Highlights
-
•
Genome organisation of QX-like IBV includes accessory protein genes 4b, 4c and 6b.
-
•
Relative variability in open reading frames across the genome is described.
-
•
Provides insights into the origins of the S1 subunit HVRs.
-
•
Describes novel features of the S2 subunit.
-
•
First model of the IBV spike monomer.
Keywords: Infectious bronchitis, Coronavirus, SNP, Spike, HVR
Abstract
Infectious bronchitis virus (IBV) is a Gammacoronavirus that causes a highly contagious respiratory disease in chickens. A QX-like strain was analysed by high-throughput Illumina sequencing and genetic variation across the entire viral genome was explored at the sub-consensus level by single nucleotide polymorphism (SNP) analysis. Thirteen open reading frames (ORFs) in the order 5′-UTR-1a-1ab-S-3a-3b-E-M-4b-4c-5a-5b-N-6b-3′UTR were predicted. The relative frequencies of missense: silent SNPs were calculated to obtain a comparative measure of variability in specific genes. The most variable ORFs in descending order were E, 3b, 5′UTR, N, 1a, S, 1ab, M, 4c, 5a, 6b. The E and 3b protein products play key roles in coronavirus virulence, and RNA folding demonstrated that the mutations in the 5′UTR did not alter the predicted secondary structure. The frequency of SNPs in the Spike (S) protein ORF of 0.67% was below the genomic average of 0.76%. Only three SNPS were identified in the S1 subunit, none of which were located in hypervariable region (HVR) 1 or HVR2. The S2 subunit was considerably more variable containing 87% of the polymorphisms detected across the entire S protein. The S2 subunit also contained a previously unreported multi-A insertion site and a stretch of four consecutive mutated amino acids, which mapped to the stalk region of the spike protein. Template-based protein structure modelling produced the first theoretical model of the IBV spike monomer. Given the lack of diversity observed at the sub-consensus level, the tenet that the HVRs in the S1 subunit are very tolerant of amino acid changes produced by genetic drift is questioned.
1. Introduction
Coronaviruses (family Coronaviridae, order Nidovirales) are enveloped, single-stranded RNA viruses with large genome sizes of ∼25–30 kb. The family is split into four genera: Alpha-, Beta, Gamma and Deltacoronaviruses, each containing pathogens of veterinary or human importance. A current evolutionary model postulates that bats are the ancestral source of Alpha- and Betacoronaviruses and birds the source of Gamma- and Deltacoronaviruses (Woo et al., 2012). The Alphacoronaviruses infect swine, cats, dogs and humans. Betacoronaviruses infect diverse mammalian species including bats, humans, rodents and ungulates. The SARS coronavirus (SARS-CoV), which verged on a pandemic in 2003 with 8273 cases in humans and 755 deaths is a Betacoronavirus. Another member of this genus, the recently-discovered Middle East Respiratory Syndrome (MERS) coronavirus (MERS-CoV) has claimed 88 human lives from 212 cases since April 2012, and dromedary camels are the suspected reservoir (Briese et al., 2014). Genus Gammacoronavirinae includes strains infecting birds and whales (Woo et al., 2012, McBride et al., 2014, Borucki et al., 2013) and deltacoronaviruses have been described in birds, swine and cats (Woo et al., 2012). The diversity of hosts and genomic features amongst CoVs have been attributed to their unique mechanism of viral recombination, a high frequency of recombination, and an inherently high mutation rate (Lai and Cavanagh, 1997).
Infectious bronchitis virus (IBV) is a gammacoronavirus which causes a highly contagious respiratory disease of economic importance in chickens (Cook et al., 2012). IBV primarily replicates in the respiratory tract but also, depending on the strain, in epithelial cells of the gut, kidney and oviduct. Clinical signs of respiratory distress, interstitial nephritis and reduced egg production are common, and the disease has a global distribution (Cavanagh, 2007, Cook et al., 2012). The IBV genome encodes at least ten open reading frames (ORFs) organised as follows: 5′ UTR-1a-1ab-S-3a-3b-E-M-5a-5b-N-3a-3′UTR. Six mRNAs (mRNA 1–6) are associated with production of progeny virus. Four structural proteins including the spike glycoprotein (S), small membrane protein (E), membrane glycoprotein (M), and nucleocapsid protein (N) are encoded by mRNAs 2, 3, 4 and 6, respectively (Casais et al., 2005, Hodgson et al., 2006). Messenger RNA (mRNA) 1 consists of ORF1a and ORF1b, encoding two large polyproteins via a ribosomal frameshift mechanism (Inglis et al., 1990). During or after synthesis, these polyproteins are cleaved into 15 non-structural proteins (nsp2–16) which are associated with RNA replication and transcription. The S glycoprotein is post-translationally cleaved at a protease cleavage recognition motif into the amino-terminal S1 subunit (92 kDa) and the carboxyl-terminal S2 subunit (84 kDa) by the host serine protease furin (de Haan et al., 2004). The multimeric S glycoprotein extends from the viral membrane, and the globular S1 subunit is anchored to the viral membrane by the S2 subunit via non-covalent bonds. Proteins 3a and 3b, and 5a and 5b are encoded by mRNA 3 and mRNA 5, respectively and are not essential to viral replication (Casais et al., 2005, Hodgson et al., 2006).
A confounding feature of IBV infection is the lack of correlation between antibodies and protection, and discrepancies between in vitro strain differentiation by virus neutralization (VN) tests and in vivo cross-protection results. Taken with the ability for high viral shedding in the presence of high titres of circulating antibodies, the involvement of other immune mechanisms are evident, and the roles of cell-mediated immunity and interferon have been experimentally demonstrated (Timms et al., 1980, Collisson et al., 2000, Pei et al., 2001, Cook et al., 2012).
Dozens of IBV serotypes that are poorly cross-protective have been discovered and studied by VN tests and molecular characterisation of the S protein gene. Most of these serotypes differ from each other by 20–25% at amino acid level in S1, but may differ by up to 50%. S1 contains the epitopes involved in the induction of neutralizing, serotype-specific and hemagglutinaton inhibiting antibodies (Cavanagh, 2007, Darbyshire et al., 1979, Farsang et al., 2002, Ignjatovic and McWaters, 1991, Meulemans et al., 2001, Gelb et al., 1997). Most of the strain differences in S1 occur in three hypervariable regions (HVRs) located between the amino acid residues 56–69 (HVR1), 117–131 (HVR2) and 274–387 (HVR3) (Moore et al., 1997, Wang and Huang, 2000). Monoclonal antibody analysis mapped the locations of many of the amino acids involved in the formation of VN epitopes to within the first and third quarters of the linear S1 polypeptide (De Wit, 2000, Kant et al., 1992, Koch et al., 1990), which is where closely-related stains (>95% amino acid identity) also differ (Bijlenga et al., 2004, Farsang et al., 2002). Cavanagh (2007) proposed that these parts of the S1 subunit are very tolerant of amino acid changes, conferring a selective advantage. Recently, the receptor-binding domain of the IBV M41 strain was mapped to residues 16–69 of the N terminus of S1, which overlaps with HVR1 (Promkuntod et al., 2014).
The S2 subunit, which drives virus-cell fusion, is more conserved between serotypes than S1, varying by only 10–15% at the amino acid level (Bosch et al., 2005, Cavanagh, 2005). Although it was initially thought that S2 played little or no role in the induction of a host immune response, it has since been shown that an immunodominant region located in the N-terminal half of the S2 subunit can induce neutralizing, but not serotype-specific, antibodies demonstrated by the ability of this subunit to confer broad protection against challenge with an unrelated serotype (Kusters et al., 1989, Toro et al., 2014).
IBVs are continuously evolving as a result of (a) frequent point mutations and (b) genomic recombination events (Cavanagh et al., 1992, Kottier et al., 1995, Jackwood et al., 2005, Zhao et al., 2013, Kuo et al., 2013, Liu et al., 2014). Multiple studies on IBV diversity have focused on inter-serotypic and inter-strain variation, and a few have focused on sub-populations within the S1 subunit in vaccine strains (Gallardo et al., 2012, Ndegwa et al., 2014). The present study aimed to explore genetic variation across the entire viral genome at the sub-consensus level. It was anticipated, based on the published literature, that certain regions, and the S1 subunit HVRs in particular, would display significant sub-genomic variation. This study focused on a QX-like strain, a serotype currently causing significant poultry health problems across Europe, Asia, South America and South Africa.
2. Materials and methods
2.1. Origin and isolation of QX-like strain ck/ZA/3665/11
Twenty-eight-day old chickens in a commercial broiler operation presented with acute lethargy, reduced feed consumption and mortality. Tracheitis and swollen kidneys were noted on post mortem, as well as a secondary Escherichia coli infection. The worst affected houses had mortality rates of 19.8%, 11.9% and 10.2%. IBV was isolated in specific pathogen free (SPF) embryonated chicken eggs (ECE) as described in Knoetze et al. (2014). After an initial two passages in ECE, the virus was passaged twice further at the University of Pretoria.
2.2. Preparation of the genome and Illumina sequencing
RNA was extracted from allantoic fluid using TRIzol® reagent (Ambion, Life Technologies, Carlsbad, USA) according to the manufacturer’s protocol. The genome was transcribed to cDNA and amplified using a TransPlex® Whole Transcriptome Amplification kit (Sigma–Aldrich, Steinheim, Germany). Illumina MiSeq sequencing on the cDNA library was performed at the ARC-Biotechnology Platform, Onderstepoort, Pretoria.
2.3. Genome assembly, RNA folding and recombination analysis
Illumina results were analysed using the CLC Genomics Workbench v 5.1.5. Paired-end reads were trimmed and a preliminary de novo assembly was performed. The larger segments were analysed by BLAST to identify the closest genomic reference strain (ITA/90254/2005, CAZ86699). This strain was retrieved and used as a scaffold for assembly-to-reference, generating a consensus sequence for 3665/11. Trimmed paired-end reads were also mapped against other IBV serotype genomes, subsequently confirming that strain 3665/11 was a pure culture of a QX-like IBV. The genome was deposited in Genbank under the accession number KP662631. RNA folding was predicted using the CLC Genomics Workbench v 5.1.5. Genetic recombination in the consensus sequence was evaluated using the recombination detection program RDP v4.31.
2.4. Genome annotation and single nucleotide polymorphism (SNP) analysis
Coding sequence and ORF prediction was carried out in VIGOR (Wang et al., 2010). Trimmed paired-end reads were re-mapped against the 3665/11 consensus sequence for SNP detection. A SNP detection table generated in the CLC Genomics Workbench was manually edited to eliminate all SNPs with a frequency of <5%. This conservative cutoff was selected to eliminate any non-specific PCR errors introduced during preparation of the transcriptome library or deep sequencing, and excluded most of the point insertions producing gaps and frameshift mutations across the genome. Nucleotide substitutions in coding regions were manually inspected for changes to the consensus amino acid (Table 1, Supplementary data). Motifs were predicted using the ELM Eukaryotic Linear Motif Resource for Functional Sites in Proteins (Dinkel et al., 2014).
2.5. Protein secondary structure prediction
Protein structures for S1 and S2 were predicted in RaptorX, a structure prediction server that predicts three dimensional (3D) structures for protein sequences without close homologs in the Protein Data Bank (PDB) (Kallberg et al., 2012). S1 and S2 3D structures were annotated and superposed in CCP4MG v2.9.0 using the Secondary Structure (SSM) superposition method. This method superimposes pairs of structures by: (1) finding the secondary structure elements (SSEs) and representing them as one simple vector spanning the length of the SSE; (2) finding equivalent SSEs in the two structures using graph-theory matching by geometric criteria of distances and angles between the vectors; (3) superimposing vectors representing equivalent SSEs; (4) finding the most likely equivalent residues in the superposed SSEs; (5) superimposing CA atoms of equivalent residues; and (6) iterating the last two steps.
3. Results and discussion
3.1. Genome assembly and annotation
The genome sequence of QX-like strain ck/ZA/3665/11 was assembled from 74, 578 IBV-specific paired-end reads of 144 bp each. The genome was 27, 388 nt in length with the 5′ UTR incomplete by ∼139 nt. Thirteen ORFS were predicted by VIGOR in the order 5′-UTR-1a-1ab-S-3a-3b-E-M-4b-4c-5a-5b-N-6b-3′UTR (Fig. 1 ). This genome organisation including 4b, 4c and 6b was similar to that of turkey coronavirus (TCoV; Cao et al., 2008), and the ORFs 4b, 4c and 6b were also predicted in Australian IBV strains (Hewson et al., 2011). When the sequences for a QX-like sequence (JQ088078) and ArkDPI (EU418976) were analysed using VIGOR, a similar genome arrangement was detected. Mass41 (AY851295) did not however contain the predicted 4b, 4c and 6b ORFs (data not shown).
Fig. 1.
Genome organisation of QX-like IBV strain 3665/11.
ORF 4b was 94 amino acids (aa) in length and no SMART domains were predicted, whereas ORF 4c was 56 aa in length and a low complexity region was identified. The 6b ORF encoded a 74 aa protein with a signal peptide predicted from residues 1 to 24 and two transmembrane domains from residues 2 to 25 and 35 to 57. No recombination was detected across the genome of QX-like strain ck/ZA/3665/11.
A gap was present between nucleotides 1666 and 1667 (Table 1, Supplementary data) (∼aa 370 in the 1a ORF). Although the gap was present in the majority (74.6%) of reads, the sequence for strain 3665/11 deposited in Genbank contains the minority adenine residue because the gap introduced a frame shift, splitting ORF 1a into two. It may be a legitimate mutation, but until further transcriptional analyses are conducted, the ORF 1a gene has been reported intact here.
3.2. Comparative frequency of mutations in ORFs
Two hundred and eight SNPs across the IBV QX-like genome were evaluated at the selected cut-off value. In Table 1 the consensus reference is juxtaposed with the allele variations, the relative frequencies of these point mutations, the actual number of counts and coverage at that position, the corresponding ORF or region and the mutational effect. Coverage ranged from 4-fold (position 11,540) up to 4587 fold (position 26,637).
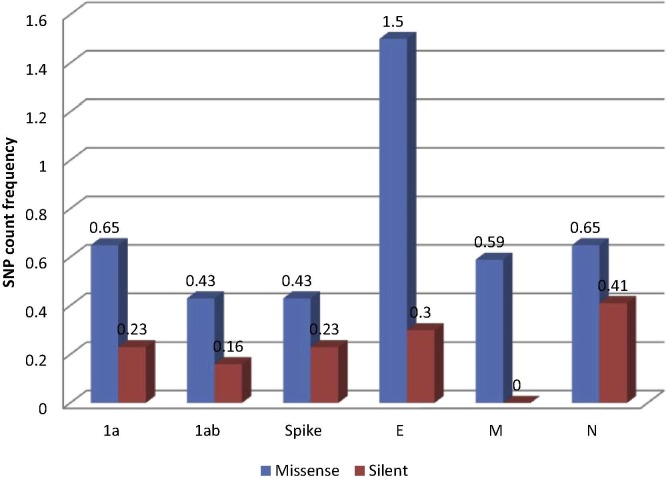
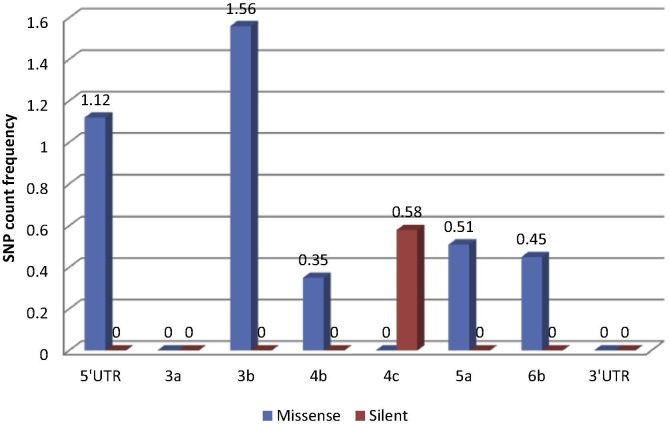
The relative frequencies of missense: silent SNPs in relation to ORF length were calculated in order to obtain a comparative measure of variability in specific genes (Table 2 ). Results for the structural genes and polymerase are illustrated in Fig. 2 , and the results for the non-structural protein ORFs and non-coding regions, which were much shorter in length, are presented in Fig. 3 . Overall the most variable ORFs in terms of total SNPs, in descending order, were: E, 3b, 5′ UTR, N, 1a, S, 1ab, M, 4c, 5a, 6b (no SNPs were detected at the 5% cut-off in the 3a and 3′ UTR regions). The most variable, as assessed by SNPs leading to missense mutations, in descending order, were: 3b, E, 5′ UTR, 1a, N, M, 5a, 1ab, S, 4b, 3a/4c/6b. These mutations presumably did not affect the tertiary protein structure and might be advantageous to the virus. The ORFs under the strongest positive selection pressure as indicated by the proportion of synonymous mutations, were, in descending order, 4c, 1ab, N, S, E, 3a/3b/M/4b/5a/6b.
Table 2.
Relative frequencies of SNPs in QX-like IBV strain 3665/11.
| Region/ORF | Length in nt (% of genome) | SNP count frequency |
||
|---|---|---|---|---|
| Total SNPs (as a% of ORF) | # mutations (as a% of ORF) | # of silent mutations (as a% of ORF) (as a% of total SNPs) | ||
| 5′UTR | 357 (1.30) | 4 (1.12) | 4 (1.12) | – |
| 1ab | 19,872 (72.55) | 152 1.47) | 112 (1.08) | 40 (0.39) [52.96] |
| S | 3438 (12.55) | 23 (0.67) | 15 (0.43) | 8 (0.23) [34.78] |
| 3a | 174 (0.64) | 0 | 0 | 0 |
| 3b | 192 (0.70) | 3 (1.56) | 3 (1.56) | 0 |
| E | 333 (1.22) | 6 (1.80) | 5 (1.50) | 1 (0.30) [16.67] |
| M | 681 (2.49) | 4 (0.59) | 4 (0.59) | 0 |
| 4b | 285 (1.04) | 1 (0.35) | 1 (0.35) | 0 |
| 4c | 171 (0.62) | 1 (0.58) | 0 | 1 (0.58) [100] |
| 5a | 198 (0.72) | 1 (0.51) | 1 (0.51) | 0 |
| N | 1230 (4.49) | 13 (1.06) | 8 (0.65) | 5 (0.41) [38.46] |
| 6b | 222 (0.81) | 1 (0.45) | 1 (0.45) | 0 |
| 3′UTR | 272 (0.99) | 0 | 0 | 0 |
| Total genome | 27,388 | 208 (0.76) | Total SNPs evaluated | |
Fig. 2.
Relative frequencies of mutations in structural and polymerase ORFs of QX-like IBV strain 3665/11.
Fig. 3.
Relative frequencies of mutations in non-structural ORFs and non-coding regions of QX-like IBV strain 3665/11.
3.2.1. E protein
The E protein ORF had significantly more missense mutations on average, at a frequency of 1.5% of the ORF, which is more than threefold higher than the average value (0.55) for the 1a, 1ab, S, M and N genes. The E protein gene was the most variable at the sub-consensus level, with 5 missense mutations and only one silent mutation across its 333 bp ORF. Despite its small size, the CoV E protein drastically influences the replication of CoVs and their pathogenicity. In the SARS-CoV, it was experimentally demonstrated that the E protein is not essential for genome replication or subgenomic mRNA synthesis, but it does affect morphogenesis, budding, assembly, intracellular trafficking and virulence. In fact, in SARS-CoV the E protein is the main antagonist associated with induction of inflammation in the lung, which causes the acute respiratory distress syndrome from which the virus derives its name (DeDiego et al., 2014). No studies have been published for the IBV E protein, but the high variability demonstrated here suggests that it may be an important virulence factor in poultry, and that a higher mutation rate possibly provides an evolutionary advantage in overcoming host cellular immune responses.
3.2.2. N protein
Although the N protein gene contained one of the highest overall frequencies of SNPs (1.06%), the N gene is evidently under greater selective pressure, since 38.9% of these mutations (0.41% as a total of the gene) were silent. The coronavirus N protein is multifunctional, playing vital roles in viral assembly and formation of the complete virion and is required for optimal viral replication. Additionally, the CoV N protein is implicated in cell cycle regulation and host translational shutoff, displays chaperone activity, activates host signal transduction and aids viral pathogenesis through the antagonism of interferon induction (reviewed by McBride et al., 2014). Given its fundamental roles in RNA binding, formation of the ribonucleoprotein complex and in the virion, it is not surprising that this structural protein is the most conserved, as evidenced by its gene having the highest ratio of silent mutations of all the genes analysed. The importance of maintaining the sequence integrity in the N protein in IBV was demonstrated by Kuo et al. (2013), who reported that two residues within the N-terminal domain of a Taiwanese IBV strain were positively selected, and that mutation of either of these significantly reduced the affinity of the N protein for the viral transcriptional regulatory sequence.
3.2.3. M protein
The glycosylated amino terminus of the M protein lies on the outside of the virion and M spans the membrane structure three times (Collisson et al., 2000). All four SNPS in the M gene resulted in missense mutations, two of which were located in the predicted transmembrane region. The M protein plays an important role in CoV virion formation. IBV M protein co-expressed with S assembled into virus-like particles (Liu et al., 2013) confirming its major role in virion formation, but CoV M proteins also interact with other proteins and perform other roles in the infected cell. For example, M together with the accessory proteins 4a, 4b and 5 were all found to prevent the synthesis of IFN-β through the inhibition of interferon promotor activation and IRF-3 function, thus influencing disease outcome (Yang et al., 2013).
3.2.4. Accessory proteins
Coronavirus accessory proteins are generally dispensable for virus replication, but they play vital roles in virulence and pathogenesis by affecting host innate immune responses, encoding pro- or anti-apoptotic activities, or by effecting other signalling pathways that influence disease outcomes (Susan & Julian, 2011). IBV was demonstrated to induce a considerable activation of the type I IFN response, but it was delayed with respect to the peak of viral replication and accumulation of viral dsRNA (Kint et al., 2014). IBV accessory proteins 3a and 3b play a role in the modulation of this delayed IFN response, by regulating interferon production at both the transcriptional and translational levels. Interestingly, IBV proteins 3a and 3b seem to have opposing effects on IFN production in infected cells: 3a seems to promote IFN production, and 3b is involved in limiting IFN production, antagonising each other to tightly regulate IFN production (Kint et al., 2014). Field isolates lacking 3a and 3b displayed reduced virulence in vitro and in vivo (Mardani et al., 2008). ORF 3a in strain 3665/11 lacked SNPs, but ORF 3b in had the highest frequency of SNPs relative to its size (n = 3; 1.56%).
ORF 4b is present in many international IBV strains (Hewson et al., 2011, Bentley et al., 2013) but is rarely mentioned in the literature since a canonical transcription regulatory sequence (TRS-B) could not be identified upstream of the encoding RNA. However, Bentley et al. (2013) demonstrated that IBV was capable of producing subgenomic mRNAs from noncanonical TRS-Bs via a template-switching mechanism with TRS-L, the conserved TRS in the leader sequence in the 5′ UTR, which may expand the Gammacoronavirus repertoire of proteins. They specifically demonstrated the transcription of the 4b ORF by this mechanism. Although no studies have been performed determining ORF 4b′s functional role in the pathogenesis of IBV, the homolog in MERs-CoV is a potent interferon antagonist (Yang et al., 2013). A single SNP causing a missense mutation was present in 11.3% of the sub-consensus population of the 4b ORF in this study. The single mutation in ORF 4c was silent, and the predicted protein contained a low complexity region. Low complexity regions are regions of protein sequences with biased amino acid composition, and may be involved in flexible binding associated with specific functions (Coletta et al., 2010).
ORF 6b, a 73 aa protein with a signal peptide and two transmembrane domains, was identified in the genome of strain 3665/11, and ORF 6b was also reported in TCoV and Australian IBV strains (Cao et al., 2008, Hewson et al., 2011). The homolog in SARS-CoV is 63 aa in length and was identified as an endoplasmic reticulum/Golgi membrane-localised protein that induces apoptosis. Apoptosis may play an important role in promoting CoV dissemination in vivo, minimising inflammation and aiding evasion of the host’s defence mechanisms (Ye et al., 2010). Protein 6 from SARS-CoV accelerated the replication of murine CoV, increasing the virulence of the original attenuated virus (Tangudu et al., 2007). Presumably, this accessory protein plays a similar role in IBV pathogenesis, although this remains to be determined experimentally.
3.2.5. Untranslated regions (UTRs)
The 3′ UTRs of CoV genomes contain conserved cis-acting sequence and structural elements that play essential roles in RNA synthesis, gene expression and virion assembly, and each sub-genomic RNA contains a 5′ leader segment that is identical to this 3′ UTR region of the genome (Goebel et al., 2004, Sola et al., 2011). No SNPs were detected in the 3′ UTR in the sub-consensus sequences of strain 3665/11, which is consistent with the vital regulatory role that this region plays. Conversely, the partial 5′ UTR sequence of strain 3665/11 was highly variable.
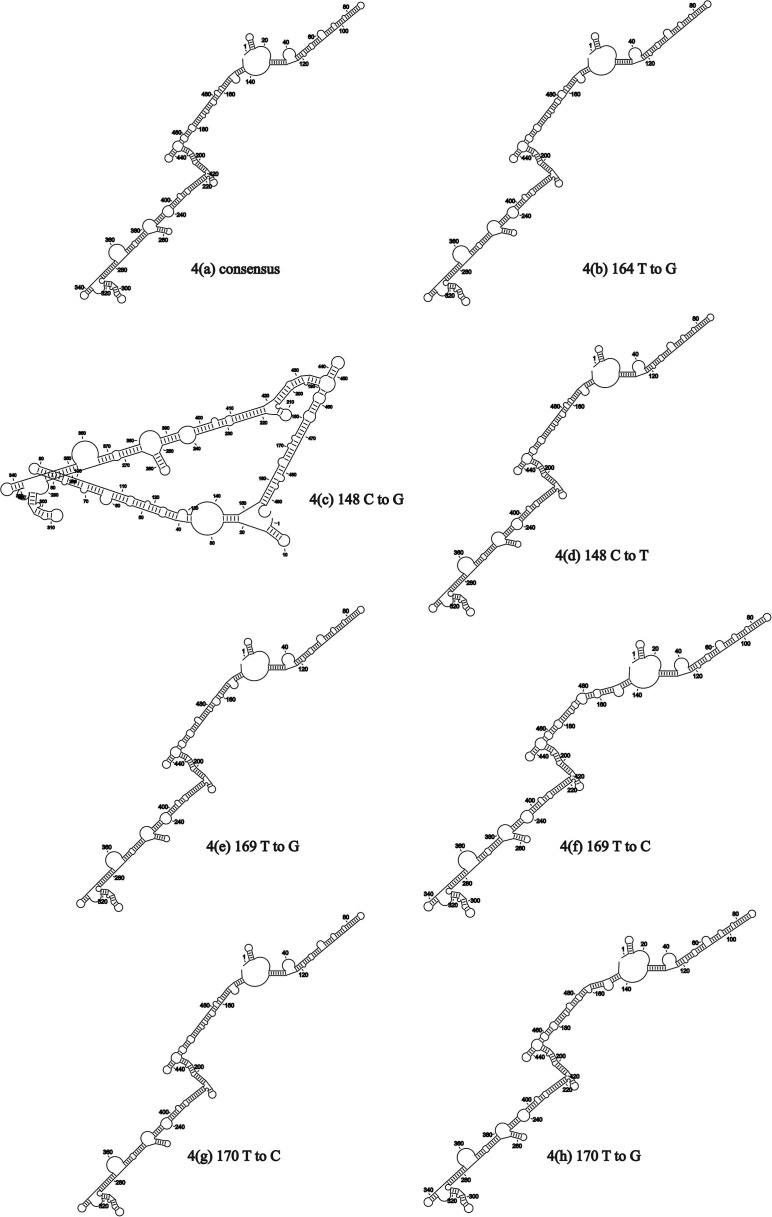
The un-sequenced 139 nucleotides from the 5′ end of the genome were extrapolated from the most similar genomic sequence, that strain ITA/90254/2005, and the secondary RNA structure of the 5′ UTR for 3665/11 was predictively folded (Fig. 5). The SNPs were then systematically substituted into the consensus sequence and RNA folding repeated. Delta G values for the predicted RNA secondary structures in Fig. 4 (a)–(h) varied from −182.9 kcal/mol to −185.5 kcal/mol. Apart from the 148C to G mutation (Fig. 4(c)), effects on RNA secondary structure were minor and the structures in Fig. 4(b) and (d)–(h) were similar. To assess the effect of combining mutations, an RNA containing 148T(U), 164T(U), 169G and 170C was folded, and this resulted in a similar stem-loop structure to those in Fig. 4(a), (b) and (d)–(h) (data not shown). Apart from the mutation 148C to G, the SNPs had little effect on the secondary RNA structure in the 5′ UTR.
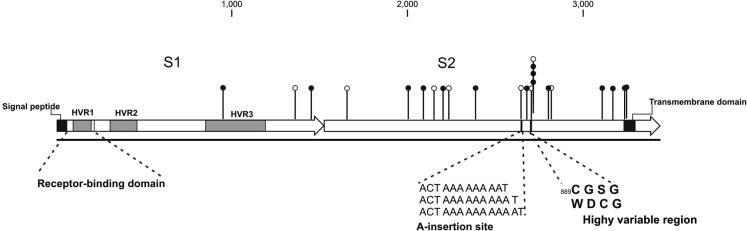
Fig. 5.
Schematic representation of the Spike protein of QX-like IBV strain 3665/11. Missense mutations are indicated as solid circles, and silent mutations as empty circles.
Fig. 4.
Effects of mutations on the predicted RNA structures in the 5′ UTR of QX-like IBV strain 3665/11.
3.3. The spike protein
Twenty-three SNPS were identified in the 3438 bp spike protein ORF; 15 of these resulted in missense mutations at the amino acid level, and 8 were silent mutations. The frequency of total SNPs in the S protein ORF was below average, at 0.67%, compared to the genome average of 0.76%. It was anticipated that the majority of mutations in the S ORF would be in the S1 gene, particularly in the HVRs, but, surprisingly, this was not the case. Only three of these SNPS (two missense and one silent) were found in the S1 gene, and all three were located in the COOH-terminal half of the S1 protein (Fig. 5). Only one mutation, a missense mutation, mapped to HVR3. No SNPS were detected in HVR1 or HVR2. The S2 subunit was considerably more variable, containing 87% of the polymorphisms detected across the entire S protein.
Two other notable features of S2 were detected: the first was a multi-A insertion site located between nucleotides 22,794 and 22,795 in the genome. The polymorphism involved the insertion of either one or two adenine nucleotides, possibly via a mechanism of polymerase stuttering. The second region of interest was located in close proximity, just downstream of the multi-A insertion site: a stretch of three consecutive mutated amino acids, namely 889C → W, 890G → D, 891S → C followed by silent mutation 892G (Fig. 5).
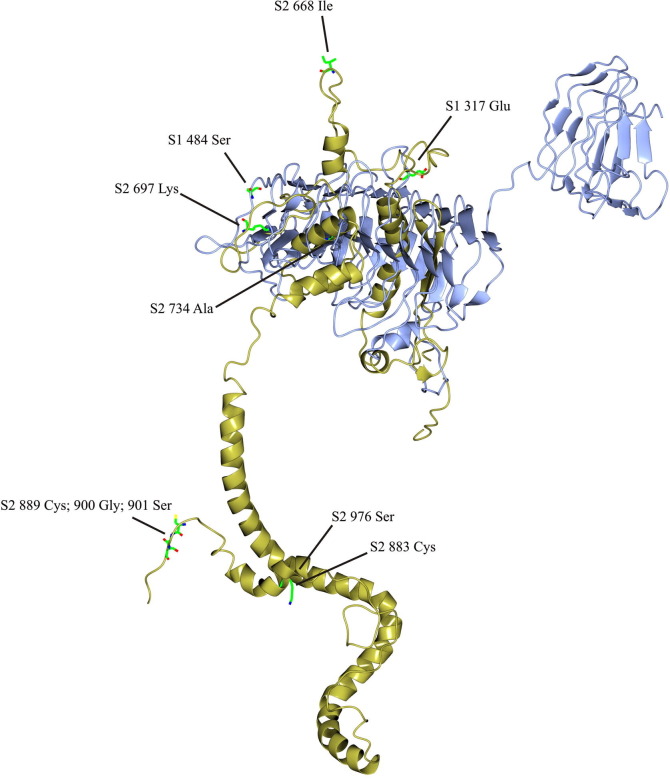
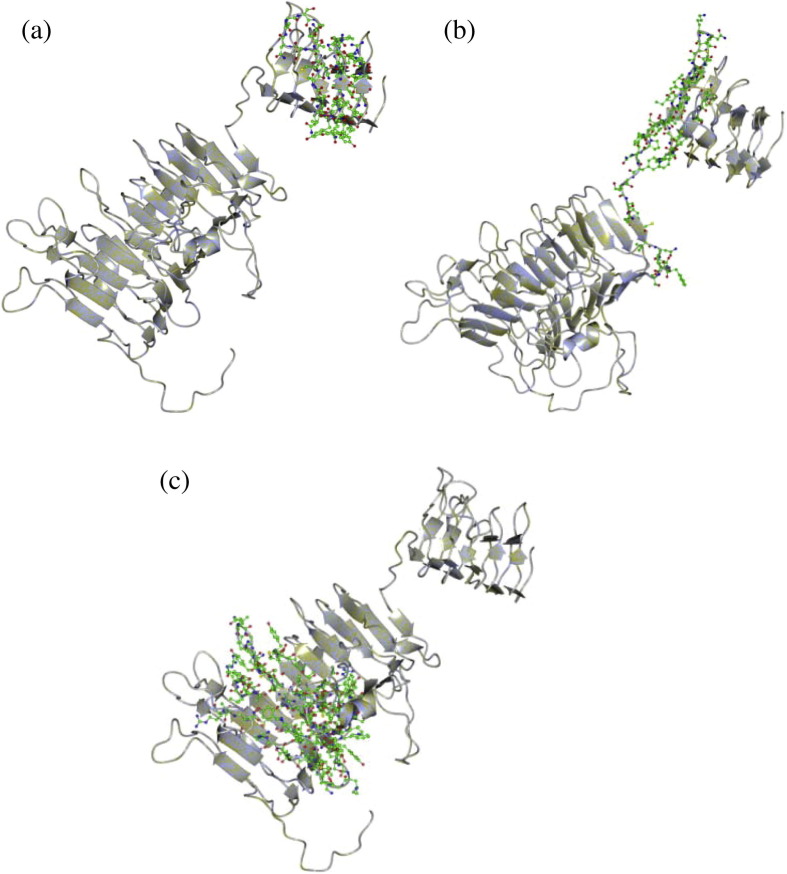
Template-based protein structure modelling was used to predict the secondary structure of the IBV spike monomer, based on the available crystal structure for the MERS-CoV S1 and S2 subunits (Fig. 6 ). S1 and S2 were modelled separately in Raptor X and then superposed. The IBV S1 structure was arranged as two beta barrels and S2 formed packed α-helices. The S2 protein was not complete and the transmembrane domain was not represented since there were no sufficiently similar structures on which to model this region, but this is the first model of the spike protein monomer for IBV. HVR1 and the putative receptor binding domain maps to the apical beta barrel (Fig. 7 (a)) and HVR2 is located on the flat plane on the base of the apical beta barrel and the peptide connecting it to the basal beta barrel (Fig. 7(b)). HVR3 maps to a region in the basal beta barrel of S1 that was predicted to contact or interact with S2 (Fig. 7(c)). The locations of the missense mutations detected by SNP analysis in the S1 and S2 subunits are indicated in Fig. 6. Many of these SNPs mapped to codons encoding amino acids on the surface of the predicted structure, but two regions were notable. Firstly, the highly variable region in S2 spanning amino acids 889–901 was exposed on the S2 stalk, although folding of the remainder of the COOH domain may have influenced this conformation. Secondly, 668Ile was exposed on a projection at the top of the monomer. This residue precedes the second furin cleavage site in the S2 subunit with the sequence 667PISSSGR/S 674. The cleavage of the S1/S2 furin motif (517RRRR/S521 in strain 3665/11) was found to be non-essential for attachment of IBV to the cell. Rather, it promotes infectivity within the cell. In studies with the Beaudette IBV strain, the second furin cleavage site in the S2 subunit was required for furin-dependent entry and syncytium formation, and the current hypothesis is that interplay between the S1 and S2 subunits determines virus attachment to specific receptors, determining tissue tropism of the virus (Promkuntod et al., 2013). The exact biological roles of these areas in S2 that are prone to mutation remain to be experimentally determined.
Fig. 6.
Predicted structure of the Spike protein monomer of QX-like IBV strain 3665/11. Missense mutations in S1 (blue) and S2 (yellow) are indicated as coloured side chains.
Fig. 7.
The predicted locations of HVR1 (7a), HVR2 (7b) and HVR3 (7c) of QX-like IBV strain 3665/11, indicated as coloured side-chains on the S1 subunit.
4. Conclusions
Archaeological remains of domestic chickens in Northeast China and the Indus Valley date back ∼8000 years (West & Zhou, 1988). The CoV group has been estimated to have arisen around 8100 BC, and the Gammacoronaviruses diverged from the CoV group around 2800 BC (Woo et al., 2012). CoVs have probably been co-evolving with their gallinaceous hosts for several thousand years. Indeed, Cook and co-authors (2012) state that “IBV is found everywhere that commercial chickens are kept”. The implication is that although IBV was only discovered some 80 years ago, the variety of serotypes we now observe are the results of hundreds if not thousands of years of genetic drift and recombination, accelerated by modern poultry farming practises where chickens are kept in high densities, and inter-regional trade in poultry and other avian species.
Studies on antigenic diversity of IBVs are heavily biased towards studies of the S1 gene, and the HVRs in particular (Cavanagh, 2007, Ducatez et al., 2009, Kant et al., 1992, Mork et al., 2014). Many of these studies cite frequent point mutations in the S1 gene, but this was not the finding of the present study. The discovery of a novel 3′-to-5′ exoribonuclease activity in CoV nsp14, which regulates replication fidelity and diversity in coronaviruses (Denison et al., 2011), lends weight to the theory that genetic drift is not primarily responsible for the degree of variation and serotypes we observe in poultry nowadays.
Instead, generation of variation by recombination is likely the main mechanism of serotypic diversity. The high frequency of RNA recombination in coronaviruses is likely caused by their unique mechanism of RNA synthesis, which involves discontinuous transcription and polymerase jumping (Jeong et al., 1996). Sequencing of many field strains has provided convincing evidence that many, possibly all, IBV strains are recombinants between different field strains (Cavanagh, 2007, Kuo et al., 2013, Liu et al., 2013, Hewson et al., 2011, Kottier et al., 1995, Wang et al., 1997).
The S1 subunit HVR1 contains the IBV receptor-binding site. Therefore despite the sequence variability in this region (which includes insertions and deletions), diverse strains must retain this critical biological function. All three HVRs may represent ancient artefacts of recombination, which have been perpetuated because they retain receptor-binding properties, with minimal permissive amino acid changes. This theory contrasts the tenet that the HVRs in the S1 subunit are very tolerant of amino acid changes produced by genetic drift, thereby conferring a selective advantage (Cavanagh, 2007, De Wit, 2000, Kant et al., 1992, Koch et al., 1990).
Whereas S1 fulfils a primary role in receptor binding (Promkuntod et al., 2014), a broader role of S2 in antigenicity and attachment to receptors is emerging. Chickens primed with a recombinant-expressed S2 subunit of a virulent ArkDPI strain and boosted with a live Mass-type vaccine were protected against challenge with live virulent ArkDPI virus (Toro et al., 2014). Although S2 subunits most likely do not contain an additional independent receptor-binding site, S2 in association with S1 forms part of a specific ectodomain which is critical to the binding of the virus to chicken tissues, which implies that both S1 and S2 contain determinants important to viral host range (Promkuntod et al., 2013). The results of the present study demonstrate that S2 is more predisposed to mutations than S1, providing an adaptive advantage and at least one other study has reported higher variability in S2 compared to S1 (Mo et al., 2012).
IBV has not been as extensively studied as other CoVs, and little progress has been made in effectively controlling or eradicating the disease in poultry. Experimental and field studies provide substantial evidence that use of a homologous IBV vaccine is best, but sometimes, intriguingly, protection can be offered by an unrelated vaccine, or by the use of two heterologous vaccines (Jones, 2010). Genotyping and phylogenetic analysis of IBV are typically focused on the S1 subunit sequence, and Liu et al. (2014) caution against drawing conclusions based on a single gene sequence, particularly a partial gene sequence. The roles of the IBV E and accessory proteins and their roles in the pathogenesis of IBV have been completely overlooked, even when the roles of the homologs in other CoVs have been proven significant. Accessory proteins of IBV and other CoVs may also offer a new generation of vaccine targets: the use of codon-deoptimization of non-structural virulence genes in influenza A virus and respiratory syncytial virus resulted in genetically stable viruses that retained immunogenicity but were attenuated (Nogales et al., 2014, Meng et al., 2014). Evidently virulence and immunogenicity in IBV is a multi-genic trait, and future studies must aim to pursue a better understanding and exploitation of the roles of various viral proteins in the host, if any advances are to be made in controlling the disease in poultry.
Acknowledgements
Adrian Knoetze and Rainbow Veterinary Laboratory are thanked for providing strain 3665/11 for this study. Funding was provided by the Poultry Section, Department of Production Animal Studies.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2015.03.033.
Appendix A. Supplementary data
THis file contains supplementary table.
References
- Bentley K., Keep S.M., Armesto M., Britton P. Identification of a noncanonically transcribed subgenomic mRNA of infectious bronchitis virus and other gammacoronaviruses. J. Virol. 2013;87(4):2128–2136. doi: 10.1128/JVI.02967-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlenga G., Cook J.K., Gelb J., Jr., Wit J.J. Development and use of the H strain of avian infectious bronchitis virus from the Netherlands as a vaccine: a review. Avian Pathol. 2004;33(6):550–557. doi: 10.1080/03079450400013154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borucki M.K., Allen J.E., Chen-Harris H., Zemla A., Vanier G., Mabery S., Torres C., Hullinger P., Slezak T. The role of viral population diversity in adaptation of bovine coronavirus to new host environments. PLoS ONE. 2013;8(1):e52752. doi: 10.1371/journal.pone.0052752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., de Haan C.A., Smits S.L., Rottier P.J. Spike protein assembly into the coronavirion: exploring the limits of its sequence requirements. Virology. 2005;334(2):306–318. doi: 10.1016/j.virol.2005.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T., Mishra N., Jain K., Zalmout I.S., Jabado O.J., Karesh W.B., Daszak P., Mohammed O.B., Alagaili A.N., Lipkin W.I. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio. 2014;5(3) doi: 10.1128/mBio.01146-14. e01146–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wu C.C., Lin T.L. Complete nucleotide sequence of polyprotein gene 1 and genome organization of turkey coronavirus. Virus Res. 2008;136(1–2):43–49. doi: 10.1016/j.virusres.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casais R., Davies M., Cavanagh D., Britton P. Gene 5 of the avian coronavirus infectious bronchitis virus is not essential for replication. J. Virol. 2005;79(13):8065–8078. doi: 10.1128/JVI.79.13.8065-8078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses in poultry and other birds. Avian Pathol. 2005;34(6):439–448. doi: 10.1080/03079450500367682. Review. [DOI] [PubMed] [Google Scholar]
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38(2):281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- Cavanagh D., Davis P.J., Cook J.K. Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype. Avian Pathol. 1992;21(3):401–408. doi: 10.1080/03079459208418858. [DOI] [PubMed] [Google Scholar]
- Coletta A., Pinney J.W., Solis D.Y., Marsh J., Pettifer S.R., Atwood T.K. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 2010;4:43. doi: 10.1186/1752-0509-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.W., Pei J., Dzielawa J., Seo S.H. Cytotoxic T lymphocytes are critical in the control of infectious bronchitis virus in poultry. Dev. Comp. Immunol. 2000;24(2–3):187–200. doi: 10.1016/s0145-305x(99)00072-5. Review. [DOI] [PubMed] [Google Scholar]
- Cook J.K., Jackwood M., Jones R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012;41(3):239–250. doi: 10.1080/03079457.2012.680432. [DOI] [PubMed] [Google Scholar]
- Darbyshire J.H., Rowell J.G., Cook J.K., Peters R.W. Taxonomic studies on strains of avian infectious bronchitis virus using neutralisation tests in tracheal organ cultures. Arch. Virol. 1979;61(3):227–238. doi: 10.1007/BF01318057. [DOI] [PubMed] [Google Scholar]
- DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014 doi: 10.1016/j.virusres.2014.07.024. pii: S0168–1702(14)00302–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M.F., Martin A.M., Owoade A.A., Olatoye I.O., Alkali B.R., Maikano I., Snoeck C.J., Sausy A., Cordioli P., Muller C.P. Characterization of a new genotype and serotype of infectious bronchitis virus in Western Africa. J. Gen. Virol. 2009;90(11):2679–2685. doi: 10.1099/vir.0.012476-0. [DOI] [PubMed] [Google Scholar]
- de Haan C.A., Stadler K., Godeke G.J., Bosch B.J., Rottier P.J. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J. Virol. 2004;78(11):6048–6054. doi: 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit J.J. Detection of infectious bronchitis virus. Avian Pathol. 2000;29(2):71–93. doi: 10.1080/03079450094108. [DOI] [PubMed] [Google Scholar]
- Dinkel H., Van Roey K., Michael S., Davey N.E., Weatheritt R.J., Born D., Speck T., Krüger D., Grebnev G., Kuban M., Strumillo M., Uyar B., Budd A., Altenberg B., Seiler M., Chemes L.B., Glavina J., Sánchez I.E., Diella F., Gibson T.J. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gkt1047. (Database issue): D259-66. http://elm.eu.org Last accessed 03/12/2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsang A., Ros C., Renström L.H., Baule C., Soós T., Belák S. Molecular epizootiology of infectious bronchitis virus in Sweden indicating the involvement of a vaccine strain. Avian Pathol. 2002;31(3):229–236. doi: 10.1080/03079450220136530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo R.A., van Santen V.L., Toro H. Effects of chicken anaemia virus and infectious bursal disease virus-induced immunodeficiency on infectious bronchitis virus replication and genotypic drift. Avian Pathol. 2012;41(5):451–458. doi: 10.1080/03079457.2012.702889. [DOI] [PubMed] [Google Scholar]
- Gelb J., Jr., Keeler C.L., Jr., Nix W.A., Rosenberger J.K., Cloud S.S. Antigenic and S-1 genomic characterization of the Delaware variant serotype of infectious bronchitis virus. Avian Dis. 1997;41(3):661–669. [PubMed] [Google Scholar]
- Goebel S.J., Hsue B., Dombrowski T.F., Masters P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78(2):669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson K.A., Ignjatovic J., Browning G.F., Devlin J.M., Noormohammadi A.H. Infectious bronchitis viruses with naturally occurring genomic rearrangement and gene deletion. Arch. Virol. 2011;156(2):245–252. doi: 10.1007/s00705-010-0850-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson T., Britton P., Cavanagh D. Neither the RNA nor the proteins of open reading frames 3a and 3b of the coronavirus infectious bronchitis virus are essential for replication. J. Virol. 2006;80(1):296–305. doi: 10.1128/JVI.80.1.296-305.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignjatovic J., McWaters P.G. Monoclonal antibodies to three structural proteins of avian infectious bronchitis virus characterization of epitopes and antigenic differentiation of Australian strains. J. Gen. Virol. 1991;72(12):2915–2922. doi: 10.1099/0022-1317-72-12-2915. [DOI] [PubMed] [Google Scholar]
- Inglis S.C., Rolley N., Brierley I. A ribosomal frameshift signal in the polymerase-encoding region of the IBV genome. Adv. Exp. Med. Biol. 1990;276:269–273. doi: 10.1007/978-1-4684-5823-7_37. [DOI] [PubMed] [Google Scholar]
- Jackwood M.W., Hilt D.A., Lee C.W., Kwon H.M., Callison S.A., Moore K.M., Moscoso H., Sellers H., Thayer S. Data from 11 years of molecular typing infectious bronchitis virus field isolates. Avian Dis. 2005;49(4):614–618. doi: 10.1637/7389-052905R.1. [DOI] [PubMed] [Google Scholar]
- Jeong Y.S., Repass J.F., Kim Y.N., Hwang S.M., Makino S. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology. 1996;217(1):311–322. doi: 10.1006/viro.1996.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.C. Viral respiratory diseases (ILT, aMPV infections, IB): are they ever under control? Br. Poult. Sci. 2010;51(1):1–11. doi: 10.1080/00071660903541378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallberg M., Wang H., Wang S., Peng J., Wang Z., Lu H., Xu J. Template-based protein structure modeling using the RaptorX web server. Nat. Protocols. 2012;7(8):1511–1522. doi: 10.1038/nprot.2012.085. http://raptorx.uchicago.edu/ last accessed 03/12/2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant A., Koch G., van Roozelaar D.J., Kusters J.G., Poelwijk F.A., van der Zeijst B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992;73(Pt 3):591–596. doi: 10.1099/0022-1317-73-3-591. [DOI] [PubMed] [Google Scholar]
- Kint J., Fernandez-Gutierrez M., Maier H.J., Britton P., Langereis M.A., Koumans J., Wiegertjes G.F., Forlenza M. Activation of the chicken type I IFN response by infectious bronchitis coronavirus. J. Virol. 2014;5 doi: 10.1128/JVI.02671-14. pii: JVI.02671-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoetze A.D., Moodley N., Abolnik C. Two genotypes of infectious bronchitis virus are responsible for serological variation in KwaZulu-Natal poultry flocks prior to 2012. Onderstepoort J. Vet. Res. 2014;81(1):1–10. doi: 10.4102/ojvr.v81i1.769. [DOI] [PubMed] [Google Scholar]
- Koch G., Hartog L., Kant A., van Roozelaar D.J. Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J. Gen. Virol. 1990;71(Pt 9):1929–1935. doi: 10.1099/0022-1317-71-9-1929. [DOI] [PubMed] [Google Scholar]
- Kottier S.A., Cavanagh D., Britton P. First experimental evidence of recombination in infectious bronchitis virus. Adv. Exp. Med. Biol. 1995;380:551–556. doi: 10.1007/978-1-4615-1899-0_88. [DOI] [PubMed] [Google Scholar]
- Kuo S.M., Kao H.W., Hou M.H., Wang C.H., Lin S.H., Su H.L. Evolution of infectious bronchitis virus in Taiwan: positively selected sites in the nucleocapsid protein and their effects on RNA-binding activity. Vet. Microbiol. 2013;162(2–4):408–418. doi: 10.1016/j.vetmic.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusters J.G., Jager E.J., Lenstra J.A., Koch G., Posthumus W.P., Meloen R.H., van der Zeijst B.A. Analysis of an immunodominant region of infectious bronchitis virus. J. Immunol. 1989;143(8):2692–2698. [PubMed] [Google Scholar]
- Lai M.M., Cavanagh D. The molecular biology of coronaviruses. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Lv L., Yin L., Li X., Luo D., Liu K., Xue C., Cao Y. Assembly and immunogenicity of coronavirus-like particles carrying infectious bronchitis virus M and S proteins. Vaccine. 2013;31(47):5524–5530. doi: 10.1016/j.vaccine.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xu Q., Han Z., Liu X., Li H., Guo H., Sun N., Shao Y., Kong X. Origin and characteristics of the recombinant novel avian infectious bronchitis coronavirus isolate ck/CH/LJL/111054. Infect. Genet. Evol. 2014;23:189–195. doi: 10.1016/j.meegid.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardani K., Noormohammadi A.H., Hooper P., Ignjatovic J., Browning G.F. Infectious bronchitis viruses with a novel genomic organization. J. Virol. 2008;82(4):2013–2024. doi: 10.1128/JVI.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride R., van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6(8):2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Lee S., Hotard A.L., Moore M.L. Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. MBio. 2014;5(5):e01704–e01714. doi: 10.1128/mBio.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans G., Boschmans M., Decaesstecker M., Berg T.P., Denis P., Cavanagh D. Epidemiology of infectious bronchitis virus in Belgian broilers: a retrospective study, 1986–1995. Avian Pathol. 2001;30(4):411–421. doi: 10.1080/03079450120066412. [DOI] [PubMed] [Google Scholar]
- Mo M., Huang B., Wei P., Wei T., Chen Q., Wang X., Li M., Fan W. Complete genome sequences of two Chinese virulent avian coronavirus infectious bronchitis virus variants. J. Virol. 2012;86(19):10903–10904. doi: 10.1128/JVI.01895-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.M., Jackwood M.W., Hilt D.A. Identification of amino acids involved in a serotype and neutralization specific epitope within the S1 subunit of avian infectious bronchitis virus. Arch. Virol. 1997;142(11):2249–2256. doi: 10.1007/s007050050239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mork A.K., Hesse M., Abd El Rahman S., Rautenschlein S., Herrler G., Winter C. Differences in the tissue tropism to chicken oviduct epithelial cells between avian coronavirus IBV strains QX and B1648 are not related to the sialic acid binding properties of their spike proteins. Vet. Res. 2014;45:67. doi: 10.1186/1297-9716-45-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndegwa E.N., Toro H., van Santen V.L. Comparison of vaccine subpopulation selection, viral loads, vaccine virus persistence in trachea and cloaca, and mucosal antibody responses after vaccination with two different Arkansas Delmarva Poultry Industry-derived infectious bronchitis virus vaccines. Avian Dis. 2014;58(1):102–110. doi: 10.1637/10609-070613-Reg.1. [DOI] [PubMed] [Google Scholar]
- Nogales A., Baker S.F., Ortiz-Riaño E., Dewhurst S., Topham D.J., Martínez-Sobrido L. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J. Virol. 2014;88(18):10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Sekellick M.J., Marcus P.I., Choi I.S., Collisson E.W. Chicken interferon type I inhibits infectious bronchitis virus replication and associated respiratory illness. J. Interferon Cytokine Res. 2001;21(12):1071–1077. doi: 10.1089/107999001317205204. [DOI] [PubMed] [Google Scholar]
- Promkuntod N., Wickramasinghe I.N., de Vrieze G., Gröne A., Verheije M.H. Contributions of the S2 spike ectodomain to attachment and host range of infectious bronchitis virus. Virus Res. 2013;177(2):127–137. doi: 10.1016/j.virusres.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promkuntod N., van Eijndhoven R.E., de Vrieze G., Gröne A., Verheije M.H. Mapping of the receptor-binding domain and amino acids critical for attachment in the spike protein of avian coronavirus infectious bronchitis virus. Virology. 2014;448:26–32. doi: 10.1016/j.virol.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susan R.W., Julian L.L. Coronavirus pathogenesis. Adv. Virus Res. 2011;81:95–164. doi: 10.1016/B978-0-12-385885-6.00009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Mateos-Gomez P.A., Almazan F., Zuñiga S., Enjuanes L. RNA-RNA and RNA-protein interactions in coronavirus replication and transcription. RNA Biol. 2011;8(2):237–248. doi: 10.4161/rna.8.2.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81(3):1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timms L.M., Bracewell C.D., Alexander D.J. Cell mediated and humoral immune response in chickens infected with avian infectious bronchitis. Br. Vet. J. 1980;36(4):346–349. doi: 10.1016/s0007-1935(17)32237-6. [DOI] [PubMed] [Google Scholar]
- Toro H., Zhao W., Breedlove C., Zhang Z., Yub Q. Infectious bronchitis virus S2 expressed from recombinant virus confers broad protection against challenge. Avian Dis. 2014;58(1):83–89. doi: 10.1637/10641-081613-Reg.1. [DOI] [PubMed] [Google Scholar]
- Yang Y., Zhang L., Geng H., Deng Y., Huang B., Guo Y., Zhao Z., Tan W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell. 2013;4(12):951–961. doi: 10.1007/s13238-013-3096-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xu L., Collison E.W. Experimental confirmation of recombination upstream of the S1 hypervariable region of infectious bronchitis virus. Virus Res. 1997;49:139–145. doi: 10.1016/S0168-1702(97)01466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.H., Huang Y.C. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch. Virol. 2000;145(2):291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Sundaram J.P., Spiro D. VIGOR, an annotation program for small viral genomes. BMC Bioinform. 2010;11(451):1–10. doi: 10.1186/1471-2105-11-451. http://www.jcvi.org/vigor Last accessed 03/12/2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West B., Zhou B.X. Did chickens go north? New evidence for domestication. J. Archeol Sci. 1988;15:515–533. [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86(7):3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Zou N., Wang F., Guo M., Liu P., Wen X., Cao S., Huang Y. Analysis of a QX-like avian infectious bronchitis virus genome identified recombination in the region containing the ORF 5a, ORF 5b, and nucleocapsid protein gene sequences. Virus Genes. 2013;46(3):454–464. doi: 10.1007/s11262-013-0884-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
THis file contains supplementary table.