Abstract
Detection of infectious viral agents has been on the increase globally with the advent and usage of more sensitive and selective novel molecular techniques in the epidemiological study of viral diseases of economic importance to the swine industry. The observation is not different for the pig-infecting member of the subfamily Parvovirinae in the family Parvoviridae as the application of novel molecular methods like metagenomics has brought about the detection of many other novel members of the group. Surprisingly, the list keeps increasing day by day with some of them possessing zoonotic potentials. In the last one decade, not less than ten novel swine-infecting viruses have been added to the subfamily, and ceaseless efforts have been in top gear to determine the occurrence and prevalence of the old and new swine parvoviruses in herds of pig-producing countries worldwide. The story, however, is on the contrary on the African continent as there is presently a dearth of information on surveillance initiatives of the viruses among swine herds of pig-producing countries in the region. Timely detection and characterization of the viral pathogens is highly imperative for the implementation of effective control and prevention of its spread. This review therefore presents a concise overview on the epidemiology of novel porcine parvoviruses globally and also provides up-to-date highlights on the reported cases of the viral agents in the African sub-region.
Keywords: Porcine parvoviruses, Diversity, Epidemiology, Africa, Pigs
1. Introduction
1.1. Diversity and classifications of parvoviruses
The prefix “parvo” in parvoviruses emanated from the Latin word “parvum” and means “small”. Parvoviruses are therefore, a group of relatively small viruses with linear, single stranded DNA (ssDNA) genomes ranging from 4 to 6.3 kilobases (kb) in size, packaged in a non-enveloped, icosahedral capsid (Tijssen et al., 2011). They are ubiquitous in nature and common infectious agents of numerous hosts ranging from non-vertebrate arthropods to vertebrates and higher mammals including human beings. Their ancestors, according to Kailasan et al. (2015), seem to have emerged several millions of years ago and have been widely distributed ever since. Although parvoviruses appear to have evolved from the same ancestor and have similar genomic features, they usually exhibit a very low relatedness at the nucleotide or protein level thereby depicting their highly extensive diversity. Their diversity seems to also influence the clinical effects exerted on their hosts which could range from non-pathogenic infections to severely lethal diseases' manifestations (Kailasan et al., 2015).
Parvoviruses belong to the family Parvoviridae which comprises two subfamilies namely: Densovirinae and Parvovirinae. Members of the family Parvoviridae were assigned into the two subfamilies based on the kind of hosts they infect. The group of parvoviruses that infect invertebrate hosts (arthropods and crustaceans) belong to the subfamily Densovirinae, whereas those that infect vertebrate hosts belong to the subfamily Parvovirinae. In the ninth edition of the taxonomical grouping that was made by the International Committee on Taxonomy of Viruses (ICTV), the subfamily Densovirinae consists of four genera namely: Densovirus, Brevidensovirus, Iteravirus and Pefudensovirus; whereas the subfamily Parvovirinae comprises five genera: Parvovirus, Erythrovirus, Dependovirus, Amdovirus and Bocavirus (Tijssen et al., 2011). However, in the latest ICTV report, series of systematic changes were made using a modified definition for classification that requires complete or nearly complete genome of the viruses to arrive at an improved taxonomic clarity of parvoviruses, and this gave rise to the introduction of new species and genera into the two subfamilies of the family Parvoviridae (Cotmore et al., 2014).
Members of the subfamily Parvovirinae are now categorized into two major groups namely: dependoparvoviruses and autonomous parvoviruses, based on their replication requirement. While the dependoparvoviruses require helper virus for a successful replication within cells, the autonomous types do not (Kailasan et al., 2015). Altogether, the subfamily Parvovirinae consists of eight genera: Amdoparvovirus (Amdovirus), Aveparvovirus, Dependoparvovirus (Dependovirus), Erythroparvovirus (Erythrovirus), Copiparvovirus, Bocaparvovirus (Bocavirus), Protoparvovirus (Parvovirus) and Tetraparvovirus. The last four genera contain the classical porcine parvovirus and other new porcine parvoviruses on which this review focuses (Cotmore et al., 2014).
1.2. The observable increase in the number of porcine parvoviruses species of the subfamily Parvovirinae
The sporadic revolutions in molecular technology which brought about the use of nucleic acid amplification techniques in pathogen detection and the use of more recent, novel molecular tools such as high-throughput sequencing have led to the detection of some novel porcine parvoviruses. The new viruses have been characterized in various research proposals with most of the classifications being considered in the latest ICTV report that is awaiting ratification. As earlier mentioned, four genera now contain the designated eight ungulate porcine parvoviruses species containing about twelve pig-infecting viruses and their variants (Cotmore et al., 2014). More recent ones are yet to be assigned into species and genus under the subfamily. These include PPVs 5, 6 and 7 (Xiao et al., 2013a; Ni et al., 2014; Schirtzinger et al., 2015; Palinski et al., 2016; Xing et al., 2018).
However, porcine parvovirus 1 (PPV1) has been the only pig-infecting virus of genus Parvovirus (i.e. Protoparvovirus) in the classification, while porcine parvovirus 2 (PPV2) and porcine parvovirus 3 (PPV3) were categorized together with human parvovirus 4 (PARV4) in the genus Tetraparvovirus due to their relatively close genomic homologies. The porcine parvovirus 4 (PPV4) belongs to the genus Copiparvovirus alongside with bovine parvovirus 2 (Cotmore et al., 2014), and the genus have been proposed to contain the novel PPVs 5 and 6 as they have constantly clustered within the group in phylogenetic analyses (Xiao et al., 2013b; Ni et al., 2014; Schirtzinger et al., 2015). In addition, eigth pig viruses and virus variants are presently grouped under the genus Bocaparvovirus to make four distinct genera of swine parvoviruses in the published classification (Cotmore et al., 2014).
Currently, many of the newly discovered porcine parvoviruses are yet to be fully studied, there is paucity of information on the importance and pathogenicity potential of the viral pathogens to global swine population (Xiao et al., 2013c). Hence, for the purpose of this current review, the classical porcine parvovirus (PPV) or porcine parvovirus type 1 (PPV1) will be briefly reviewed as a representative of other porcine parvoviruses, while an overview on the detection and prevalence of other viruses will be duly stressed. Also, the later part of this review will bring to the fore the epidemiology of the porcine parvoviruses in swine herds of African countries taking cognizance of the past, present and future. This becomes highly imperative as the region is observed to be moving at a relatively low pace in the use of new technologies for the detection of infectious agents; this abnormal trend could undermine the global efforts in prevention and control of myriads of emerging and re-emerging animal and human diseases that are ravaging our world.
2. Classical porcine parvovirus: a known pathogen of swine reproductive disorder
2.1. Brief history of porcine parvovirus type 1 (PPV1)
Early in the 1960s, cases of reproductive failure were rampant in commercial swine farms due to unknown causes which experts thought could be attributed to nutritional and environmental factors among many others (Lawson, 1961). PPV1 was subsequently isolated in Germany as a contaminant of pig cell cultures used for the cultivation of classical swine fever virus in mid 1960s (Mayr and Mahnel, 1964) and was later confirmed to be associated with reproductive losses in swine (Dunne et al., 1965; Cartwright and Huck, 1967). In the subsequent years, the clinical manifestations of PPV1 reproductive disease were stated as reoccurring of oestrus in sows, abortion and farrowing of mummified or stillborn foetuses, generally regarded as SMEDI (stillbirth, mummification, embryonic death and infertility) (Thomson and Prozesky, 1994; Mengeling et al., 2000).
The pathologic effect of the virus in a pregnant sow and its foetuses is due to the tropism it has for actively replicating cells such as foetal myocardiocytes, which makes foetal infection often results in death, depending on the stage of sow gestation (Opriessnig et al., 2017). Many data from previous studies on the determination of major etiologic agents of reproductive failure in swine have shown PPV as important cause of porcine foetal death. Mengeling et al. (1991) earlier implicated PPV1 in the death of about 35% (105 of 302) of dead foetuses collected and tested in the USA. In some relatively recent studies, Zhang et al. (2010) found out only PPV1 as the causal agent of an acute outbreak of abortions in a domesticated herd of 500 wild boar females in Heilongjiang province, China; while, Tummaruk and Tantilertcharoen (2012) observed that 86% (143/166) of gilts culled due to reproductive failure in Thai swine herds were seropositive for PPV1. The virus is considered to be extremely stable in the environment, endemic in many parts of the world and capable of infecting pig herds of all categories (Almond et al., 2006; Truyen and Streck, 2012).
2.2. Genomic structure and organization in PPV1
PPV1 has a small, single-stranded, negative-sense DNA genome of approximately 5 kb which is packaged in a non-enveloped viral capsid (Molitor et al., 1984). The genome has a unique feature of distinct palindromic hairpin termini and contains two major open reading frames (ORFs) (Bergeron et al., 1993; Bergeron et al., 1996). The ORF1 found at the 5′ end of the viral genome codes for non-structural proteins 1 (NS1) which could be spliced alternatively to obtain two additional non-structural proteins (NS2 and NS3). The non-structural proteins have some enzymatic functions that are important in the viral replication and packaging (Bergeron et al., 1996). The ORF2 which codes for capsid proteins (VP1, 2 and 3) is located at the 3′ end of the viral genome. The VP1 and 2 proteins are formed from differently spliced mRNAs, while VP3 is formed by proteolytic cutting of VP2 (Bergeron et al., 1996). About sixty copies of the capsid proteins are used in the assembly of the virus' icosahedral capsid, as previously reviewed by Streck et al. (2015a).
2.3. Epidemiology of PPV1
PPV1 is the earliest known swine parvovirus of veterinary importance (Dunne et al., 1965). The virus is highly ubiquitous and has been found in swine herds of different categories including domestic and wild pigs, breading age females and boars, young piglets and fattening pigs (Dea et al., 1985; Duhamel et al., 1991; Lager and Mengeling, 1994; Drolet et al., 2002; Cadar et al., 2012; Truyen and Streck, 2012). It has also been discovered among pigs of different health statuses including vaccinated and non-vaccinated pigs, healthy and sick pigs (Jóźwik et al., 2009).
The exceptional stability of the virus in the surroundings influences its infectivity and its spread. It is highly thermo-stable, having the ability to survive dry heat to an extent of 90 °C (Eterpi et al., 2009). Furthermore, the virus can resist up to 70% ethanol and disinfectants like sodium hypochlorite at a low concentration of 2500 ppm. Hence, the viral pathogens can remain infectious in a contaminated pen, farm tools and clothings for months thereby enhancing its transmission from one farm to another (Truyen and Streck, 2012). That is the reason why strict observation of biosecurity measures are necessary in piggery operations, so as to prevent the farm to farm spread of the pathogens and other related ones. Farm to farm transmission can also be enhanced through gilt replacement when asymptomatic gilts are acquired. This is because infected pigs may not necessarily show any clinical manifestation if well vaccinated (Truyen and Streck, 2012; Foerster et al., 2016). The virus can also be introduced into a vulnerable farm when an infected boar or sperm is used for breeding purposes; a seronegative boar can also be infected during mating from vaginal mucus of an infected sow (Szelei et al., 2006). Furthermore, the viral pathogen can spread within a herd through faecal droppings, nasal and oral secretions from infected pigs (Truyen and Streck, 2012).
2.4. Viral pathogenesis and clinical manifestations
The PPV1 strains have varying levels of pathogenicity as depicted by the two variants namely porcine parvovirus Kresse (PPV-Kresse) and NADL-2 (PPV-NADL2) (Tijssen et al., 1995). The disparity in their pathogenic potential is believed to be as a result of some amino acid residues substitution in their VP2 structural protein. While the former is known to be virulent variant, the later is non-pathogenic and generally employed in producing the inactivated and modified live vaccines (Paul and Mengeling, 1984; Mengeling et al., 1984; Bergeron et al., 1996; Simpson et al., 2002). Although the avirulent PPV-NADL2 has a history of causing limited viraemia without crossing the placenta barrier (Mengeling et al., 1984), the PPV-Kresse is exceptionally pathogenic, having a strange capacity to cause mortality not only in a susceptible foetuses, but also in the immuno-competent ones in cases of maternal infections (Zeeuw et al., 2007) and induce dermatitis in immuno-competent foetuses and young pigs (Choi et al., 1987; Whitaker et al., 1990; Lager and Mengeling, 1994).
The main clinical manifestations of PPV1 infections in swine are the reproductive disorders in breeding sows. When a seronegative pregnant sow is infected at the early stage of gestation, its conceptus is usually not affected due to an initial protection received (Mengeling et al., 2000). However, during the embryonic stage, occurrence of PPV1 infection normally results to transplacental infections of the embryo leading to embryonic death and resorption. At a gestation period of 35 days and above when bone formation begins, the viral infection gives rise to foetal death and mummification. Finally, if a pregnant sow is infected at the advanced stage of gestation of 70 days and above, its foetus has already become immuno-competent at that point; hence, the foetus can independently resist the viral infection and becomes seropositive at birth with likelihood of having a subclinical infection status (Lenghaus et al., 1978; Mengeling et al., 2000; Zeeuw et al., 2007).
Although, the viral pathogen has been linked to many other disease conditions including non-suppurative myocarditis in suckling piglets, diarrhoea, interstitial nephritis in slaughter-aged pigs and skin lesions (Kresse et al., 1985; Whitaker et al., 1990; Lager and Mengeling, 1994; Bolt et al., 1997; Drolet et al., 2002), its etiological role in the disease manifestations is not fully elucidated. Also, PPV1 is a common co-infecting pathogen that aggravates the disease conditions in porcine circovirus type 2 (PCV2) infections by facilitating the onset of postweaning multisystemic wasting syndrome (PMWS) which is presently one of the viral swine diseases of huge economic importance globally (Allan et al., 1999; Krakowka et al., 2000; Segalés et al., 2005).
2.5. Prevention and control of PPV1 infection
The control of PPV1 infection has been a difficult task due to the stable nature of the virus, which normally enhances the persistence of the pathogen in swine herds (Truyen and Streck, 2012). However, reproductive system operation failure due to PPV1 infection is usually prevented through vaccination of the gilts prior to insemination and this has proven effective and economical over the years for successful farm operations (Wrathall et al., 1984; Pye et al., 1990; Mengeling et al., 1991; Parke and Burgess, 1993). The recorded success, however, has been a function of effective vaccination regimes in terms of timing and consistency (Truyen and Streck, 2012). Firstly, it has been shown that a trans-placental passage of maternal antibodies does not occur in pigs (Salmon et al., 2009); hence, a surviving piglet is expected to obtain antibodies from its mother against PPV1 through colostrum intake at its early stage of life (Devillers et al., 2011). However, the depletion of the maternally acquired antibodies is a normal phenomenon that determines when vaccination of pigs can be carried out in order for it to be effective as vaccination at a high level of maternal antibodies brings about negative interference (Truyen and Streck, 2012). An appropriate timing has been recently determined to be after three months of age for a gilt to be inseminated prior to five or six months old (Gava et al., 2017).
The commercially available PPV1 vaccines are made through whole-virus inactivation of the avirulent PPV-NADL2 strains and have been effective in preventing reproductive disorders in sows (Mengeling et al., 1980; Paul et al., 1980; Mengeling et al., 1984). However, it has been proven experimentally that both homologous and heterologous inactivated PPV1 vaccines can not prevent viral infection and shedding, thereby making total immunization and subsequent eradication of the virus impossible. This, therefore, necessitates the need for a consistent follow-up vaccinations of all breeders appropriately (Jóźwik et al., 2009; Foerster et al., 2016). This limitation, coupled with seeming emergence of new strains of PPV1 that show lower homology with the vaccine strains, therefore necessitates futher development of more efficient vaccines (Jóźwik et al., 2009; Ren et al., 2013; Streck et al., 2015a).
3. General epidemiology of other known porcine parvoviruses
Since about five decades ago that PPV1 was first detected in Germany (Mayr et al., 1968), many more porcine parvoviruses other than those in bocavirus group have been detected in swine population with the recent advancement in the use of molecular techniques in pathogen discovery and epidemiology (Table 1 ) (Ni et al., 2014). Generally, the new development has brought about a continuing increase in the number of new parvoviruses grouped under the subfamily Parvovirinae including those that have been found in other animal species such as Ovine hokovirus (O-PARV4) and also a group of novel porcine bocaviruses (Lau et al., 2011; Tse et al., 2011; Huang et al., 2014). The human parvovirus 4 (PARV4) is also a good example of human parvoviruses that have recently been detected with a close relationship with O-PARV4 and porcine hokovirus (PPV3) and as such they have been currently grouped in the same genus Tetraparvovirus (Cotmore et al., 2014). In this section, porcine parvoviruses other than PPV1 will be concisely discussed including the very latest PPV7.
Table 1.
Detected porcine parvoviruses till date.
| Virus | Abbreviation | Year first detected | Place | Source | Reference |
|---|---|---|---|---|---|
| Porcine parvovirus 1 | PPV, PPV1 | 1965 | Germany | Porcine primary cell culture used for the propagation of classical swine fever (CSF) virus | Cartwright and Huck, 1967 |
| Porcine parvovirus 2 | PPV2, CnP-PARV4 | 2001 | Myanmar | Serum samples from pigs obtained for HEV screening | Hijikata et al., 2001 |
| Porcine parvovirus 3 | PPV3, P-PARV4, porcine hokovirus | 2008 | Hong Kong | Multiple samples from healthy, sick and dead pigs | Lau et al., 2008 |
| Porcine parvovirus 4 | PPV4 | 2010 | USA | Lung lavage of a diseased pig co-infected with PCV2 | Cheung et al., 2010 |
| Porcine parvovirus 5 | PPV5 | 2013 | USA | Lung tissues of grower | Xiao et al., 2013a |
| Porcine parvovirus 6 | PPV6 | 2014 | China | Aborted pig foetuses | Ni et al., 2014 |
| Porcine parvovirus 7 | PPV7 | 2016 | USA | Rectal swabs of healthy adult pigs | Palinski et al., 2016 |
3.1. Porcine parvovirus type 2 (PPV2)
Hijikata et al. (2001) first detected the genome of PPV2 accidentally during an epidemiological survey for hepatitis E virus in Myanmar pigs. The viral genome was detected in 8 out of the 86 (10%) screened sera from Myanmar pigs in the study. About a decade later, highly related parvoviruses were detected from serum samples obtained from commercial farms with severe field outbreaks of “high fever” suggested to be as a result of porcine reproduction and respiratory syndrome virus (PRRSV) and PCV2 infections of pigs in Southeastern China (Wang et al., 2010). The novel Chinese strains formed a clade together with the initial one from Myanmar and were designated as Cnvirus sub-lineage under Parvovirinae. However, they are presently grouped as Ungulate tetraparvovirus 3 under the genus Tetraparvovirus (Cotmore et al., 2014).
Subsequently, PPV2 has been reported in many other countries including Hungary, USA, Japan, Germany and Thailand with prevalences of 6%, 21%, 58%, 78% and 83% respectively (Cságola et al., 2012; Xiao et al., 2013d; Streck et al., 2013; Saekhow et al., 2014; Saekhow and Ikeda, 2015). Higher prevalences ranging from 78% (Germany) to 100% (Japan) were however obtained when tonsils samples were used in the PPV2 screening as compared to when other samples including blood, faeces and lungs were used (Streck et al., 2013; Saekhow et al., 2014). The prevalence rates that could be obtained for the virus was therefore assumed to be dependent on the type of organ assayed, although the virus tissue tropism and infection routes are yet to be elucidated (Saekhow and Ikeda, 2015).
3.2. Porcine parvovirus type 3 (PPV3)
The PPV3, also referred to as porcine hokovirus (PHoV), belongs to the Ungulate tetraparvovirus 2 species and it is presently grouped together with PPV2 in the genus Tetraparvovirus, one of the two genera that comprise human and pig pathogens together under the subfamily Parvovirinae. The virus was also accidentally detected together with bovine parvoviruses (BHoV) in Hong Kong when an effort to determine the relationship that exists between the newly identified human parvovirus types 4 and 5 (PARVs 4 and 5) and animal parvoviruses was made. Although the PHoV and BHoV show a genetic similarity to the human parvovirus types 4 and 5, however according to Lau et al. (2008), the risk of the animal hokoviruses to human health is not likely to be significant since the viruses were absent in human samples examined in the study. Nevertheless, they further asserted that there is possibility of further mutation and/or recombination in the viral strains, leading to overcoming the interspecies barrier as previously observed in many emerging infectious zoonotic diseases that cause epidemics in humans; such like the severe acute respiratory syndrome coronavirus (SARS-CoV) that was responsible for the SARS epidemic in 2003, which originated from wild animals (Lau et al., 2008). The virus is believed to have a global distribution status as it has been found in domestic pigs in Asia (Pan et al., 2012; Li et al., 2013; Saekhow and Ikeda, 2015), North America (Xiao et al., 2012), Europe (Cságola et al., 2012) and Africa (Adlhoch et al., 2013). It has also been found in wild boars in Germany and Romania with prevalence of 33% and 50% respectively (Adlhoch et al., 2010; Cadar et al., 2011).
3.3. Porcine parvovirus type 4 (PPV4)
The first detection of PPV4 occurred in North Carolina from lung lavage of PCV2-infected pigs (Cheung et al., 2010). Ever since then, the virus has been detected in other countries including China, Hungary, Germany and Thailand with prevalence of 2%, 6%, 7% and 44% respectively (Huang et al., 2010; Cságola et al., 2012; Streck et al., 2013; Saekhow and Ikeda, 2015). Although the virus was initially detected from samples obtained from pigs with multiple clinical manifestations including PRRS and PCV2-associated diseases, the involvement of the viral pathogen in the observed disease conditions is yet to be resolved (Cheung et al., 2010; Saekhow and Ikeda, 2015). Phylogenetically, PPV4 has a closer relationship with bovine parvovirus 2 (BPV2). However, in terms of genomic organization and structure, the virus is quite similar to other porcine viruses under the genus Bocavirus due to its possession of an additional ORF3 gene situated at the center of its genome. The deduced amino acids of the PPV4 ORF3 gene are however quite different from those of bocaviruses (Cheung et al., 2010). This is suggested to be the basis of its separate clustering outside the bocavirus group. The virus is presently assigned to the genus Copiparvovirus together with BPV2 in the ICTV latest report (Cotmore et al., 2014).
3.4. Novel unassigned porcine parvoviruses
In recent years, some other novel porcine parvoviruses have been detected, but yet to be assigned to species and genera in the latest ICTV classification report. The viruses have been tentatively designated as porcine parvovirus types 5, 6 and 7 (PPV5, PPV6 and PPV7). Xiao et al. (2013a) detected a novel porcine parvovirus with the closest genomic sequence identity of 64.1–67.3% to PPV4 from swine lung samples during a study to ascertain the prevalence of PPV4 in the USA. The viral genome was further characterized and was found to be different from PPV4 as its genome lacks the extra ORF3 that is peculiar to the PPV4 and bocaviruses (Xiao et al., 2013b). The findings of phylogenetic analyses on the virus depicted its grouping with PPV4 and BPV2 as it formed a separate cluster with them under the genus Copiparvovirus. The virus was provisionally named as PPV5 and its prevalence in pigs from the USA was 6.6% (32 out of 483) (Xiao et al., 2013b).
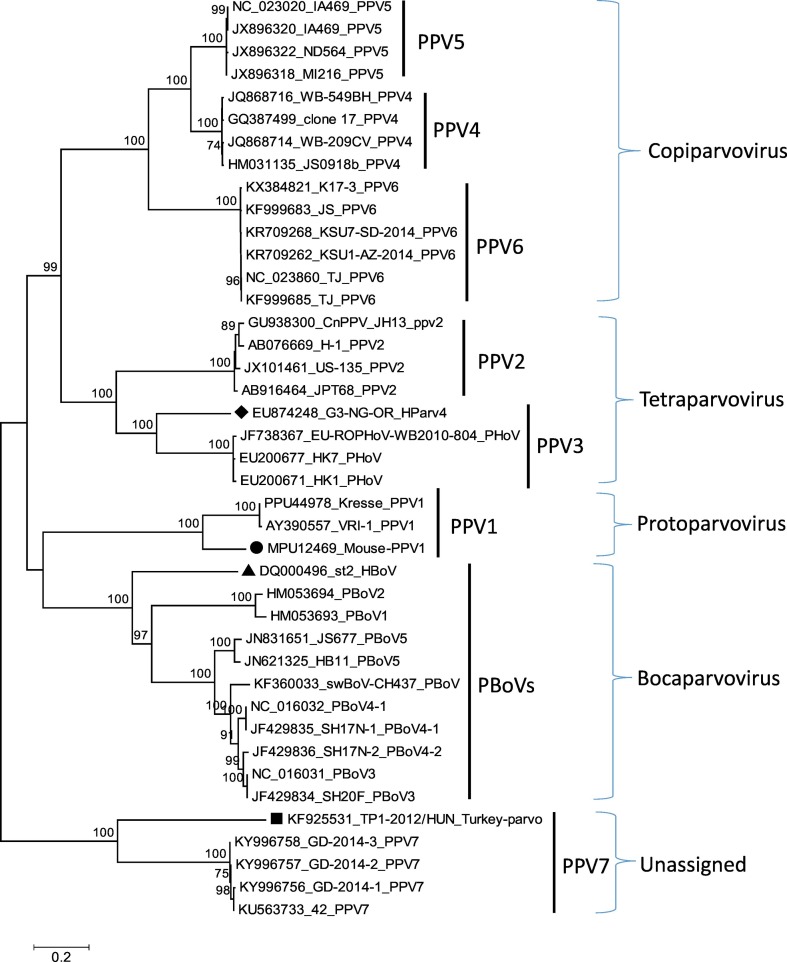
In the following year, Ni et al. (2014) for the first time identified another novel pig parvovirus, tentatively named as PPV6, from aborted swine foetuses which showed negative results for other swine's reproductive failure pathogens in pigs in China. Subsequently, the virus has been detected through a high throughput sequencing of PRRSV positive serum samples from swine in the USA and Mexico, with a very high similarity to the PPV6 discovered in China (Schirtzinger et al., 2015). Reports from the two investigations have unanimously proposed the classification of the novel PPV6 under the genus Copiparvovirus alongside the PPVs 4 and 5, as they all formed the same cluster in their phylogenetic analyses and as it could be seen in Fig. 1 .
Fig. 1.
Phylogenetic analysis base on the NS1 gene of all porcine parvoviruses detected till date. While the black diamond sign indicates the human parvovirus 4 (PARV4) clustering with porcine hokovirus (PHoV), the black triangle depicts the reference genome of human bocavirus 2 (HBoV) clustering with other porcine bocaviruses. The black rectangle shows the turkey parvovirus (TuPV) that forms clade with the novel PPV7. The unrooted phylogenetic tree was done in Mega 6 using the Neighbour-joining algorithms and 1000 bootstrap replicates (Tamura et al., 2013).
More recently, another novel porcine parvovirus was discovered through metagenomic sequencing of pooled samples which include serum, lung lavage, nasal swabs and rectal swab samples from pigs in the USA, having an overall prevalence of 16/182 (8.6%) (Palinski et al., 2016). The viral pathogen was also recently found in serum samples from pigs in China with an overall prevalence of 21/64 (32.8%), and a higher value of 19/29(65.5%) in pigs from PCV2-positive farms compared with those from PCV2-negative farms which had a value of 2/35 (5.7%) (Xing et al., 2018). The phylogenetic analysis of the viral genomes in the latest study showed 98.7–99.7% nucleotide identity between the viral strains from the two countries (Xing et al., 2018), whereas the previous studies revealed closer identity of 42.4% and 37.9% at the NS1 proteins for the fruit bat (EhPV2) and turkey parvoviruses (TuPV) respectively (Palinski et al., 2016). The novel viral agent has been provisionally named PPV7, and a proposal was made for the formation of another genus Chapparvovirus (coined from Chiroptera, Avian and Porcine) under the subfamily Parvovirinae to contain PPV7, EhPV2 and TuPV as they have shown less than 30% identity to other species in subfamily Parvovirinae, which is far below the stipulated species inclusion value of at least 85% identity in NS1. The phylogenetic analysis based on the NS1 gene of all the known porcine parvoviruses in this study also confirmed a separate grouping and clustering for PPV7 and TuPV (Fig. 1).
3.5. Porcine bocaviruses (PBoVs)
The bovine parvovirus (BPV) is the first known member of the genus Bocavirus (now Bocaparvovirus) and the only virus in the ungulate bocaparvovirus 1 species. The virus is the first member of the genus that was detected about six decades ago in the USA from calves with a clinical symptom of diarroea (Abixanti and Warfield, 1961). Presently, there are about eight porcine viruses and virus variants that have been grouped into four species, namely ungulate bocaparvovirus 2, 3, 4 and 5 under the genus Bocaparvovirus with the BPV (Cotmore et al., 2014). The genus Bocaparvovirus is the second group in the subfamily Parvovirinae that consists of human and swine viruses together. The genus also comprises of feline bocavirus, canine bocaviruses, gorilla bocavirus and California sea lion bocaviruses (Cotmore et al., 2014).
The first porcine bocavirus was discovered in 2009 from lymph node samples collected from piglets diagnosed to have PCV2 wasting syndrome (Blomström et al., 2009). The viral pathogen was detected by using the random amplification technique and high-throughput sequencing in a bid to study other co-infecting pathogens in PMWS-affected Swedish pigs. Partial genome (1879 bp) that comprises the entire NP1 and partial VP1/VP2 of the Swedish porcine boca-like virus (PBo-likeV) was amplified and analyzed alongside with the genomes of Torque Teno virus (TTV) and PCV2, thereby confirming the presence of the viruses in the PCV2-associated wasting syndrome case, although the contribution of PBo-likeV to the development of PMWS is yet to be elucidated (Blomström et al., 2009).
Subsequently, the detection of many other porcine bocaviruses which are different from the Swedish strain have been on the rise globally, and there have been vigorous efforts to characterize and classify them (Cheng et al., 2010; McKillen et al., 2011; Li et al., 2012; Yang et al., 2012; Xiao et al., 2013c). However, there have been a lot of disparities and confusions along the process (Gunn et al., 2015). In this review, the grouping according to the latest ICTV report is adopted in which eight porcine bocaviruses were identified and assigned into four species namely: ungulate bocaparvovirus 2 (consisting of porcine bocavirus 1, 2 and 6 i.e. PBoV1, PBoV2 and PBoV6), ungulate bocaparvovirus 3 (having only porcine bocavirus 5 i.e. PBoV5), ungulate bocaparvovirus 4 (consisting of porcine bocavirus 3 and 7 i.e. PBoV3 and PBoV7), and Ungulate bocaparvovirus 5 (comprising of porcine bocavirus 4–1and 4–2 i.e. PBoV4–1 and PBoV4–2) (Cotmore et al., 2014).
PBoVs have been detected and characterized by using molecular techniques from arrays of samples which include faeces, serum, lung, lymph nodes and nasopharyngeal samples (Zhai et al., 2010; Lau et al., 2011; Cságola et al., 2012; Blomström et al., 2013; Choi et al., 2014). Although the viral agents have been detected in both symptomatic and asymptomatic pigs, many questions about their pathogenicity are yet to be answered (Gunn et al., 2015). However, more details about the epidemiology and pathogenesis of PBoVs have been previously reviewed (Zhou et al., 2014).
4. Epidemiology of porcine parvoviruses in African countries
Pig farming systems in most African countries, as typified by South African system, include partially to fully free range system in rural communities, small and semi-commercial units and commercial scales. In the free ranging system, pigs normally move around in the community, feeding on scraps and wastes from household with zero biosecurity control, thereby increasing the risk of diseases transmission and spread (Mather and Marshall, 2011; Mokoele et al., 2015). Although pig production in African countries, regarding the frequency of reared pigs, is insignificant compared to other pig-producing countries of the world such as China, USA and Brazil; the importance of pig production cannot be overemphasized in the region considering its economic value in alleviating poverty and solving hunger challenge in developing countries of the world (FAO, 2012). However, little or no effort is geared towards detecting and studying most of the swine viral pathogens of global economic importance in the region. This assertion is strongly corroborated in a recent review analysis on PCV2 and its associated diseases in the sub-Saharan Africa region in which only 2 out of the 49 (4%) countries represented in the region have meager documentation about the detection of the swine viral pathogen in the region (Afolabi et al., 2017a).
As is the case of PCV2 earlier stated, investigations on porcine parvoviruses in the entire African continent is relatively low when compared with vast research efforts that have been tailored towards the detection and characterization of the circulating viral strains in other countries of the world. Nevertheless, this section is dedicated to highlighting the past and current reported cases of porcine parvovirus infection in some African countries, in a bid to serve as awakening call for meaningful future proactive research activities in unraveling the occurrence and prevalence of the viral pathogens in the region. This will go a long way in ameliorating the plight of peasant farmers in the region whose livelihood is dependent on the piggery business.
4.1. Porcine parvoviruses in South African swine
The first reported detection of porcine parvovirus in South African pigs took place in 1975 during an outbreak of swine reproductive failure that occurred at a pig farm in which PPV1 was isolated from aborted foetuses (Pini, 1975). Subsequently, the virus was detected as the etiologic agent of similar clinical feature at some other farms. This prompted Prozesky et al. (1980) to launch a pathological investigation on the isolated local strain of PPV1, so as to elucidate its effect on foetuses and pregnant sows for the purpose of comparing their findings with other documented cases. Experimental reproduction of typical PPV1-associated reproductive failure characterized by resorption, abortion and mummification of foetuses was successfully carried out through the in-utero inoculation of 15 sows at different levels of gestation with the local viral strain. Vascular lesions were equally observed in the endometrium of the sows, which the authors assumed could contribute to the reproductive disorders observed in the inoculated sows (Prozesky et al., 1980).
Apart from the initial case study and the subsequent experimental evaluation carried out about four decades ago, there has been no documented research work on porcine parvoviruses in South African pigs till date, except a very recent molecular epidemiological study that unravelled the prevalence of some porcine parvoviruses in selected swine herds with a background of PCV2 infection. The prevalence of PPV1-4, PBo-likeV and PBoV1/2 were shown to be 32/110 (29.1%), 24/110 (21.8%), 6/110 (5.5%), 48/110 (43.6%), 24/110 (21.8%) and 49/110 (44.6%) respectively (Afolabi et al., 2018a). All the selected porcine parvoviruses were detected from swine samples obtained from confirmed PCV2-infected farms (Afolabi et al., 2017b, Afolabi et al., 2018b), indicating that South African pigs are not free from parvovirus infections. These insightful findings call for an urgent large scale epidemiological survey in the country and the entire Southern African region for effective control of the viral pathogens.
4.2. Detection of porcine parvoviruses in East African countries of Uganda and Kenya
Relatively, more reported cases of porcine parvoviruses are available from the East African countries of Uganda and Kenya. In a study aimed at determining the presence of viral pathogens from serum samples obtained from bush pigs that roam around between the national park and farmland in Uganda, PPV4 alongside Torque teno sus viruses 1 and 2 (TTSuV1 and TTSuV2) were detected through a metagenomic approach (Blomström et al., 2012). The PPV4 from the serum of bush pigs showed a sequence identity of 75.9–77.1% with other PPV4 sequences in the GenBank which cluster together with them in the phylogenetic analysis, with some display of divergence from other reference sequences used. The detection was the first report on PPV4 in bush pigs from Africa (Blomström et al., 2012). Related efforts were made to screen for the virus in samples from the domestic pigs using the primers designed from the PPV4 sequence from the bush pig, however, no positive result was obtained (Brink, 2011).
Also, with the use of conventional PCR specific for the PBoV types previously detected in Sweden and China, Blomström et al. (2013) went further to screen for the possible presence of porcine bocaviruses in 95 serum samples collected from six different districts previously used for the detection of TTSuV-1 and 2 in Ugandan pigs (Brink et al., 2012). They recorded a very low detection rate of 2/95 (2.1%) compared to 16.8% and 48.4% previously obtained for the TTSuV-1 and TTSuV-2 respectively using the same samples. However, from the study, the first PBoV genome from Africa was assembled, having all the coding regions of the virus. The viral strain BuK8 (JX854557) from Uganda had an amino acid sequence similarity of 98–99% to the two Chinese strains H18 (HQ291308) and SX (HQ223038) from only one group of presently known porcine bocaviruses. Future investigation was suggested to determine the occurrence of other PBoV species in the country (Blomström et al., 2013).
Subsequently, in a more recent study, Amimo et al. (2016) screened for the presence of viral pathogens from the faecal samples obtained from healthy pigs from Kenya and Uganda using metagenomics approach. Virome from 12 pigs were analyzed and sequence contigs belonging to six (6) viral families including Parvoviridae were identified, constituting 11% of the total reads. Further analysis of the contig sequences to determine the individual virus present in the samples showed the presence of astroviruses in 10 out of the 12 samples (83.3%), followed by porcine bocaviruses which was detected in 7 out of the 12 samples, representing 58.3%. Subsequently, a commendable effort was made by the authors to sequence the full genomes of the two porcine bocavirus strains JOA_011 and JOA_015 obtained from healthy pigs in farms from the two countries. The two sequences were further analyzed and showed high relatedness with the strain KU14 from South Korea (Amimo et al., 2017). It is expected that future large scale epidemiological studies that will unravel the presence and prevalence of the viral pathogens in pigs from the region will be conducted, considering the fact that two (PPV4 and PBoV) out of the numerous members of the viral group have already been found in the reported studies (Blomström et al., 2012, Blomström et al., 2013; Amimo et al., 2017).
4.3. Porcine parvoviruses in West African countries of Cameroun and Nigeria
Interestingly, almost at the same time that Blomström et al. (2013) detected the first PBoV in Uganda, Ndze et al. (2013) in a separate study also reported the occurrence, diversity and prevalence of some DNA viruses including those of the Parvoviridae family in Cameroonian pigs. While other viral groups were absent in the samples from the asymptomatic pigs, some porcine parvoviruses in the subfamily Parvovirinae were detected including PPV4 with prevalence of 10/50 (20%). Others are bocaviruses including PBoV1/2 with the prevalence of 3/50 (6%), 9/50 (18%) for PBoV3, 9/50 (18%) for PBoV4 and 8/50 (16%) for the combination of PBoV5 and PBoV6V/7 V (Ndze et al., 2013). High co-infection rates between PPV4 and the various species of bocaviruses were equally noted in the study. Future efforts are required to amplify and sequence the complete genomes of the detected PBoVs for an adequate genetic characterization of the viral strains in circulation within the country.
In another study, Adlhoch et al. (2013) used 282 samples collected in the year 2012 for the purpose of investigating the presence of hepatitis E virus (HEV) in Cameroonian pigs to screen for the presence of porcine hokovirus (PPV3), a related virus to human parvovirus 4 (PARV4), in the swine herds of the country. The DNA samples obtained were pooled, with each pool containing 3 different samples. From a total of 94 pooled samples, 69% (65/94) samples from Doula, Yaoundé, and Bamenda districts of Cameroon were positive for PHoV. Subsequently, effort was made to determine the prevalence based on the individual samples apart from the one obtained for the pooled samples, however, 12 samples were reported missing by the authors and no reason was given to that effect. Hence, a general prevalence of 47% (128/270) was estimated from the remaining individual samples obtained from the Cameroonian pigs. Phylogenetic analysis was subsequently performed on the partial genomes generated from some of the pooled samples and the Cameroonian sequences had close homology of 98–99% to other reference sequences from the Europe, USA and China. The authors, however, suggested further studies on the virus at other premises of Africa in order to determine the circulating viral strains in the region (Adlhoch et al., 2013).
In Nigeria, according to Aiki-Raji et al. (2018), the status of PPV1 in Nigerian pigs is largely unknown. In their sero-prevalence study recently done to unravel the prevalence of PRRSV and PPV1 in pigs from the southwestern region of the country, a prevalence of 36.1% was obtained for the classical porcine parvovirus in the studied area which comprised only two states, Oyo and Lagos (Aiki-Raji et al., 2018). This finding also calls for proactive surveillance of the pathogen in pigs from the country in order to determine its true status in swine herds of the country so as to facilitate effective control measures.
5. Conclusion
The detection of parvoviruses in pigs has rapidly increased in recent years. Although the classical porcine parvovirus has been the most studied member of the viral group globally, efforts in various parts of the world in detecting and analyzing the prevalence of other novel porcine parvoviruses is highly commendable compared to a lethargic approach that is being used in the African region. Considering the economic importance of parvoviruses to the piggery business as highlighted in this review, most especially the continuous emergence of new strains/species with presumed zoonotic potentials due to their relatively close genetic relatedness with human parvoviruses, the need for effective large-scale surveillance and characterization of the porcine parvoviruses and many other neglected swine pathogens in the region becomes imperative. This will go a long way in enhancing the implementation of the globally acclaimed “One Health, One World” initiative for combating zoonotic diseases in the region.
Competing interests
The authors declare that there is no competing interest regarding the publication of this paper.
Acknowledgement
The authors acknowledge the financial supports of National Research Foundation (NRF) and South African Medical Research Council (SAMRC/UFH/P790). Dr. Afolabi received scholarship from National Research Foundation (109622) and also a tuition bursary support from Govan Mbeki Research and Development Centre (GMRDC) of University of Fort Hare. Thanks to Dr. Titilawo for proof reading the article.
References
- Abixanti F.R., Warfield M.S. Recovery of a hemadsorbing virus (HADEN) from the gastrointestinal tract of calves. Virol. 1961;14:288–289. doi: 10.1016/0042-6822(61)90206-9. [DOI] [PubMed] [Google Scholar]
- Adlhoch C., Kaiser M., Ellerbrok H., Pauli G. High prevalence of porcine Hokovirus in German wild boar populations. Virol. J. 2010;7:171. doi: 10.1186/1743-422X-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlhoch C., Kaiser M., Kingsley M.T., Schwarz N.G., Ulrich M., de Paula V.S., Ehlers J., Löwa A., Daniel A.M., Poppert S., Schmidt-Chanasit J. Porcine hokovirus in domestic pigs, Cameroon. Emerg. Infect. Dis. 2013;19:2060. doi: 10.3201/eid1912.130891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afolabi K.O., Iweriebor B.C., Okoh A.I., Obi L.C. Global status of porcine circovirus type 2 and its associated diseases in sub-Saharan Africa. Adv. Virol. 2017;2017:6807964. doi: 10.1155/2017/6807964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afolabi K.O., Iweriebor B.C., Obi L.C., Okoh A.I. Molecular detection of porcine circovirus type 2 in swine herds of eastern Cape Province South Africa. BMC Microbiol. 2017;17:212. doi: 10.1186/s12866-017-1121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afolabi K.O., Iweriebor B.C., Obi L.C., Okoh A.I. Prevalence of porcine parvoviruses in some south African swine herds with confirmed high farm level occurrence of porcine circovirus type 2. Acta Trop. 2018 doi: 10.1016/j.actatropica.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Afolabi K.O., Iweriebor B.C., Obi C.L., Okoh A.I. Genetic characterization and diversity of porcine circovirus type 2 in non-vaccinated south African swine herds. Transbound. Emerg. Dis. 2018 doi: 10.1111/tbed.13036. [DOI] [PubMed] [Google Scholar]
- Aiki-Raji C.O., Adebiyi A.I., Abiola J.O., Oluwayelu D.O. Prevalence of porcine reproductive and respiratory syndrome virus and porcine parvovirus antibodies in commercial pigs, Southwest Nigeria Beni Suef University. Aust. J. Basic Appl. Sci. 2018;7:80–83. [Google Scholar]
- Allan G.M., Kennedy S., McNeilly F., Foster J.C., Ellis J.A., Krakowka S.J., Meehan B.M., Adair B.M. Experimental reproduction of severe wasting disease by co-infection of pigs with porcine circovirus and porcine parvovirus. J. Comp. Pathol. 1999;121:1–11. doi: 10.1053/jcpa.1998.0295. [DOI] [PubMed] [Google Scholar]
- Almond G.W., Flowers W.L., Batista L., D'Allaire S. In: Diseases of Swine. Straw B.E., Zimmerman J.J., D'Allaire S., Taylor D.J., editors. Blackwell Publishing; Ames, Iowa: 2006. Diseases of the reproductive system; pp. 113–147. [Google Scholar]
- Amimo J.O., El Zowalaty M.E., Githae D., Wamalwa M., Djikeng A., Nasrallah G.K. Metagenomic analysis demonstrates the diversity of the fecal virome in asymptomatic pigs in East Africa. Arch. Virol. 2016;161:887–897. doi: 10.1007/s00705-016-2819-6. [DOI] [PubMed] [Google Scholar]
- Amimo J.O., Njuguna J., Machuka E., Okoth E., Djikeng A. First complete genome sequences of porcine Bocavirus strains from East Africa. Gen. Announc. 2017;5 doi: 10.1128/genomeA.00093-17. (e00093–17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron J., Menezes J., Tijssen P. Genomic organization and mapping of transcription and translation products of the NADL-2 strain of porcine parvovirus. Virol. 1993;197:86–98. doi: 10.1006/viro.1993.1569. [DOI] [PubMed] [Google Scholar]
- Bergeron J., Hebert B., Tijssen P. Genome organization of the Kresse strain of porcine parvovirus: identification of the allotropic determinant and comparison with those of NADL-2 and field isolates. J. Virol. 1996;70:2508–2515. doi: 10.1128/jvi.70.4.2508-2515.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström A.L., Belák S., Fossum C., McKillen J., Allan G., Wallgren P., Berg M. Detection of a novel porcine boca-like virus in the background of porcine circovirus type 2 induced postweaning multisystemic wasting syndrome. Virus Res. 2009;146:125–129. doi: 10.1016/j.virusres.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Blomström A.L., Ståhl K., Masembe C., Okoth E., Okurut A.R., Atmnedi P., Kemp S., Bishop R., Belák S., Berg M. Viral metagenomic analysis of bushpigs (Potamochoerus larvatus) in Uganda identifies novel variants of porcine parvovirus 4 and torque Teno sus virus 1 and 2. Virol. J. 2012;9:192. doi: 10.1186/1743-422X-9-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomström A.L., Ståhl K., Okurut A.R., Masembe C., Berg M. Genetic characterisation of a porcine bocavirus detected in domestic pigs in Uganda. Virus Genes. 2013;47:370–373. doi: 10.1007/s11262-012-0855-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt D.M., Häni H., Müller E., Waldvogel A.S. Non-suppurative myocarditis in piglets associated with porcine parvovirus infection. J. Comp. Pathol. 1997;117:107–118. doi: 10.1016/s0021-9975(97)80027-8. [DOI] [PubMed] [Google Scholar]
- Brink M. Swedish University of Agricultural Sciences; Uppsala: 2011. Porcine Viruses in Uganda: A Study of TTSuV and PPV4 in Wild and Domestic Pigs. [Dissertation]http://urn.kb.se/resolve?urn=urn:nbn:se:slu:epsilon-s-341 [Google Scholar]
- Brink M., Ståhl K., Masembe C., Okurut A.R., Berg M., Blomström A.L. First time molecular detection and phylogenetic relationships of torque Teno sus virus 1 and 2 in domestic pigs in Uganda: further evidence for a global distribution. Virol. J. 2012;9:39. doi: 10.1186/1743-422X-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadar D., Cságola A., Lőrincz M., Tombácz K., Spînu M., Tuboly T. Distribution and genetic diversity of porcine hokovirus in wild boars. Arch. Virol. 2011;156:2233–2239. doi: 10.1007/s00705-011-1125-6. [DOI] [PubMed] [Google Scholar]
- Cadar D., Dán Á., Tombácz K., Lőrincz M., Kiss T., Becskei Z., Spînu M., Tuboly T., Cságola A. Phylogeny and evolutionary genetics of porcine parvovirus in wild boars. Infect. Genet. Evol. 2012;12:1163–1171. doi: 10.1016/j.meegid.2012.04.020. [DOI] [PubMed] [Google Scholar]
- Cartwright S.F., Huck R.A. Viruses isolated in association with herd infertility absortions and stillbirths in pigs. Vet. Rec. 1967;81:196–197. [Google Scholar]
- Cheng W.X., Li J.S., Huang C.P., Yao D.P., Liu N., Cui S.X., Jin Y., Duan Z.J. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.K., Wu G., Wang D., Bayles D.O., Lager K.M., Vincent A.L. Identification and molecular cloning of a novel porcine parvovirus. Arch. Virol. 2010;155:801–806. doi: 10.1007/s00705-010-0646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C.S., Molitor T.W., Joo H.S., Gunther R. Pathogenicity of a skin isolate of porcine parvovirus in swine fetuses. Vet. Microbiol. 1987;15:19–29. doi: 10.1016/0378-1135(87)90125-8. [DOI] [PubMed] [Google Scholar]
- Choi M.G., Park S.J., Nguyen V.G., Chung H.C., Kim A.R., Park B.K. Molecular detection and genetic analysis of porcine bocavirus in Korean domestic swine herds. Arch. Virol. 2014;159:1487–1492. doi: 10.1007/s00705-013-1944-8. [DOI] [PubMed] [Google Scholar]
- Cotmore S.F., Agbandje-McKenna M., Chiorini J.A., Mukha D.V., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A.J. The family parvoviridae. Arch. Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cságola A., Lőrincz M., Cadar D., Tombácz K., Biksi I., Tuboly T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch. Virol. 2012;157:1003–1010. doi: 10.1007/s00705-012-1257-3. [DOI] [PubMed] [Google Scholar]
- Dea S., Elazhary M.A., Martineau G.P., Vaillancourt J. Parvovirus-like particles associated with diarrhea in unweaned piglets. Can. J. Comp. Med. 1985;49:343. [PMC free article] [PubMed] [Google Scholar]
- Devillers N., Le Dividich J., Prunier A. Influence of colostrum intake on piglet survival and immunity. Anim. 2011;5:1605–1612. doi: 10.1017/S175173111100067X. [DOI] [PubMed] [Google Scholar]
- Drolet R., D'Allaire S., Larochelle R., Magar R., Ribotta M., Higgins R. Infectious agents identified in pigs with multifocal interstitial nephritis at slaughter. Vet. Rec. 2002;150:139–142. doi: 10.1136/vr.150.5.139. [DOI] [PubMed] [Google Scholar]
- Duhamel G.E., Bargar T.W., Schmitt B.J., Molitor T.W., Lu W. Identification of parvovirus-like virus particles in intestinal crypt epithelial cells of pigs with diarrhea. J. Vet. Diagn. Investig. 1991;3:96–98. doi: 10.1177/104063879100300126. [DOI] [PubMed] [Google Scholar]
- Dunne H.W., Gobble J.L., Hokanson J.F., Kradel D.C., Bubash G.R. Porcine reproductive failure associated with a newly identified" SMEDI" group of picorna viruses. Amer. J. Vet. Res. 1965;26:1284. [PubMed] [Google Scholar]
- Eterpi M., McDonnell G., Thomas V. Disinfection efficacy against parvoviruses compared with reference viruses. J. Hospl. Infect. 2009;73:64–70. doi: 10.1016/j.jhin.2009.05.016. [DOI] [PubMed] [Google Scholar]
- FAO . 2012. Pig Sector Kenya, FAO Animal Production and Health Livestock Country Reviews, No. 3, Rome. [Google Scholar]
- Foerster T., Streck A.F., Speck S., Selbitz H.J., Lindner T., Truyen U. An inactivated whole-virus porcine parvovirus vaccine protects pigs against disease but does not prevent virus shedding even after homologous virus challenge. J. Gen. Virol. 2016;97:1408–1413. doi: 10.1099/jgv.0.000446. [DOI] [PubMed] [Google Scholar]
- Gava D., Souza C.K., Mores T.J., Argenti L.E., Streck A.F., Canal C.W., Bortolozzo F.P., Wentz I. Dynamics of vanishing of maternally derived antibodies of ungulate protoparvovirus 1 suggests an optimal age for gilts vaccination. Trop. Anim. Health Prod. 2017;49:1085–1088. doi: 10.1007/s11250-017-1301-0. [DOI] [PubMed] [Google Scholar]
- Gunn L., Collins P.J., Fanning S., McKillen J., Morgan J., Staines A., O'Shea H. Detection and characterisation of novel bocavirus (genus Bocaparvovirus) and gastroenteritis viruses from asymptomatic pigs in Ireland. Infect. Ecol. Epidemiol. 2015;5:27270. doi: 10.3402/iee.v5.27270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Abe K., Win K.M., Shimizu Y.K., Keicho N., Yoshikura H. Identification of new parvovirus DNA sequence in swine sera from Myanmar. Jpn. J. Infect. Dis. 2001;54:244–245. [PubMed] [Google Scholar]
- Huang L., Zhai S.L., Cheung A.K., Zhang H.B., Long J.X., Yuan S.S. Detection of a novel porcine parvovirus, PPV4, in chinese swine herds. Virol. J. 2010;7:333. doi: 10.1186/1743-422X-7-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Mor S.K., Erber J., Voss E., Goyal S.M. Detection and characterization of porcine bocavirus in the United States. Arch. Virol. 2014;159:1797–1801. doi: 10.1007/s00705-013-1972-4. [DOI] [PubMed] [Google Scholar]
- Jóźwik A., Manteufel J., Selbitz H.J., Truyen U. Vaccination against porcine parvovirus protects against disease, but does not prevent infection and virus shedding after challenge infection with a heterologous virus strain. J. Gen. Virol. 2009;90:2437–2441. doi: 10.1099/vir.0.012054-0. [DOI] [PubMed] [Google Scholar]
- Kailasan S., Agbandje-McKenna M., Parrish C.R. Parvovirus family conundrum: what makes a killer? Ann. Rev. Virol. 2015;2:425–450. doi: 10.1146/annurev-virology-100114-055150. [DOI] [PubMed] [Google Scholar]
- Krakowka S., Ellis J.A., Meehan B., Kennedy S., McNeilly F., Allan G. Viral wasting syndrome of swine: experimental reproduction of postweaning multisystemic wasting syndrome in gnotobiotic swine by coinfection with porcine circovirus 2 and porcine parvovirus. Vet. Pathol. 2000;37:254–263. doi: 10.1354/vp.37-3-254. [DOI] [PubMed] [Google Scholar]
- Kresse J.I., Taylor W.D., Stewart W.W., Eernisse K.A. Parvovirus infection in pigs with necrotic and vesicle-like lesions. Vet. Microbiol. 1985;10:525–531. doi: 10.1016/0378-1135(85)90061-6. [DOI] [PubMed] [Google Scholar]
- Lager K.M., Mengeling W.L. Porcine parvovirus associated with cutaneous lesions in piglets. J. Vet. Diagn. Investig. 1994;6:357–359. doi: 10.1177/104063879400600313. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Tse H., Fu C.T., Au W.K., Chen X.C., Tsoi H.W., Tsang T.H., Chan J.S., Tsang D.N., Li K.S. Identification of novel porcine and bovine parvoviruses closely related to human parvovirus 4. J. Gen. Virol. 2008;89:1840–1848. doi: 10.1099/vir.0.2008/000380-0. [DOI] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Yip C.C., Li K.S., Fu C.T., Huang Y., Chan K.H., Yuen K.Y. Co-existence of multiple strains of two novel porcine bocaviruses in the same pig, a previously undescribed phenomenon in members of the family Parvoviridae, and evidence for inter-and intra-host genetic diversity and recombination. J. Gen. Virol. 2011;92:2047–2059. doi: 10.1099/vir.0.033688-0. [DOI] [PubMed] [Google Scholar]
- Lawson J.R. Reports of the Meeting of the Expert Panel of Livestock Infertility, Rome, Italy. 1961. Infectious infertility of swine; p. 67. [Google Scholar]
- Lenghaus C., Forman A.J., Hale C.J. Experimental infection of 35, 50 and 60 day old pig foetuses with porcine parvovirus. Austr. Vet. J. 1978;54:418–422. doi: 10.1111/j.1751-0813.1978.tb05565.x. [DOI] [PubMed] [Google Scholar]
- Li B., Ma J., Xiao S., Fang L., Zeng S., Wen L., Zhang X., Ni Y., Guo R., Yu Z., Zhou J. Complete genome sequence of a novel species of porcine bocavirus, PBoV5. J. Virol. 2012;86:1286–1287. doi: 10.1128/JVI.06589-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wei Y., Liu J., Tang Q., Liu C. Prevalence of porcine hokovirus and its co- infection with porcine circovirus 2 in China. Arch. Virol. 2013;158:1987–1991. doi: 10.1007/s00705-013-1690-y. [DOI] [PubMed] [Google Scholar]
- Mather C., Marshall A. Biosecurity's unruly spaces. Geogr. J. 2011;177:300–310. doi: 10.1111/j.1475-4959.2010.00392.x. [DOI] [PubMed] [Google Scholar]
- Mayr A., Mahnel H. Cultivation of hog cholera virus in pig kidney cultures with cytopathogenic effect. Zentralbl. Bakteriol. (Orig A) 1964;195:157. [PubMed] [Google Scholar]
- Mayr A., Bachmann P.A., Siegl G., Mahnel H., Sheffy B.E. Characterization of a small porcine DNA virus. Arch. Virol. 1968;25:38–51. doi: 10.1007/BF01243088. [DOI] [PubMed] [Google Scholar]
- McKillen J., McNeilly F., Duffy C., McMenamy M., McNair I., Hjertner B., Millar A., McKay K., Lagan P., Adair B., Allan G. Isolation in cell cultures and initial characterisation of two novel bocavirus species from swine in Northern Ireland. Vet. Microbiol. 2011;152:39–45. doi: 10.1016/j.vetmic.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Mengeling W.L., Paul P.S., Brown T.T. Transplacental infection and embryonic death following maternal exposure to porcine parvovirus near the time of conception. Arch. Virol. 1980;65:55–62. doi: 10.1007/BF01340540. [DOI] [PubMed] [Google Scholar]
- Mengeling W.L., Pejsak Z., Paul P.S. Biological assay of attenuated strain NADL-2 and virulent strain NADL-8 of porcine parvovirus. Amer. J. Vet. Res. 1984;45:2403–2407. [PubMed] [Google Scholar]
- Mengeling W.L., Lager K.M., Zimmerman J.K., Samarikermani N., Beran G.W. A current assessment of the role of porcine parvovirus as a cause of fetal porcine death. J. Vet. Diagn. Investig. 1991;3:33–35. doi: 10.1177/104063879100300107. [DOI] [PubMed] [Google Scholar]
- Mengeling W.L., Lager K.M., Vorwald A.C. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim. Reprod. Sci. 2000;60:199–210. doi: 10.1016/s0378-4320(00)00135-4. [DOI] [PubMed] [Google Scholar]
- Mokoele J.M., van Rensburg L.J., Van Lochem S., Bodenstein H., Du Plessis J., Carrington C.A., Spencer B.T., Fasina F.O. Overview of the perceived risk of transboundary pig diseases in South Africa. J. South Afric. Vet. Assoc. 2015;86:1–9. doi: 10.4102/jsava.v86i1.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor T.W., Joo H.S., Collett M.S. Porcine parvovirus DNA: characterization of the genomic and replicative form DNA of two virus isolates. Virol. 1984;137:241–254. doi: 10.1016/0042-6822(84)90216-2. [DOI] [PubMed] [Google Scholar]
- Ndze V.N., Cadar D., Cságola A., Kisfali P., Kovács E., Farkas S., Ngu A.F., Esona M.D., Dán Á., Tuboly T., Bányai K. Detection of novel porcine bocaviruses in fecal samples of asymptomatic pigs in Cameroon. Infect. Genet. Evol. 2013;17:277–282. doi: 10.1016/j.meegid.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Ni J., Qiao C., Han X., Han T., Kang W., Zi Z., Cao Z., Zhai X., Cai X. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol. J. 2014;11:203. doi: 10.1186/s12985-014-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opriessnig T., Gerber P.F., Matzinger S.R., Meng X.J., Halbur P.G. Markedly different immune responses and virus kinetics in littermates infected with porcine circovirus type 2 or porcine parvovirus type 1. Vet. Immunol. Immunopathol. 2017;191:51–59. doi: 10.1016/j.vetimm.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Palinski R.M., Mitra N., Hause B.M. Discovery of a novel Parvovirinae virus, porcine parvovirus 7, by metagenomic sequencing of porcine rectal swabs. Virus Genes. 2016;52:564–567. doi: 10.1007/s11262-016-1322-1. [DOI] [PubMed] [Google Scholar]
- Pan Y., Zeng Q., Zhu C., Hua X., Wang M., Pan K., Cui L. Frequency and characterization of porcine hokovirus (PHoV) in domestic pigs in eastern China. Arch. Virol. 2012;157:1785–1788. doi: 10.1007/s00705-012-1350-7. [DOI] [PubMed] [Google Scholar]
- Parke C.R., Burgess G.W. An economic assessment of porcine parvovirus vaccination. Austr. Vet. J. 1993;70:177–180. doi: 10.1111/j.1751-0813.1993.tb06124.x. [DOI] [PubMed] [Google Scholar]
- Paul P.S., Mengeling W.L. Oronasal and intramuscular vaccination of swine with a modified live porcine parvovirus vaccine: multiplication and transmission of the vaccine virus. Amer. J. Vet. Res. 1984;45:2481–2485. [PubMed] [Google Scholar]
- Paul P.S., Mengeling W.L., Brown J.T. Effect of vaccinal and passive immunity on experimental infection of pigs with porcine parvovirus. Amer. J. Vet. Res. 1980;41:1368–1371. [PubMed] [Google Scholar]
- Pini A. Porcine parvovirus in pig herds in southern Africa. J. South Afri. Vet. Assoc. 1975;46:241–244. [PubMed] [Google Scholar]
- Prozesky L., Thomson G.R., Gainaru M.D., Herr S., Kritzinger L.J. Lesions resulting from inoculation of porcine foetuses with porcine parvovirus. Onderstepoort. J. Vet. Res. 1980;47:269–274. [PubMed] [Google Scholar]
- Pye D., Bates J., Edwards S.J., Hollingworth J. Development of a vaccine preventing parvovirus-induced reproductive failure in pigs. Austr. Vet. J. 1990;67:179–182. doi: 10.1111/j.1751-0813.1990.tb07750.x. [DOI] [PubMed] [Google Scholar]
- Ren X., Tao Y., Cui J., Suo S., Cong Y., Tijssen P. Phylogeny and evolution of porcine parvovirus. Virus Res. 2013;178:392–397. doi: 10.1016/j.virusres.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Saekhow P., Ikeda H. Prevalence and genomic characterization of porcine parvoviruses detected in Chiangmai area of Thailand in 2011. Microbiol. Immunol. 2015;59:82–88. doi: 10.1111/1348-0421.12218. [DOI] [PubMed] [Google Scholar]
- Saekhow P., Mawatari T., Ikeda H. Coexistence of multiple strains of porcine parvovirus 2 in pig farms. Microbiol. Immunol. 2014;58:382–387. doi: 10.1111/1348-0421.12159. [DOI] [PubMed] [Google Scholar]
- Salmon H., Berri M., Gerdts V., Meurens F. Humoral and cellular factors of maternal immunity in swine. Dev. Comp. Immunol. 2009;33:384–393. doi: 10.1016/j.dci.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Schirtzinger E.E., Suddith A.W., Hause B.M., Hesse R.A. First identification of porcine parvovirus 6 in North America by viral metagenomic sequencing of serum from pigs infected with porcine reproductive and respiratory syndrome virus. Virol. J. 2015;12:170. doi: 10.1186/s12985-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés J., Allan G.M., Domingo M. Porcine circovirus diseases. Anim. Health Res. Rev. 2005;6:119–142. doi: 10.1079/ahr2005106. [DOI] [PubMed] [Google Scholar]
- Simpson A.A., Hébert B., Sullivan G.M., Parrish C.R., Zádori Z., Tijssen P., Rossmann M.G. The structure of porcine parvovirus: comparison with related viruses. J. Mol. Biol. 2002;315:1189–1198. doi: 10.1006/jmbi.2001.5319. [DOI] [PubMed] [Google Scholar]
- Streck A.F., Homeier T., Foerster T., Fischer S., Truyen U. Analysis of porcine parvoviruses in tonsils and hearts from healthy pigs reveals high prevalence and genetic diversity in Germany. Arch. Virol. 2013;158:1173–1180. doi: 10.1007/s00705-013-1603-0. [DOI] [PubMed] [Google Scholar]
- Streck A.F., Canal C.W., Truyen U. Molecular epidemiology and evolution of porcine parvoviruses. Infect. Genet. Evol. 2015;36:300–306. doi: 10.1016/j.meegid.2015.10.007. [DOI] [PubMed] [Google Scholar]
- Szelei J., Zadori Z., Tijssen P. In: Parvoviruses. Kerr J.R., Cotmore S.F., Bloom M.E., Linden R.M., Parrish C.R., editors. Hodder Arnold; London, UK: 2006. Porcine parvovirus; pp. 435–445. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson G.R., Prozesky L. In: Infectious Diseases of Livestock. Coetzer J.A.W., Thomson G.R., Tustin R.C., editors. Oxford University Press; South Africa: 1994. Porcine parvovirus infection; pp. 888–894. [Google Scholar]
- Tijssen P., Bergeron J., Dubuc R., Hébert B. Seminars in Virology. vol. 6. Academic Press; 1995. Minor genetic changes among porcine parvovirus groups are responsible for major distinguishing biological properties; pp. 319–328. [Google Scholar]
- Tijssen P., Agbandje-McKenna M., Almendral J.M., Bergoin M., Flegel T.W., Hedman K., Kleinschmidt J., Li Y., Pintel D.J., Tattersall P. In: Virus Taxonomy. 9th Report of the International Committee on Taxonomy of Viruses. King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Academic Press; London: 2011. Family Parvoviridae; pp. 405–425. [Google Scholar]
- Truyen U., Streck A.F. In: Diseases of Swine. Zimmerman J.K., Karriker L., Ramirez A., Schwartz K., Stevenson G., editors. John Wiley & Sons; Chichester, United Kingdom: 2012. Porcine parvovirus; pp. 447–455. [Google Scholar]
- Tse H., Tsoi H.W., Teng J.L., Chen X.C., Liu H., Zhou B., Zheng B.J., Woo P.C., Lau S.K., Yuen K.Y. Discovery and genomic characterization of a novel ovine partetravirus and a new genotype of bovine partetravirus. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tummaruk P., Tantilertcharoen R. Seroprevalence of porcine reproductive and respiratory syndrome, Aujeszky's disease, and porcine parvovirus in replacement gilts in Thailand. Trop. Anim. Health Prod. 2012;44:983–989. doi: 10.1007/s11250-011-9999-6. [DOI] [PubMed] [Google Scholar]
- Wang F., Wei Y., Zhu C., Huang X., Xu Y., Yu L., Yu X. Novel parvovirus sublineage in the family of Parvoviridae. Virus Genes. 2010;41:305–308. doi: 10.1007/s11262-010-0506-3. [DOI] [PubMed] [Google Scholar]
- Whitaker H.K., Neu S.M., Pace L.W. Parvovirus infection in pigs with exudative skin disease. J. Vet. Diagn. Investig. 1990;2:244–246. doi: 10.1177/104063879000200322. [DOI] [PubMed] [Google Scholar]
- Wrathall A.E., Wells D.E., Cartwright S.F., Frerichs G.N. An inactivated, oil-emulsion vaccine for the prevention of porcine parvovirus-induced reproductive failure. Res. Vet. Sci. 1984;36:136–143. [PubMed] [Google Scholar]
- Xiao C.T., Giménez-Lirola L.G., Halbur P.G., Opriessnig T. Increasing porcine PARV4 prevalence with pig age in the US pig population. Vet. Microbiol. 2012;160:290–296. doi: 10.1016/j.vetmic.2012.05.038. [DOI] [PubMed] [Google Scholar]
- Xiao C.T., Halbur P.G., Opriessnig T. Complete genome sequence of a novel porcine parvovirus (PPV) provisionally designated PPV5. Gen. Announc. 2013;1 doi: 10.1128/genomeA.00021-12. (e00021–12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C.T., Gimenez-Lirola L.G., Jiang Y.H., Halbur P.G., Opriessnig T. Characterization of a novel porcine parvovirus tentatively designated PPV5. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0065312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C.T., Halbur P.G., Opriessnig T. Molecular evolutionary genetic analysis of emerging parvoviruses identified in pigs. Infect. Genet. Evol. 2013;16:369–376. doi: 10.1016/j.meegid.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Xiao C.T., Gerber P.F., Giménez-Lirola L.G., Halbur P.G., Opriessnig T. Characterization of porcine parvovirus type 2 (PPV2) which is highly prevalent in the USA. Vet. Microbiol. 2013;161:325–330. doi: 10.1016/j.vetmic.2012.07.038. [DOI] [PubMed] [Google Scholar]
- Xing X., Zhou H., Tong L., Chen Y., Sun Y., Wang H., Zhang G. First identification of porcine parvovirus 7 in China. Arch. Virol. 2018;163:209–213. doi: 10.1007/s00705-017-3585-9. [DOI] [PubMed] [Google Scholar]
- Yang W.Z., Yu J.M., Li J.S., Cheng W.X., Huang C.P., Duan Z.J. Genome characterization of a novel porcine bocavirus. Arch. Virol. 2012;157:2125–2132. doi: 10.1007/s00705-012-1407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeuw E.J.L., Leinecker N., Herwig V., Selbitz H.J., Truyen U. Study of the virulence and cross-neutralization capability of recent porcine parvovirus field isolates and vaccine viruses in experimentally infected pregnant gilts. J. Gen. Virol. 2007;88:420–427. doi: 10.1099/vir.0.82302-0. [DOI] [PubMed] [Google Scholar]
- Zhai S.L., Yue C., Wei Z.Z., Ran D.L., Long J.X., Lin T., Deng Y., Sun L.C., Huang L., Yuan S.S. Development and application of a PCR assay for detection of porcine bocavirus. Chin. J. Anim. Inf. Dis. 2010;2:14–17. [Google Scholar]
- Zhang C.F., Song C.P., Chen C.M., Cui S.J., Miao L.F. Reproductive failure in wild boars associated to porcine parvovirus infection and in vivo and in vitro characterization of the causal isolate. Trop. Anim. Health Prod. 2010;42:1611–1613. doi: 10.1007/s11250-010-9644-9. [DOI] [PubMed] [Google Scholar]
- Zhou F., Sun H., Wang Y. Porcine bocavirus: achievements in the past five years. Viruses. 2014;6:4946–4960. doi: 10.3390/v6124946. [DOI] [PMC free article] [PubMed] [Google Scholar]