Wilson W., Morozov I., Young A., Richt J.A. A glycoprotein subunit vaccine elicits a strong Rift Valley fever virus neutralizing antibody response in sheep. Vector Borne Zoonotic Dis. 2014 doi: 10.1089/vbz.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faburay B., Wilson W.C., Gaudreault N.N., Davis A.S., Shivanna V., Bawa B., Sunwoo S.Y., Ma W., Drolet B.S., Morozov I., McVey D.S., Richt J.A. A recombinant Rift Valley fever virus glycoprotein subunit vaccine confers full protection against Rift Valley fever challenge in sheep. Sci Rep. 2016 doi: 10.1038/srep27719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firbas C., Jilma B., Tauber E., Buerger V., Jelovcan S., Lingnau K., Buschle M., Frisch J., Klade C.S. Immunogenicity and safety of a novel therapeutic hepatitis C virus (HCV) peptide vaccine: a randomized, placebo controlled trial for dose optimization in 128 healthy subjects. Vaccine. 2006;24:4343–4353. doi: 10.1016/j.vaccine.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Garcia K.C., Teyton L., Wilson I.A. Structural basis of T cell recognition. Annu. Rev. Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- Hartman A. Rift Valley fever. Clin. Lab. Med. 2017;37:285–301. doi: 10.1016/j.cll.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan O.A., Ahlm C., Sang R., Evander M. The 2007 Rift Valley fever outbreak in Sudan. PLoS Negl. Trop. Dis. 2011 doi: 10.1371/journal.pntd.0001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Cheng Y., Kong L., Azadnia P., Giang E., Kim J., Wood M.R., Wilson I.A., Law M., Zhu J. Approaching rational epitope vaccine design for hepatitis C virus with meta-server and multivalent scaffolding. Sci Rep. 2015 doi: 10.1038/srep12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T. 2009. Rift Valley Fever Virus NSs Protein Promotes Post-Transcriptional Downregulation of Protein Kinase PKR and Inhibits eIF2α Phosphorylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T. Rift Valley fever vaccines: an overview of the safety and efficacy of the live-attenuated MP-12 vaccine candidate. Expert. Rev. Vaccines. 2017;16:601–611. doi: 10.1080/14760584.2017.1321482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T., Makino S. The pathogenesis of Rift Valley fever. Viruses. 2011 doi: 10.3390/v3050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T., Peters C.J., Makino S. Rift Valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a Minigenome system. J. Virol. 2005 doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam R., Sakib M.S., Aubhishek Z. A computational assay to design an epitope-based peptide vaccine against chikungunya virus. Futur. Virol. 2012;7:1029–1042. doi: 10.2217/fvl.12.95. [DOI] [Google Scholar]
- Jin X., Newman M.J., De-Rosa S., Cooper C., Thomas E., Keefer M., Fuchs J., Blattner W., Livingston B.D., McKinney D.M., Noonan E., deCamp A., Defawe O.D., Wecker M. A novel HIV T helper epitope-based vaccine elicits cytokine-secreting HIV-specific CD4 + T cells in a Phase I clinical trial in HIV-uninfected adults. Vaccine. 2009;27:7080–7086. doi: 10.1016/j.vaccine.2009.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap M., Jaiswal V., Farooq U. Prediction and analysis of promiscuous T cell-epitopes derived from the vaccine candidate antigens of Leishmania donovani binding to MHC class-II alleles using in silico approach. Infect. Genet. Evol. 2017;53:107–115. doi: 10.1016/j.meegid.2017.05.022. [DOI] [PubMed] [Google Scholar]
- Kori P., Sajjan S.S., Madagi S.B. In silico prediction of epitopes for Chikungunya viral strains. J. Pharm. Investig. 2015;45:579–591. doi: 10.1007/s40005-015-0205-0. [DOI] [Google Scholar]
- Kortekaas J., Antonis A.F.G., Kant J., Vloet R.P.M., Vogel A., Oreshkova N., de Boer S.M., Bosch B.J., Moormann R.J.M. Efficacy of three candidate Rift Valley fever vaccines in sheep. Vaccine. 2012;30:3423–3429. doi: 10.1016/j.vaccine.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes11 Edited by F. Cohen. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lagerqvist N., Näslund J., Lundkvist Å., Bouloy M., Ahlm C., Bucht G. Characterisation of immune responses and protective efficacy in mice after immunisation with Rift Valley fever virus cDNA constructs. Virol. J. 2009 doi: 10.1186/1743-422X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal G., Shaila M.S., Nayak R. Activated mouse T cells downregulate, process and present their surface TCR to cognate anti-idiotypic CD4 + T cells. Immunol. Cell Biol. 2006;84:145–153. doi: 10.1111/j.1440-1711.2005.01405.x. [DOI] [PubMed] [Google Scholar]
- Larsen M.V., Lundegaard C., Lamberth K., Buus S., Lund O., Nielsen M. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinforma. 2007;8:424. doi: 10.1186/1471-2105-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux-Roels G., Bonanni P., Tantawichien T., Zepp F. Vaccine development. Perspect. Vaccinol. 2011;1:115–150. [Google Scholar]
- Linthicum K.J., Britch S.C., Anyamba A. Rift Valley fever: an emerging mosquito-borne disease. Annu. Rev. Entomol. 2016;61:395–415. doi: 10.1146/annurev-ento-010715-023819. [DOI] [PubMed] [Google Scholar]
- Lohia N., Baranwal M. Conserved peptides containing overlapping CD4 + and CD8 + T-cell epitopes in the H1N1 influenza virus: an immunoinformatics approach. Viral Immunol. 2014;27:225–234. doi: 10.1089/vim.2013.0135. [DOI] [PubMed] [Google Scholar]
- Lohia N., Baranwal M. Identification of conserved peptides comprising multiple T cell epitopes of matrix 1 protein in H1N1 influenza virus. Viral Immunol. 2015;28:570–579. doi: 10.1089/vim.2015.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Gil E., Lorenzo G., Hevia E., Borrego B., Eiden M., Groschup M., Gilbert S.C., Brun A. A single immunization with MVA expressing GnGc glycoproteins promotes epitope-specific CD8 +-T cell activation and protects immune-competent mice against a lethal RVFV infection. PLoS Negl. Trop. Dis. 2013;7:1–14. doi: 10.1371/journal.pntd.0002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell R.B., Flick R. Rift Valley fever virus: an unrecognized emerging threat? Hum. Vaccin. 2010;6:597–601. doi: 10.4161/hv.6.7.11761. [DOI] [PubMed] [Google Scholar]
- Mandell R.B., Flick R. Rift Valley fever virus: a real bioterror threat. J. Bioterror. Biodef. 2011;2:108. doi: 10.4172/2157-2526.1000108. [DOI] [Google Scholar]
- Mandell R.B., Koukuntla R., Mogler L.J.K., Carzoli A.K., Freiberg A.N., Holbrook M.R., Martin B.K., Staplin W.R., Vahanian N.N., Link C.J., Flick R. A replication-incompetent Rift Valley fever vaccine: chimeric virus-like particles protect mice and rats against lethal challenge. Virology. 2010;397:187–198. doi: 10.1016/j.virol.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield K.L., Banyard A.C., McElhinney L., Johnson N., Horton D.L., Hernández-Triana L.M., Fooks A.R. Rift Valley fever virus: a review of diagnosis and vaccination, and implications for emergence in Europe. Vaccine. 2015;33:5520–5531. doi: 10.1016/j.vaccine.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Métras R., Porphyre T., Pfeiffer D.U., Kemp A., Thompson P.N., Collins L.M., White R.G. Exploratory space-time analyses of Rift Valley fever in South Africa in 2008–2011. PLoS Negl. Trop. Dis. 2012 doi: 10.1371/journal.pntd.0001808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller S., Croning M.D., Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Monaco F., Pinoni C., Cosseddu G.M., Khaiseb S., Calistri P., Molini U., Bishi A., Conte A., Scacchia M., Lelli R. Rift Valley fever in Namibia, 2010. Emerg. Infect. Dis. 2013 doi: 10.3201/eid1912.130593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muh H.C., Tong J.C., Tammi M.T. AllerHunter: a SVM-pairwise system for assessment of allergenicity and allergic cross-reactivity in proteins. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R., Saluzzo J.F., Lopez N., Dreier T., Turell M., Smith J., Bouloy M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am. J. Trop. Med. Hyg. 1995;53:405–411. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- Näslund J., Lagerqvist N., Habjan M., Lundkvist Å., Evander M., Ahlm C., Weber F., Bucht G. Vaccination with virus-like particles protects mice from lethal infection of Rift Valley fever virus. Virology. 2009;385:409–415. doi: 10.1016/j.virol.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Naylor P.H., Egan J.E., Berinstein N.L. Peptide based vaccine approaches for cancer—a novel approach using a WT-1 synthetic long peptide and the IRX-2 immunomodulatory regimen. Cancers (Basel). 2011 doi: 10.3390/cancers3043991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguku P.M., Sharif S.K., Mutonga D., Amwayi S., Omolo J., Mohammed O., Farnon E.C., Gould L.H., Lederman E., Rao C., Sang R., Schnabel D., Feikin D.R., Hightower A., Njenga M.K., Breiman R.F. An investigation of a major outbreak of Rift Valley fever in Kenya: 2006–2007. Am. J. Trop. Med. Hyg. 2010 doi: 10.4269/ajtmh.2010.09-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M., Lundegaard C., Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinforma. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nii-Trebi N.I. Emerging and neglected infectious diseases: insights, advances, and challenges. Biomed. Res. Int. 2017 doi: 10.1155/2017/5245021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminform. 2011 doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzún P., Kobe B. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum. Vaccin. Immunother. 2016;12:763–767. doi: 10.1080/21645515.2015.1094595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B., Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinforma. 2005 doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Habjan M., Unger H., Weber F. Virus-like particles expressing the nucleocapsid gene as an efficient vaccine against Rift Valley fever virus. Vector Borne Zoonotic Dis. 2010;10:701–703. doi: 10.1089/vbz.2009.0248. [DOI] [PubMed] [Google Scholar]
- Pickett B.E., Sadat E.L., Zhang Y., Noronha J.M., Squires R.B., Hunt V., Liu M., Kumar S., Zaremba S., Gu Z., Zhou L., Larson C.N., Dietrich J., Klem E.B., Scheuermann R.H. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R., Binn L.N., Harrison V.R. Immunization against rift valley fever virus. Studies on the immunogenicity of lyophilized formalin-inactivated vaccine. J. Immunol. 1964;93:293–299. [PubMed] [Google Scholar]
- Rolin A.I., Berrang-Ford L., Kulkarni M.A. The risk of Rift Valley fever virus introduction and establishment in the United States and European Union. Emerg. Microbes Infect. 2013 doi: 10.1038/emi.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloniec E.F., Ivey R.A., 3rd, Whittington K.B., Kang A.H., Park H.-W. Crystallographic structure of a rheumatoid arthritis MHC susceptibility allele, HLA-DR1 (DRB1*0101), complexed with the immunodominant determinant of human type II collagen. J. Immunol. 2006;177:3884–3892. doi: 10.4049/jimmunol.177.6.3884. [DOI] [PubMed] [Google Scholar]
- Rusnak J.M., Gibbs P., Boudreau E., Clizbe D.P., Pittman P. Immunogenicity and safety of an inactivated Rift Valley fever vaccine in a 19-year study. Vaccine. 2011;29:3222–3229. doi: 10.1016/j.vaccine.2011.02.037. [DOI] [PubMed] [Google Scholar]
- Saha S., Raghava G.P.S. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins Struct. Funct. Bioinforma. 2006;65:40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- Sakib M.S., Islam M.R., Hasan A.K.M.M., Nabi A.H.M.N. Prediction of epitope-based peptides for the utility of vaccine development from fusion and glycoprotein of Nipah virus using in silico approach. Adv. Bioinforma. 2014 doi: 10.1155/2014/402492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn C.S., Parker M.D., Ennis W.H., Dalrymple J.M., Collett M.S., Slizich J.A., Schmaljohn A.L. Baculovirus expression of the M genome segment of Rift Valley fever virus and examination of antigenic and immunogenic properties of the expressed proteins. Virology. 1989;170:184–192. doi: 10.1016/0042-6822(89)90365-6. [DOI] [PubMed] [Google Scholar]
- Shen Y., Maupetit J., Derreumaux P., Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theory Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- Shi J., Zhang J., Li S., Sun J., Teng Y., Wu M., Li J., Li Y., Hu N., Wang H., Hu Y. Epitope-based vaccine target screening against highly pathogenic MERS-CoV: an in silico approach applied to emerging infectious diseases. PLoS One. 2015;10:1–16. doi: 10.1371/journal.pone.0144475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sominskaya I., Skrastina D., Dislers A., Vasiljev D., Mihailova M., Ose V., Dreilina D., Pumpens P. Construction and immunological evaluation of multivalent hepatitis B virus (HBV) core virus-like particles carrying HBV and HCV epitopes. Clin. Vaccine Immunol. 2010;17:1027–1033. doi: 10.1128/CVI.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sow A., Faye O., Ba Y., Ba H., Diallo D., Faye O., Loucoubar C., Boushab M., Barry Y., Diallo M., Sall A.A. Rift Valley fever outbreak, southern Mauritania, 2012. Emerg. Infect. Dis. 2014;20:296–299. doi: 10.3201/eid2002.131000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sow A., Faye O., Ba Y., Diallo D., Fall G., Faye O., Bob N.S., Loucoubar C., Richard V., Dia A.T., Diallo M., Malvy D., Sall A.A. Widespread Rift Valley fever emergence in Senegal in 2013–2014. Open Forum Infect. Dis. 2016 doi: 10.1093/ofid/ofw149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spik K., Shurtleff A., McElroy A.K., Guttieri M.C., Hooper J.W., Schmaljohn C. Immunogenicity of combination DNA vaccines for Rift Valley fever virus, tick-borne encephalitis virus, Hantaan virus, and Crimean Congo hemorrhagic fever virus. Vaccine. 2006;24:4657–4666. doi: 10.1016/j.vaccine.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Staneková Z., Varečková E. Conserved epitopes of influenza A virus inducing protective immunity and their prospects for universal vaccine development. Virol. J. 2010 doi: 10.1186/1743-422X-7-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry F.E., Moise L., Martin R.F., Torres M., Pilotte N., Williams S.A., de Groot A.S. Time for T? Immunoinformatics addresses vaccine design for neglected tropical and emerging infectious diseases. Expert Rev. Vaccines. 2015;14:21–35. doi: 10.1586/14760584.2015.955478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenet P., Shen Y., Maupetit J., Guyon F., Derreumaux P., Tufféry P. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40:W288–93. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Higgins D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. (Chapter 2, Unit 2.3) [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivona S., Gardy J.L., Ramachandran S., Brinkman F.S.L., Raghava G.P.S., Flower D.R., Filippini F. Computer-aided biotechnology: from immuno-informatics to reverse vaccinology. Trends Biotechnol. 2008;26:190–200. doi: 10.1016/j.tibtech.2007.12.006. [DOI] [PubMed] [Google Scholar]
- van Vuren P.J., Potgieter A.C., Paweska J.T., van Dijk A.A. Preparation and evaluation of a recombinant Rift Valley fever virus N protein for the detection of IgG and IgM antibodies in humans and animals by indirect ELISA. J. Virol. Methods. 2007;140:106–114. doi: 10.1016/j.jviromet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Wallace D.B., Ellis C.E., Espach A., Smith S.J., Greyling R.R., Viljoen G.J. Protective immune responses induced by different recombinant vaccine regimes to Rift Valley fever. Vaccine. 2006;24:7181–7189. doi: 10.1016/j.vaccine.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Won S., Ikegami T., Peters C.J., Makino S. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J. Virol. 2006 doi: 10.1128/JVI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S., Ikegami T., Peters C.J., Makino S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J. Virol. 2007 doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Watts D.M., Costanzo M.C., Tang X., Venegas L.A., Jiao F., Sette A., Sidney J., Sewell A.K., Wooldridge L., Makino S., Morrill J.C., Peters C.J., Kan-Mitchell J. The Nucleocapsid protein of Rift Valley fever virus is a potent human CD8(+) T cell antigen and elicits memory responses. PLoS One. 2013 doi: 10.1371/journal.pone.0059210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables