Highlights
-
•
The genetic basis of host responses to infectious bronchitis virus is unclear.
-
•
We detected 20 significant markers for the antibody response to infectious bronchitis virus in chicken.
-
•
Loci on chromosomes 1 and 5 explained 12% and 13% of phenotypic variation.
-
•
The host immune response cluster had 13 beta-defensin and interleukin-17F genes.
-
•
Our results will contribute to the control of outbreaks of infectious bronchitis.
Keywords: Chicken, Coronavirus, Infectious bronchitis virus, Immune response, Genome-wide association study, Single nucleotide polymorphism
Abstract
Coronaviruses are a hot research topic because they can cause severe diseases in humans and animals. Infectious bronchitis virus (IBV), belonging to gamma-coronaviruses, causes a highly infectious respiratory viral disease and can result in catastrophic economic losses to the poultry industry worldwide. Unfortunately, the genetic basis of the host immune responses against IBV is poorly understood. In the present study, the antibody levels against IBV post-immunization were measured by an enzyme-linked immunosorbent assay in the serum of 511 individuals from a commercial chicken (Gallus gallus) population. A genome-wide association study using 43,211 single nucleotide polymorphism markers was performed to identify the major loci affecting the immune response against IBV. This study detected 20 significant (P < 1.16 × 10−6) effect single nucleotide polymorphisms for the antibody level against IBV. These single nucleotide polymorphisms were distributed on five chicken chromosomes (GGA), involving GGA1, GGA3, GGA5, GGA8, and GGA9. The genes in the 1-Mb windows surrounding each single nucleotide polymorphism with significant effect for the antibody level against IBV were associated with many biological processes or pathways related to immunity, such as the defense response and mTOR signaling pathway. A genomic region containing a cluster of 13 beta-defensin (GAL1–13) and interleukin-17F genes on GGA3 probably plays an important role in the immune response against IBV. In addition, the major loci significantly associated with the antibody level against IBV on GGA1 and GGA5 could explain about 12% and 13% of the phenotypic variation, respectively. This study suggested that the chicken genome has several important loci affecting the immune response against IBV, and increases our knowledge of how to control outbreaks of infectious bronchitis.
1. Introduction
Since severe acute respiratory syndrome (SARS), caused by a coronavirus (CoV), emerged and caused human mortality in China in 2002, CoVs have received relatively much attention as human pathogens (Saif, 2004). The emergence of SARS-CoV also suggested that the evolution of CoVs has broken through the species barrier (Woo et al., 2006, Dong et al., 2007). Consequently, research has focused on CoV infections in animals, including wild and domestic animals.
Avian infectious bronchitis virus (IBV) belongs to the gamma-CoVs. Infectious bronchitis (IB), which is caused by IBV, was first reported in the United States in the 1930s (Cavanagh, 2007). IB began as a disease of young birds, but as time progressed, the disease spread to older birds worldwide (Sjaak et al., 2011). IBV can cause a severe respiratory viral disease of chickens and shows high mortality. IBV can also replicate in some epithelial cells of the gut, kidney and oviduct, resulting in poor weight gain in broilers, and serious egg yield drop and poor egg quality in layers and breeders (Ignjatovic and Sapats, 2000, Cavanagh, 2001, Cavanagh, 2007). Thus, IB has a significant economic impact on the modern poultry industry. In addition, the CoV causing SARS most likely recombined genomic sequences from mammalian-like and avian-like CoVs (e.g. IBV), according to the molecular genetic data (Stavrinides and Guttman, 2004). The impact of SARS on public health has led to the epidemiology of IB receiving much attention in many countries (Bourogaa et al., 2009, Pohuang et al., 2009, Rimondi et al., 2009, Kulkarni and Resurreccion, 2010, Villarreal et al., 2010, Chacon et al., 2011, Ji et al., 2011, Abdel-Moneim et al., 2012, Acevedo et al., 2012, Ma et al., 2012a, Ma et al., 2012b). Studying IBV alone to develop IB vaccines is insufficient to control IB outbreaks because IBV is highly variable (Sjaak et al., 2011). Viruses and hosts show coevolution, so increasing our knowledge of host responses against IBV is helpful to protect the poultry industry from IB. However, there are few studies of the host responses against IBV, especially their genetic basis.
Fortunately, host immune responses to pathogens, such as Marek’s disease virus and Escherichia coli, can be heritable in chickens (Yonash et al., 1996, Pitcovski et al., 2001, Sarson et al., 2008). Thus, we believed that it would be useful to explore the immune response of chicken to IBV by high-resolution mapping of loci affecting the antibody levels against IBV. Genome-wide association studies (GWASs) have become one of the most commonly used strategies for identifying genes for complex traits in humans, as well as in animals. In chickens, some major loci associated with growth (Gu et al., 2011, Xie et al., 2012), egg production (Liu et al., 2011, Wolc et al., 2012), resistance to Marek’s disease (Li et al., 2012) and immune response to Newcastle disease virus (Luo et al., 2013) were identified by GWASs.
This study aimed to identify major genomic regions associated with the immune response against IBV using a GWAS based on a 60 k single nucleotide polymorphism (SNP) chip (Groenen et al., 2011) in chickens. We hoped to increase our knowledge of methods for controlling outbreaks of IB from a host perspective.
2. Materials and methods
2.1. Animals and phenotypic measurements
The Animal Care Committee of Institute of Animal Science, Guangdong Academy of Agricultural Sciences (Guangzhou, People’s Republic of China) approved the study (Approval No. GAAS-IAS-2009-73).
All experimental birds were from an F2 population, which was built from the full-sib intercross of two divergent lines (23 P and 51 F1). The first line was a fast-growing Chinese yellow broiler, which had undergone more than 10 generations of selection for high growth rate. The second line was the Huiyang bearded chicken, a Chinese local breed with a low growth rate and high meat quality tailored to Chinese tastes. The resistance to IBV of the second line is better than that of the first line (unpublished results). The population included 511 individuals from six hatches. All experimental birds were weighed at 28 and 91 days, and were immunized with a commercial IBV live attenuated vaccine of the H120 strain (Intervet International B.V., Boxmeer, Netherlands), using the standard dose given in the instructions of the vaccine, by eye drop at day 30 and serum was collected at day 91. Venous blood was collected into centrifuge tubes containing anticoagulant and stored in −80 °C for SNP genotyping. The antibody levels against IBV (S/P values) were determined using an indirect enzyme-linked immunosorbent assay (ELISA) test according to the instructions of a commercial ELISA kit (BioChek, Inc., Foster City, CA, USA). The ELISA kit measures IBV-specific immunoglobulin Y, but not immunoglobulin A or immunoglobulin M. All of the experimental birds were positive for the IBV-specific immunoglobulin Y post-immunization.
2.2. SNP genotyping and selection
Genomic DNA of all birds was extracted from venous blood by a phenol–chloroform method. DNA LandMarks Inc. (Quebec, Canada) helped with the genotyping of the chicken 60 k SNP chips from Illumina Inc. (Groenen et al., 2011) using 75 μL of approximately 50 ng/μL genomic DNA. Six of 511 samples were excluded because more than 5% of the SNP genotypes were missing. Of 57,636 SNPs in the 60 k chip, 43,211 SNPs with a minor allele frequency (MAF) of 5% or greater, a call rate of 95% or greater, and having exact chromosome position in the 505 samples were selected for use in the current study. The mean distance between adjacent SNPs was 23.39 kb.
2.3. Statistical analysis
Population stratification can influence accuracy of a GWAS; therefore, we conducted a principal component analysis (PCA), which approximately explains population structure, in the experimental population using the validated information SNPs by GAPIT (Lipka et al., 2012). Simultaneously, the kinship for the 505 birds was estimated by program TASSEL version 3.0 (Bradbury et al., 2007). The phenotypic data for the antibody levels against IBV were initially adjusted for sex and hatch effects by the general linear model Yij = μ + SEXi + Hj + eij, where Yij is the phenotypic value for the antibody response against IBV, μ is the overall mean, SEXi is the effect of the ith sex, Hj is the effect of the jth hatch, and eij is the residual effect. The adjusted phenotypic value was calculated as μ + eij. The adjusted phenotypic values, including the PCA and the kinship information were then used to perform the GWAS, as well as genetic parameters estimation for the antibody response against IBV, with a mixed linear model implemented in TASSEL version 3.0 (Bradbury et al., 2007). The model was described as follows:
where y is the vector of the adjusted phenotypic values; b is the vector of fixed effects including the SNP and population structure (PCA); a is the vector of random additive genetic effects from multiple background quantitative trait loci (QTLs) for individuals; e is the vector of random residuals; and X and Z are the corresponding incidence matrices. The variance of random effects were and , and the distributions for the random effects were assumed as: and , where K is the kinship matrix; is the animal additive genetic variance; I is an identity matrix; and is the residual variance.
The threshold P-value for declaring genome-wide significance was 0.05/43,211 = 1.16 × 10−6 (−log10(P) = 5.94), according to Bonferroni correction. Haploview (Barrett et al., 2005) was used to calculate the linkage disequilibrium (LD), whose parameter was r 2 in this study, for SNPs on each chicken (Gallus gallus) chromosome (GGA). The extent of LD was calculated under the standard with r 2 = 0.1. Each candidate genomic region related to the antibody response against IBV was predicted to cover a length of the genome-wide significant SNP position ± the extent of LD in the corresponding GGA.
2.4. Bioinformatics analysis
Information on the genes in the candidate genomic regions was obtained from Ensembl (Ensembl Genome Browser, 2011) and NCBI (National Center for Biotechnology Information, 2011). The Database for Annotation, Visualization and Integrated Discovery (DAVID) (Dennis et al., 2003, Huang et al., 2009) was used to conduct pathway analysis for the candidate genes. Further global analysis for the identified pathways was performed through the KEGG pathway database and the KEGG Atlas (Kanehisa and Goto, 2000, Okuda et al., 2008, Kanehisa et al., 2012, Kotera et al., 2012), including comparisons with other species.
3. Results and discussion
3.1. Population structure
As shown in Fig. 1 , PCA identified the population stratification in this experimental population. The population could be divided into at least six subpopulations, although the differences were not very large among those subpopulations. Population stratification can interfere with reliability of GWAS (Cardon and Palmer, 2003, Tian et al., 2008, Epstein et al., 2012, Ma et al., 2012a, Ma et al., 2012b); therefore, the statistical model of the GWAS needed to include population stratification information. The first three principal components, as three covariates of the mixed model, were used to correct for stratification in the GWAS, according to Price et al. (2006). The ratios of variance of the first three principal components relative to the total variance in the antibody levels against IBV, the body weight at 28 days and the body weight at 91 days were 0.65%, 2.60% and 2.67%, respectively. The effect size of the principal components for the antibody levels against IBV was smaller than that for the body weights; however, the effect of the principal components could not be neglected for the GWAS of the antibody levels against IBV.
Fig. 1.
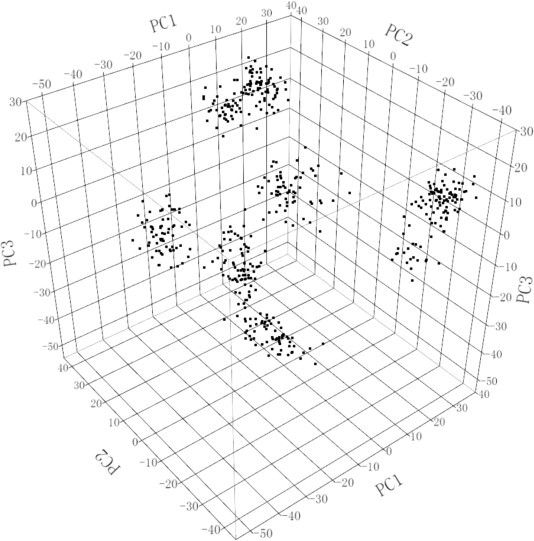
A three-dimensional plot of principal component analysis (PCA) of the experimental population. PC1, PC2 and PC3 values are, respectively, from the first, second and third principal component for each individual chicken. The PCA is based on previously validated SNP information.
Besides population stratification, the cryptic relationships between individuals in the sample can increase the false-positive rate in a GWAS. To reduce the effect of cryptic relationships, the kinships between birds in this study were calculated using 43,211 SNPs (Supplementary Table 1). The average kinship was 0.08, but the maximum kinship reached 0.63, and the estimated effect of the kinship could reach 8.79%, suggesting that it was necessary to perform the GWAS with the kinship information in this study. In addition, QTL mapping cannot be achieved by GWAS if miss heritability occurs for the target trait in the experimental population (Maher, 2008, Slatkin, 2009). Fortunately, the estimated heritability of the antibody levels against IBV was approximately 0.22 in the current experimental population. In general, the power of the design population is higher than that of a random population for QTL mapping. For an F2 population, the power of detecting QTLs could reach 0.90 when the sample size is 500 and the heritability exceeds 0.04 (Satagopan et al., 2007). If the heritability reaches 0.18, the power of detecting QTLs would be close to one in an F2 population including 200 individuals from two outbred lines crossing by 50 k SNP panels (Ledur et al., 2010). In fact, the heritability of body weight is generally no more than 0.25 in chickens (Wolc et al., 2009, Chen et al., 2011); however, many studies have successfully detected QTLs for body weights by GWAS (Gu et al., 2011, Xie et al., 2012). Thus, it was feasible to map QTLs for the antibody levels against IBV using the F2 experimental population by GWAS with 43,211 SNPs.
The extent of LD for the whole genome was considered as 1 Mb because the LD declined to r 2 = 0.1 at a distance of about 1 Mb in the experimental population. Different chromosomes had different extents of LD, and the largest extent of LD, almost 2 Mb, appeared on GGA1 (Supplementary Fig. 1). Traditional QTL mapping can identify a confidence interval for the corresponding QTL locations using genetic distances by bootstrap or other statistical methods (Jongjoo et al., 2002, Manichaikul et al., 2006), whereas confidence intervals cannot be calculated in GWAS at present. We tried to predict candidate genomic regions containing the significantly associated SNPs and the extent of LD in the corresponding genomic regions to mine candidate genes affecting the antibody level against IBV.
3.2. Significant SNP effects related to the immunity response against IBV
Twenty SNP effects were detected with genome-wide significance for the antibody level against IBV (P < 1.16 × 10−6, Table 1 and Supplementary Fig. 2). These significantly associated SNPs were distributed in five chicken chromosomes, including GGA1 (n = 8), GGA3 (n = 4), GGA5 (n = 5), GGA8 (n = 1) and GGA9 (n = 2). Notably, no significantly associated SNP was detected on GGA16, which harbors the chicken major histocompatibility complex (cluster of immune function genes). The most likely reason was that very few SNP markers (n = 14) on GGA16 could be used for the GWAS of the antibody level against IBV in this study, and GGA16 has very high recombination rates. Alternatively, the specific nature of the current experimental population might be an important reason. Further studies are necessary to detect QTLs for the antibody level against IBV on GGA16.
Table 1.
Twenty SNP markers with genome-wide significant effects for the antibody level against infectious bronchitis virus (P < 1.16 × 10−6).
| SNP | Allele | Chromosomea | Physical position (bp) | Nearest gene | P-value | R2 |
|---|---|---|---|---|---|---|
| GGaluGA287132 | T/C | 5 | 49,506,678 | 386.0 Kb D VRK1 | 2.66 × 10−14 | 0.132 |
| rs16505398 | C/T | 5 | 49,565,972 | 445.3 Kb D VRK1 | 2.69 × 10−14 | 0.132 |
| GGaluGA287070 | G/A | 5 | 49,185,847 | 65.2 Kb D VRK1 | 3.11 × 10−14 | 0.131 |
| rs13623466 | C/T | 1 | 132,771,759 | 7.7 Kb U DHRSX | 9.98 × 10−14 | 0.126 |
| GGaluGA325321 | T/C | 8 | 7,838,819 | LAMC1 | 1.8 × 10−13 | 0.124 |
| rs13939926 | A/G | 1 | 135,455,921 | 9.6 Kb U GABRB3 | 1.98 × 10−13 | 0.123 |
| GGaluGA043691 | T/C | 1 | 133,232,594 | ASMTL | 2.19 × 10−13 | 0.123 |
| GGaluGA058751 | G/A | 1 | 183,219,141 | 7.9 Kb D CRYL1 | 2.22 × 10−13 | 0.123 |
| GGaluGA042002 | G/A | 1 | 126,146,475 | 16.4 Kb U ENSGALG00000012431 | 2.22 × 10−13 | 0.123 |
| GGaluGA239405 | G/A | 3 | 110,204,493 | GAL12 | 3.12 × 10−7 | 0.061 |
| GGaluGA288788 | G/A | 5 | 54,119,882 | 2.8 Kb D AKT1 | 4.67 × 10−7 | 0.060 |
| GGaluGA336811 | G/A | 9 | 7,003,679 | NMNAT3 | 5.27 × 10−7 | 0.059 |
| rs15227869 | T/C | 1 | 34,551,034 | 10.7 Kb D FAM19A2 | 5.3 × 10−7 | 0.059 |
| rs13985175 | T/G | 1 | 185,026,688 | 13.8 Kb D CWF19L2 | 5.85 × 10−7 | 0.059 |
| rs16340667 | A/G | 3 | 109,648,277 | PINX1 | 6.52 × 10−7 | 0.058 |
| rs14549067 | T/C | 5 | 54,301,518 | KIAA0284 | 8.16 × 10−7 | 0.057 |
| GGaluGA334666 | G/A | 9 | 3,567,680 | YEATS2 | 8.30 × 10−7 | 0.057 |
| rs16340706 | T/C | 3 | 109,693,525 | 13.2 Kb U PINX1 | 8.34 × 10−7 | 0.057 |
| rs14409849 | C/T | 3 | 108,819,850 | INTS9 | 8.51 × 10−7 | 0.057 |
| GGaluGA059344 | T/C | 1 | 184,710,676 | 5.8 Kb D RAB39A | 8.55 × 10−7 | 0.057 |
Chromosome and physical position refer to galGal3.
Eight significantly associated SNPs were in functional genes, and the other 12 SNPs were in intergenic regions (Table 1). GGA1 had the most significantly associated SNPs, while the SNP with the greatest effect on the antibody level against IBV was at position 49,506,678 bp on GGA5. This SNP could explain 13.2% of phenotypic variation for the antibody level against IBV, but was positioned in a gene desert. The gene desert also included the other two SNPs with the top significant effect on the antibody level against IBV. The Vaccinia related kinase 1 (VRK1) gene was the nearest functional gene to the gene desert (Table 1 and Fig. 2 ). VRK1 is ubiquitously expressed in T cells and the genomic region covering VRK1 and the gene desert was observed to play an important role in T-cell acute lymphoblastic leukemia in humans (Roh et al., 2005, Nagel et al., 2007). Single-cell transcriptomics also revealed VRK1 expression changed in mouse bone-marrow-derived dendritic cells after immunizing with lipopolysaccharide (Shalek et al., 2013). Given the genomic synteny among chickens, humans and mice, the nature of the genomic region covering the three SNPs with the top significant effect on the antibody level against IBV might be very important for adaptive immunity in chickens.
Fig. 2.
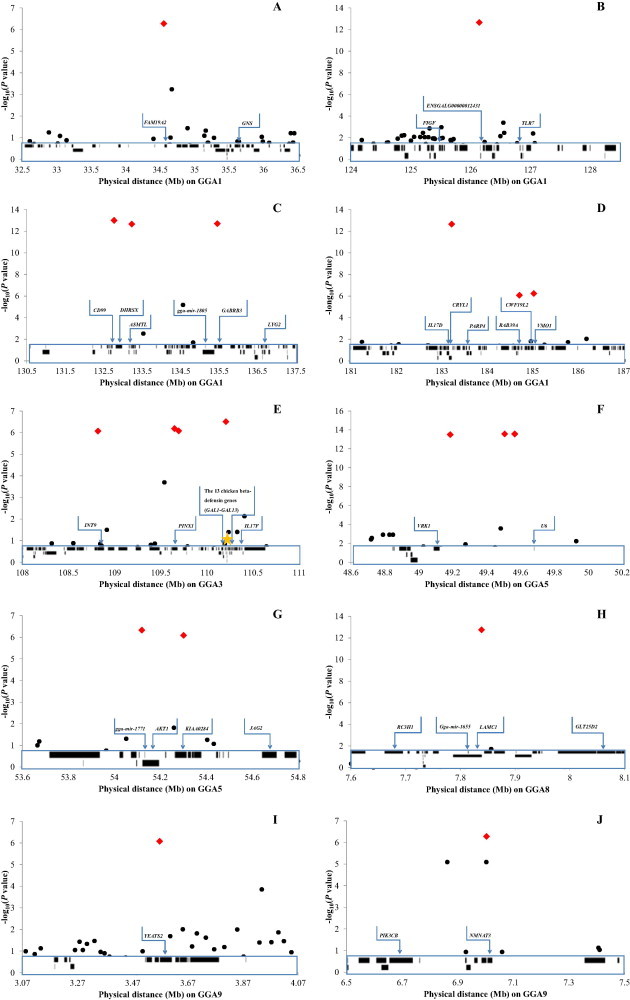
Candidate regions for the antibody level against infectious bronchitis virus (IBV). Rhombuses and points indicate significant and the other SNP effects for the antibody level against IBV, respectively, at the 5% Bonferroni genome-wide significance threshold (P = 1.16 × 10−6). Blocks indicate the location of genes. Arrows highlight genes, and a pentagram represents the cluster of 13 chicken beta-defensin genes (GAL1–GAL13).
Each of the other significantly associated SNPs explained over 5% of the phenotypic variation. The sum of all the significant SNP effects relative to the phenotypic variation was more than one (Table 1), which indicated that several significant SNPs must represent the same QTLs because of the strong LD between SNPs. Given the extent of LD in the experimental population (Supplementary Fig. 1), we identified 10 genomic regions that could be major QTLs related to the antibody level against IBV. As shown in Supplementary Table 2, the major QTLs for the antibody level against IBV covered 453 genes or non-coding RNAs according to the Ensembl database (Ensembl Genome Browser, 2011). Gene ontology was used to evaluate the information on the corresponding genes. As shown in Table 2 , given a significance threshold of P < 0.05, these genes were enriched in one molecular function, one cellular component and four biological processes. Interestingly, most of the statistically significant biological processes were related to the immune response, such as defense response and polysaccharide metabolic process, which corresponded to their association with the antibody level against IBV.
Table 2.
Gene ontology (GO) analysis of genes in the candidate regions related to the antibody level against infectious bronchitis virus (P < 1.16 × 10−6).
| Category | GO term accession | GO term description | Number of genes | Percent of all genes | Involved genes | DAVID P-value |
|---|---|---|---|---|---|---|
| Biological process | GO:0042742 | Defense response to bacterium | 13 | 2.9 | LYG2, GAL1, GAL2, GAL3, GAL4, GAL5, GAL6, GAL7, GAL8, GAL9, GAL10, GAL12, GAL13 | 5.6 × 10−13 |
| Biological process | GO:0009617 | Response to bacterium | 14 | 3.1 | BDKRB1, LYG2, GAL1, GAL2, GAL3, GAL4, GAL5, GAL6, GAL7, GAL8, GAL9, GAL10, GAL12, GAL13 | 5.0 × 10−11 |
| Biological process | GO:0009617 | Defense response | 18 | 4 | AKT1, IL17D, ENSGALG00000016677, IL17F, LYG2, GAL1, GAL2, GAL3, GAL4, GAL5, GAL6, GAL7, GAL8, GAL9, GAL10, GAL12, GAL13, TLR7 | 1.2 × 10−10 |
| Biological process | GO:0005976 | Polysaccharide metabolic process | 4 | 0.9 | AKT1, GNS, GLT25D2, LYG2 | 4.4 × 10−2 |
| Cellular component | GO:0005576 | Extracellular region | 31 | 6.9 | T6GAL1, ADIPOQ, ANGPTL7, FIGF, CLU, GPC1, LAMC1, LAMC2, MMP27, MMP7, MMP13, POMC, TNN, TNR, VMO1, LYG2, GAL1, GAL2, GAL3, GAL4, GAL5, GAL6, GAL7, GAL8, GAL9, GAL10, GAL12, GAL13 | 2.30 × 10−5 |
| Molecular function | GO:0004947 | Bradykinin receptor activity | 2 | 0.4 | BDKRB1, BDKRB2 | 4.80 × 10−2 |
A cluster of 13 beta-defensin genes (GAL1–13) contributing to the defense response was precisely located in a QTL related to the antibody level against IBV on GGA3. Previous studies revealed the GAL1–13 genes could regulate the humoral immune response (Breuilh et al., 2007, Anginot et al., 2013). Surprisingly, previous reports stated that the positional candidate gene cluster mainly regulated the response to bacterium, not viruses. Hasenstein and Lamont (2007) reported that the SNPs in the cluster of GAL11–13 genes were associated with Salmonella response in an advanced intercross line. Nevertheless, earlier evidence proved that infection with IBV was close to bacterium (e.g. E. coli) invasion in chicken (Springer et al., 1974, Nakamura et al., 1992, Yunis et al., 2002a, Yunis et al., 2002b). This reinforced the importance of the cluster of GAL11–13 genes for regulating the antibody level against IBV.
In addition, lysozyme G like protein 2 (LYG2), interleukin-17D (IL17D) and toll-like receptor 7 (TLR7) genes, like GAL1–13 genes, involve defense response and should be key genes associated with the antibody level against IBV (Table 2). These three functional genes were in the strongly significant (P < 2.5 × 10−13) QTLs on GGA1 (Fig. 2B–D). Expression of the LYG2 gene was upregulated significantly after pathogen challenge (Matulova et al., 2013). The IL17D gene was strongly associated with immune response to a polysaccharide vaccine for typhoid (Majumder et al., 2009). Interleukin-17F (IL17F), in the same gene family as IL17D, was also on one of the QTLs for the antibody level against IBV (Fig. 2E). The IL17 family plays a key role in the induction and stimulation of chemotaxis and the function of T helper 17 lymphocytes (Iwakura et al., 2008, Mabuchi et al., 2012). More interestingly, the interleukin 17 pathway was activated in cells infected with SARS-CoV or the other new human coronavirus (Josset et al., 2013). This suggests that the interleukin 17 pathway plays an important role in host responses to rapidly evolving CoVs. In addition to the interleukin 17 pathway, we found that the mTOR signaling pathway involving TLR7 was likely to be important for host responses to CoVs. TLR7 is a key factor for optimal B cell responses during chronic viral infection of mice (Clingan and Matloubian, 2013). The Toll-like receptor family has been linked to the pathogen recognition of immunity (Kagan and Iwasaki, 2012). Therefore, these genes were all closely linked to immune responses and important candidate genes for the antibody level against IBV. However, obtaining more evidence of candidate genes related to the antibody response against IBV requires further study. This might include another association study based on a random population or a large population of advanced intercross lines with large size, and exploring the relationships between the expressions of the predicted candidate genes and the antibody response against IBV in intro and in vivo.
3.3. Comparison with other association studies for immune responses in chickens
The current study focused on QTL mapping for the immune response to IBV in chickens and detected 10 QTLs (Fig. 2). IB and Newcastle disease both belong to respiratory infectious diseases; therefore, the host immune responses against IBV and Newcastle disease virus might involve similar molecular networks. Indeed, two out of the 10 QTLs for the immune response against IBV overlapped with QTLs for the immune responses against lipopolysaccharides and Newcastle disease virus from the experiment with 1022 SNPs and 583 hens from nine different lines, at the threshold of P < 0.05 (Biscarini et al., 2010). One region was at the end of the short arm of GGA3, and Biscarini et al. (2010) thought that IL17F in this region was one of the most important candidate genes for adaptive immune responses to lipopolysaccharides and Newcastle disease virus. In addition, a recent study found that the GAL1–13 genes in the QTL were also candidate genes for immune responses against IBV (Fig. 2E). This suggested that this region has an extensive impact on the immune responses to pathogens, including bacteria and virus. The other overlapping QTL region was at ∼3.6 Mb (galGal3) from the proximal end of GGA9. The region was significantly associated with the antibody response to Newcastle disease virus (Biscarini et al., 2010). In addition, a suggestive QTL (P < 2.31 × 10−5, −log10(P) = 4.64) for the antibody response against IBV was at ∼100 Mb (galGal3) from the proximal end of GGA1 (Supplementary Fig. 2), which corresponded to a QTL detected by 60 k SNPs and 511 F2 birds for the antibody response against Newcastle disease virus (Luo et al., 2013).
The remaining eight QTLs for the antibody response against IBV were novel and specific QTLs in this study. They were not only different from the QTLs for the antibody response against Newcastle disease virus, they also differed from the QTLs for immune responses against other stimuli, such as Brucella abortus (Zhou et al., 2003), E. coli (Yunis et al., 2002a, Yunis et al., 2002b), keyhole lympet hemocyanin (Siwek et al., 2003), sheep red blood cells (Dorshorst et al., 2011), infectious bursal disease virus (Ewald et al., 2007) and lipoteichoic acid (Slawinska et al., 2011). Nevertheless, all the QTLs linked to the antibody response against IBV were not independent of body weights in chickens (Supplementary Table 3), and birds with the favored genotype of the QTLs linked to the antibody response against IBV had significantly lower body weights (unpublished results). This observation requires further study.
4. Conclusions
This study found more than 10 major QTLs related to the antibody response against IBV. The contributions of GGA1, 3, 5, 8 and 9 are the most important for the antibody response against IBV. The findings lay a preliminary foundation for controlling outbreaks of IB, even SARS, from a host perspective.
Competing interests
The authors have no competing interests to declare.
Acknowledgments
The authors would like to thank Dr. Zheya Sheng (College of Biological Science, China Agricultural University, Beijing, China) for her technical assistance in SNP genotyping data handling. This study was supported by the Major State Basic Research Development (973) Program of China (Grant No. 2012CB723701), the Research Project of Core Technologies of New Strategic Industries in Guangdong Province (Grant No. 2012A020800005) and the Earmarked Fund for Modern Agro-industry Technology Research System (Grant No. nycytx-42).
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2013.12.004.
Contributor Information
Chenglong Luo, Email: chenglongluo1981@163.com.
Hao Qu, Email: qhw03@163.com.
Jie Ma, Email: zzitmj@126.com.
Jie Wang, Email: wangjie030@126.com.
Xiaoxiang Hu, Email: huxx@cau.edu.cn.
Ning Li, Email: ninglcau@cau.edu.cn.
Dingming Shu, Email: shudm@263.net.
Appendix A. Supplementary data
Supplementary Fig. 1.
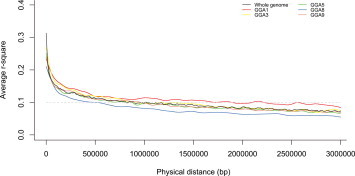
Linkage disequilibrium pattern of the experimental population. The gray dashed line indicates the threshold (r2 = 0.1) for identifying the extent of linkage disequilibrium.
Supplementary Fig. 2.
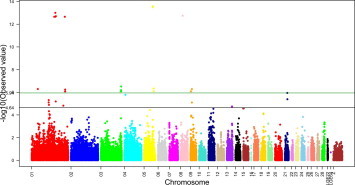
Manhattan plots for SNP effects for the antibody level against infectious bronchitis virus. The green solid line indicates genome-wide significance (P < 1.16 × 10−6) with Bonferroni correction. The black solid line indicates suggestive genome-wide significance (P < 2.31 × 10−5).
Kinships between individuals in the experimental population.
Information on genes in the QTLs for the antibody level against infectious bronchitis virus.
Relationship with growth traits of the most significant SNP in each of 10 QTLs for the antibody level against infectious bronchitis virus.
References
- Abdel-Moneim A.S., Afifi M.A., El-Kady M.F. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch. Virol. 2012;157:2453–2457. doi: 10.1007/s00705-012-1445-1. [DOI] [PubMed] [Google Scholar]
- Acevedo A.M., Diaz D.A.H., Brandao P.E., Colas M., Oliveira S., Perez L.J. First evidence of the emergence of novel putative infectious bronchitis virus genotypes in Cuba. Res. Vet. Sci. 2012;93:1046–1049. doi: 10.1016/j.rvsc.2012.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anginot A., Espeli M., Chasson L., Mancini S.J., Schiff C. Galectin 1 modulates plasma cell homeostasis and regulates the humoral immune response. J. Immunol. 2013;190:5526–5533. doi: 10.4049/jimmunol.1201885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Biscarini F., Bovenhuis H., van Arendonk J.A., Parmentier H.K., Jungerius A.P., van der Poel J.J. Across-line SNP association study of innate and adaptive immune response in laying hens. Anim. Genet. 2010;41:26–38. doi: 10.1111/j.1365-2052.2009.01960.x. [DOI] [PubMed] [Google Scholar]
- Bourogaa H., Miled K., Gribaa L., El B.I., Ghram A. Characterization of new variants of avian infectious bronchitis virus in Tunisia. Avian Dis. 2009;53:426–433. doi: 10.1637/8666-022609-Reg.1. [DOI] [PubMed] [Google Scholar]
- Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Breuilh L., Vanhoutte F., Fontaine J., van Stijn C.M., Tillie-Leblond I., Capron M., Faveeuw C., Jouault T., van Die I., Gosset P., Trottein F. Galectin-3 modulates immune and inflammatory responses during helminthic infection: impact of galectin-3 deficiency on the functions of dendritic cells. Infect. Immun. 2007;75:5148–5157. doi: 10.1128/IAI.02006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon L.R., Palmer L.J. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. A nomenclature for avian coronavirus isolates and the question of species status. Avian Pathol. 2001;30:109–115. doi: 10.1080/03079450120044506. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Chacon J.L., Rodrigues J.N., Assayag J.M., Peloso C., Pedroso A.C., Ferreira A.J. Epidemiological survey and molecular characterization of avian infectious bronchitis virus in Brazil between 2003 and 2009. Avian Pathol. 2011;40:153–162. doi: 10.1080/03079457.2010.544641. [DOI] [PubMed] [Google Scholar]
- Chen C.Y., Misztal I., Aguilar I., Tsuruta S., Meuwissen T.H., Aggrey S.E., Wing T., Muir W.M. Genome-wide marker-assisted selection combining all pedigree phenotypic information with genotypic data in one step: an example using broiler chickens. J. Anim. Sci. 2011;89:23–28. doi: 10.2527/jas.2010-3071. [DOI] [PubMed] [Google Scholar]
- Clingan J.M., Matloubian M. B cell-intrinsic TLR7 signaling is required for optimal B cell responses during chronic viral infection. J. Immunol. 2013;191:810–818. doi: 10.4049/jimmunol.1300244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G.J., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dong B.Q., Liu W., Fan X.H., Vijaykrishna D., Tang X.C., Gao F., Li L.F., Li G.J., Zhang J.X., Yang L.Q., Poon L.L., Zhang S.Y., Peiris J.S., Smith G.J., Chen H., Guan Y. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J. Virol. 2007;81:6920–6926. doi: 10.1128/JVI.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorshorst B.J., Siegel P.B., Ashwell C.M. Genomic regions associated with antibody response to sheep red blood cells in the chicken. Anim. Genet. 2011;42:300–308. doi: 10.1111/j.1365-2052.2010.02146.x. [DOI] [PubMed] [Google Scholar]
- Ensembl Genome Browser, 2011. Ensembl release 63. Available from: <http://useast.ensembl.org/index.html> (accessed 30.06.11).
- Epstein M.P., Duncan R., Broadaway K.A., He M., Allen A.S., Satten G.A. Stratification-score matching improves correction for confounding by population stratification in case–control association studies. Genet. Epidemiol. 2012;36:195–205. doi: 10.1002/gepi.21611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S.J., Ye X., Avendano S., McLeod S., Lamont S.J., Dekkers J.C. Associations of BF2 alleles with antibody titres and production traits in commercial pure line broiler chickens. Anim. Genet. 2007;38:174–176. doi: 10.1111/j.1365-2052.2007.01574.x. [DOI] [PubMed] [Google Scholar]
- Groenen M.A., Megens H.J., Zare Y., Warren W.C., Hillier L.W., Crooijmans R.P., Vereijken A., Okimoto R., Muir W., Cheng H.H. The development and characterization of a 60 K SNP chip for chicken. BMC Genomics. 2011;12:274. doi: 10.1186/1471-2164-12-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Feng C., Ma L., Song C., Wang Y., Da Y., Li H., Chen K., Ye S., Ge C., Hu X., Li N. Genome-wide association study of body weight in chicken F2 resource population. PLoS One. 2011;6:e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenstein J.R., Lamont S.J. Chicken gallinacin gene cluster associated with Salmonella response in advanced intercross line. Avian Dis. 2007;51:561–567. doi: 10.1637/0005-2086(2007)51[561:CGGCAW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ignjatovic J., Sapats S. Avian infectious bronchitis virus. Rev. Sci. Technol. 2000;19:493–508. doi: 10.20506/rst.19.2.1228. [DOI] [PubMed] [Google Scholar]
- Iwakura Y., Nakae S., Saijo S., Ishigame H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol. Rev. 2008;226:57–79. doi: 10.1111/j.1600-065X.2008.00699.x. [DOI] [PubMed] [Google Scholar]
- Ji J., Xie J., Chen F., Shu D., Zuo K., Xue C., Qin J., Li H., Bi Y., Ma J., Xie Q. Phylogenetic distribution and predominant genotype of the avian infectious bronchitis virus in China during 2008–2009. Virol. J. 2011;8:184. doi: 10.1186/1743-422X-8-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongjoo K., Davis S.K., Taylor J.F. Application of non-parametric bootstrap methods to estimate confidence intervals for QTL location in a beef cattle QTL experimental population. Genet. Res. 2002;79:259–263. doi: 10.1017/s001667230200561x. [DOI] [PubMed] [Google Scholar]
- Josset L., Menachery V.D., Gralinski L.E., Agnihothram S., Sova P., Carter V.S., Yount B.L., Graham R.L., Baric R.S., Katze M.G. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. MBio. 2013;4:e113–e165. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J.C., Iwasaki A. Phagosome as the organelle linking innate and adaptive immunity. Traffic. 2012;13:1053–1061. doi: 10.1111/j.1600-0854.2012.01377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera M., Hirakawa M., Tokimatsu T., Goto S., Kanehisa M. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol. Biol. 2012;802:19–39. doi: 10.1007/978-1-61779-400-1_2. [DOI] [PubMed] [Google Scholar]
- Kulkarni A.B., Resurreccion R.S. Genotyping of newly isolated infectious bronchitis virus isolates from northeastern Georgia. Avian Dis. 2010;54:1144–1151. doi: 10.1637/9358-040510-Reg.1. [DOI] [PubMed] [Google Scholar]
- Ledur M.C., Navarro N., Perez-Enciso M. Large-scale SNP genotyping in crosses between outbred lines: how useful is it? Heredity (Edinb.) 2010;105:173–182. doi: 10.1038/hdy.2009.149. [DOI] [PubMed] [Google Scholar]
- Li D.F., Lian L., Qu L.J., Chen Y.M., Liu W.B., Chen S.R., Zheng J.X., Xu G.Y., Yang N. A genome-wide SNP scan reveals two loci associated with the chicken resistance to Marek’s disease. Anim. Genet. 2012;44:217–222. doi: 10.1111/j.1365-2052.2012.02395.x. [DOI] [PubMed] [Google Scholar]
- Lipka A.E., Tian F., Wang Q., Peiffer J., Li M., Bradbury P.J., Gore M.A., Buckler E.S., Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- Liu W., Li D., Liu J., Chen S., Qu L., Zheng J., Xu G., Yang N. A genome-wide SNP scan reveals novel loci for egg production and quality traits in white leghorn and brown-egg dwarf layers. PLoS One. 2011;6:e28600. doi: 10.1371/journal.pone.0028600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Qu H., Ma J., Wang J., Li C., Yang C., Hu X., Li N., Shu D. Genome-wide association study of antibody response to Newcastle disease virus in chicken. BMC Genet. 2013;14:42. doi: 10.1186/1471-2156-14-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Shao Y., Sun C., Han Z., Liu X., Guo H., Liu X., Kong X., Liu S. Genetic diversity of avian infectious bronchitis coronavirus in recent years in China. Avian Dis. 2012;56:15–28. doi: 10.1637/9804-052011-Reg.1. [DOI] [PubMed] [Google Scholar]
- Ma L., Wiggans G.R., Wang S., Sonstegard T.S., Yang J., Crooker B.A., Cole J.B., Van Tassell C.P., Lawlor T.J., Da Y. Effect of sample stratification on dairy GWAS results. BMC Genomics. 2012;13:536. doi: 10.1186/1471-2164-13-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabuchi T., Chang T.W., Quinter S., Hwang S.T. Chemokine receptors in the pathogenesis and therapy of psoriasis. J. Dermatol. Sci. 2012;65:4–11. doi: 10.1016/j.jdermsci.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- Majumder P.P., Staats H.F., Sarkar-Roy N., Varma B., Ghosh T., Maiti S., Narayanasamy K., Whisnant C.C., Stephenson J.L., Wagener D.K. Genetic determinants of immune-response to a polysaccharide vaccine for typhoid. Hugo J. 2009;3:17–30. doi: 10.1007/s11568-010-9134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A., Dupuis J., Sen S., Broman K.W. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics. 2006;174:481–489. doi: 10.1534/genetics.106.061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matulova M., Varmuzova K., Sisak F., Havlickova H., Babak V., Stejskal K., Zdrahal Z., Rychlik I. Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Vet. Res. 2013;44:37. doi: 10.1186/1297-9716-44-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel S., Scherr M., Kel A., Hornischer K., Crawford G.E., Kaufmann M., Meyer C., Drexler H.G., MacLeod R.A. Activation of TLX3 and NKX2-5 in t(5; 14)(q35; q32) T-cell acute lymphoblastic leukemia by remote 3′-BCL11B enhancers and coregulation by PU.1 and HMGA1. Cancer Res. 2007;67:1461–1471. doi: 10.1158/0008-5472.CAN-06-2615. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Cook J.K., Frazier J.A., Narita M. Escherichia coli multiplication and lesions in the respiratory tract of chickens inoculated with infectious bronchitis virus and/or E. coli. Avian Dis. 1992;36:881–890. [PubMed] [Google Scholar]
- National Center for Biotechnology Information, 2011. Entrez gene. Available from: <http://www.ncbi.nlm.nih.gov> (accessed 06.07.11).
- Okuda S., Yamada T., Hamajima M., Itoh M., Katayama T., Bork P., Goto S., Kanehisa M. KEGG atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcovski J., Cahaner A., Heller E.D., Zouri T., Gutter B., Gotfried Y., Leitner G. Immune response and resistance to infectious bursal disease virus of chicken lines selected for high or low antibody response to Escherichia coli. Poult. Sci. 2001;80:879–884. doi: 10.1093/ps/80.7.879. [DOI] [PubMed] [Google Scholar]
- Pohuang T., Chansiripornchai N., Tawatsin A., Sasipreeyajan J. Detection and molecular characterization of infectious bronchitis virus isolated from recent outbreaks in broiler flocks in Thailand. J. Vet. Sci. 2009;10:219–223. doi: 10.4142/jvs.2009.10.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rimondi A., Craig M.I., Vagnozzi A., Konig G., Delamer M., Pereda A. Molecular characterization of avian infectious bronchitis virus strains from outbreaks in Argentina (2001–2008) Avian Pathol. 2009;38:149–153. doi: 10.1080/03079450902737821. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif L.J. Animal coronaviruses: what can they teach us about the severe acute respiratory syndrome? Rev. Sci. Tech. 2004;23:643–660. doi: 10.20506/rst.23.2.1513. [DOI] [PubMed] [Google Scholar]
- Sarson A.J., Parvizi P., Lepp D., Quinton M., Sharif S. Transcriptional analysis of host responses to Marek’s disease virus infection in genetically resistant and susceptible chickens. Anim. Genet. 2008;39:232–240. doi: 10.1111/j.1365-2052.2008.01710.x. [DOI] [PubMed] [Google Scholar]
- Satagopan J.M., Sen S., Churchill G.A. Sequential quantitative trait locus mapping in experimental crosses. Stat. Appl. Genet. Mol. Biol. 2007;6:e12. doi: 10.2202/1544-6115.1264. [DOI] [PubMed] [Google Scholar]
- Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D., Trombetta J.J., Gennert D., Gnirke A., Goren A., Hacohen N., Levin J.Z., Park H., Regev A. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwek M., Buitenhuis A.J., Cornelissen S.J., Nieuwland M.G., Bovenhuis H., Crooijmans R.P., Groenen M.A., de Vries-Reilingh G., Parmentier H.K., van der Poel J.J. Detection of different quantitative trait loci for antibody responses to keyhole lympet hemocyanin and Mycobacterium butyricum in two unrelated populations of laying hens. Poult. Sci. 2003;82:1845–1852. doi: 10.1093/ps/82.12.1845. [DOI] [PubMed] [Google Scholar]
- Sjaak D.W.J., Cook J.K., van der Heijden H.M. Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol. 2011;40:223–235. doi: 10.1080/03079457.2011.566260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. Epigenetic inheritance and the missing heritability problem. Genetics. 2009;182:845–850. doi: 10.1534/genetics.109.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawinska A., Witkowski A., Nieuwland M., Minozzi G., Bednarczyk M., Siwek M. Quantitative trait loci associated with the humoral innate immune response in chickens were confirmed in a cross between Green-Legged Partridgelike and White Leghorn. Poult. Sci. 2011;90:1909–1915. doi: 10.3382/ps.2011-01465. [DOI] [PubMed] [Google Scholar]
- Springer W.T., Luskus C., Pourciau S.S. Infectious bronchitis and mixed infections of Mycoplasma synoviae and Escherichia coli in gnotobiotic chickens. I. Synergistic role in the airsacculitis syndrome. Infect. Immun. 1974;10:578–589. doi: 10.1128/iai.10.3.578-589.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides J., Guttman D.S. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:76–82. doi: 10.1128/JVI.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Gregersen P.K., Seldin M.F. Accounting for ancestry: population substructure and genome-wide association studies. Hum. Mol. Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarreal L.Y., Sandri T.L., Souza S.P., Richtzenhain L.J., de Wit J.J., Brandao P.E. Molecular epidemiology of avian infectious bronchitis in Brazil from 2007 to 2008 in breeders, broilers, and layers. Avian Dis. 2010;54:894–898. doi: 10.1637/9218-121709-Reg.1. [DOI] [PubMed] [Google Scholar]
- Wolc A., White I.M., Avendano S., Hill W.G. Genetic variability in residual variation of body weight and conformation scores in broiler chickens. Poult. Sci. 2009;88:1156–1161. doi: 10.3382/ps.2008-00547. [DOI] [PubMed] [Google Scholar]
- Wolc A., Arango J., Settar P., Fulton J.E., O’Sullivan N.P., Preisinger R., Habier D., Fernando R., Garrick D.J., Hill W.G., Dekkers J.C. Genome-wide association analysis and genetic architecture of egg weight and egg uniformity in layer chickens. Anim. Genet. 2012;43(Suppl. 1):87–96. doi: 10.1111/j.1365-2052.2012.02381.x. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Yip C.C., Huang Y., Tsoi H.W., Chan K.H., Yuen K.Y. Comparative analysis of 22 coronavirus HKU1 genomes reveals a novel genotype and evidence of natural recombination in coronavirus HKU1. J. Virol. 2006;80:7136–7145. doi: 10.1128/JVI.00509-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L., Luo C., Zhang C., Zhang R., Tang J., Nie Q., Ma L., Hu X., Li N., Da Y., Zhang X. Genome-wide association study identified a narrow chromosome 1 region associated with chicken growth traits. PLoS One. 2012;7:e30910. doi: 10.1371/journal.pone.0030910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonash N., Leitner G., Waiman R., Heller E.D., Cahaner A. Genetic differences and heritability of antibody response to Escherichia coli vaccination in young broiler chicks. Poult. Sci. 1996;75:683–690. doi: 10.3382/ps.0750683. [DOI] [PubMed] [Google Scholar]
- Yunis R., Ben-David A., Heller E.D., Cahaner A. Antibody responses and morbidity following infection with infectious bronchitis virus and challenge with Escherichia coli, in lines divergently selected on antibody response. Poult. Sci. 2002;81:149–159. doi: 10.1093/ps/81.2.149. [DOI] [PubMed] [Google Scholar]
- Yunis R., Heller E.D., Hillel J., Cahaner A. Microsatellite markers associated with quantitative trait loci controlling antibody response to Escherichia coli and Salmonella enteritidis in young broilers. Anim. Genet. 2002;33:407–414. doi: 10.1046/j.1365-2052.2002.00890.x. [DOI] [PubMed] [Google Scholar]
- Zhou H., Li H., Lamont S.J. Genetic markers associated with antibody response kinetics in adult chickens. Poult. Sci. 2003;82:699–708. doi: 10.1093/ps/82.5.699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kinships between individuals in the experimental population.
Information on genes in the QTLs for the antibody level against infectious bronchitis virus.
Relationship with growth traits of the most significant SNP in each of 10 QTLs for the antibody level against infectious bronchitis virus.


