Abstract
Coactivating and inhibitory receptors that share at least one ligand interact with a wide variety of ligands, indicating their importance in a range of situations. Here, we discuss principles of mainly human paired receptor function and ligand recognition, and possible therapeutic implications of targeting these receptors in cancer, autoimmune diseases, and allergy. We summarise and emphasise the idea that these receptors, which have evolved in part in response to pathogen pressure, fine-tune the immune response, preserve homeostasis, and that pathogens and tumours use the dominance of the inhibitory receptors over the coactivating receptors to avoid immune elimination. Finally, we discuss the options of using paired receptors and their ligand for immune cell education and therapy.
Trends
Paired receptors were initially described as activating and inhibitory receptors homologous in their extracellular domains and located in close proximity on the genome.
Here we give a broader definition, as a pair of receptors that includes an inhibitory and a coactivating receptor; where each member has at least one common cellular ligand, and upon binding, exerts contrasting effects (inhibition or co-stimulation).
In these pairs, the inhibitory receptor binds the shared ligand with greater affinity than does the coactivating receptor.
This high-affinity binding of the inhibitory receptor results also in competitive binding. Some of the inhibitory receptors can also exert their function by physically interacting with the activating receptor and preventing its activity.
Inhibitory receptors and ligands for paired receptors are used by pathogens and tumours for immune evasion.
Introduction
Current models of immunological activation have expanded our knowledge from single ligand–receptor interactions to integrated models in which several receptors have a combinatorial role in the activation of a specific type of immune cell. The model of T cell activation is a clear example of this; whereas the canonical T cell receptor (TCR) is thought to provide the primary activating signal, co-stimulatory receptors and soluble factors have become integral parts of a more complex picture [1]. Similarly, mast cells are primarily activated by the IgE receptors (such as FcεRI), but soluble factors, inhibitory, activating, and coactivating receptors can together modulate the activities of mast cells [2]. The complexity of immunological signalling is best exemplified in natural killer (NK) cells. In these cells, no single receptor is ultimately responsible for cellular activation; rather, a balance of signals from a wide variety of activating receptors, coactivating receptors, and inhibitory receptors (again, together with soluble factors) determines NK cell activity [3]. A more comprehensive understanding of these integrative signalling processes has recently become of great clinical significance owing to the development of immune checkpoint inhibitors.
Inhibitory and coactivating receptors that recognise the same cellular ligands – which we refer to here as paired receptors (for a broader definition of the previous one, see Trends) – are the focus of this review (Figure 1 A); see also a brief description of these receptors in [4] and selected examples in 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16. We first discuss how paired receptors are defined and how they may have evolved. We describe their complex expression patterns and determine what controls their activities. We hypothesise that the paired receptors that have probably evolved mainly in response to pathogens are now also used to maintain immune cells in a responsive state. We further suggest that they also limit overzealous inflammation by dampening the activation signals provided by the coactivating receptors that could (at least in some cases) lead to autoimmunity. Furthermore, we discuss how pathogens manipulate paired receptors, to infect cells and subsequently evade immune attack. Finally, we examine the potential use of the paired receptors as therapeutic targets in various clinical situations and propose that the paired receptors might be involved in immune cell education; the process by which immune cells acquire functional capabilities.
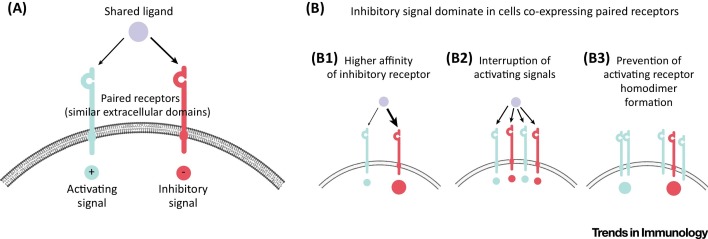
Figure 1.
Paired Receptors Mediate a Balanced Form of Inhibition. (A) Schematic representation of a receptor pair. The shared ligand can bind either receptor, but the receptors themselves have opposite downstream signals, either activating or inhibitory. (B) Several mechanisms cause the inhibitory signal to be dominant over the activating signal when both paired receptors are expressed on the same cell. (B1). The inhibitory receptor (marked in red), has a higher affinity for the shared ligand than the activating receptor in all receptor pairs (for example, TIGIT versus DNAM1 8, 38). (B2) As a result of its higher affinity for ligand, the inhibitory receptor competes with its activating partner for ligand binding such that the inhibitory receptor physically interrupts the binding of the activating receptor to the ligand 8, 38. (B3) There is also evidence that inhibitory receptors can interfere with the homodimerization of their corresponding activating receptor, thereby preventing signalling through the activating receptor (e.g., TIGIT can interfere with the homodimerisation of DNAM1 [20]). Abbreviations: DNAM1, DNAX accessory molecule 1; TIGIT, T cell immunoreceptor with immunglobulin and ITIM domains.
Historically, studies of mouse receptors, such as the Ly49 receptor family on NK cells [also known as the killer cell lectin-like receptor subfamily A (KLRA) family], significantly contributed to the concept of paired receptors (reviewed in [17]). Due to text limitations we are not able to elaborate on all possible aspects of all of the paired receptors and we instead highlight several examples. Furthermore, although there are many known paired receptors, both in humans and in mice, we focus mainly on a few well-characterised human pairs that enable us to best present the general principles we outline. In that regard, we do not discuss here paired receptors in which the inhibitory and the activating proteins that recognise the same ligand are not well defined [such as the leukocyte immunoglobulin-like receptor (LILR) family], or receptors in which one of the pair seemed not to function in humans (such as paired immunoglobulin-like type 2 receptor α; PILRA).
Definition and Evolution of Paired Receptors
Paired receptors were initially described as activating and inhibitory receptors that are highly homologous in their extracellular domains and that are located in close proximity on the genome. These receptors, which recognise the same cellular ligands, seemed to evolve together through gene duplication [18]. Paired receptors belong to many different families of molecules, and they include members of the immunoglobulin superfamily, lectin family, and lectin-like protein family. Today, receptors that are not located next to each other on the genome, such as the inhibitory receptor TIGIT (T cell immunoreceptor with immunglobulin and ITIM domains), the coactivating receptor DNAM1 (DNAX accessory molecule 1; also known as CD226), and the inhibitory receptor CD112R (also known as PVRIG), can also be considered to be paired receptors as they recognise the same cellular ligands (mainly the poliovirus receptor PVR), but have opposing effects (inhibition and coactivation) [19]. For the purpose of this article, we therefore define paired receptors as a pair of receptors that includes an inhibitory and a coactivating receptor; each member of the pair has at least one common cellular ligand (Table 1 ), and upon binding of this ligand, each member of the pair exerts contrasting effects (inhibition or co-stimulation) (Figure 1A). Note that paired receptors in the context of this article do not need to be functionally linked and can signal independently of each other – in other words, the inhibitory receptor does not need to directly inhibit signalling through the activating receptor.
Table 1.
Human Paired Receptors and Their Shared Ligands
| Paired receptors: coactivating inhibitory | Paired receptors mainly expressed on | Shared cellular ligands for paired receptors | Cellular ligands mainly expressed on | Ligand–receptor affinitya | Pathogen ligands for paired receptors | Refs |
|---|---|---|---|---|---|---|
| CD28 CTLA4 |
T cells | CD80 and CD86 | DCs, monocytes, B cells, neutrophils, T cells, mesenchymal stem cells, and activated NK cells | CD28: 4 μM (CD80), 20 μM (CD86) CTLA4: 0.4 μM (CD80), 4 μM (CD86) |
SEB (for CD28) | 6, 27, 48 |
| KIR2DL and KIR3DL KIR2DS and KIR3DS |
NK cells | Intact classical MHC class I molecules (particularly for KIR2DL and KIR3DL) HLA-F and MHC class I open conformers (particularly for KIR2DS and KIR3DS) |
All nucleated cells | KIR2DL and KIR3DL: weak (could not be determined in most cases) KIR2DS and KIR3DS: 10 μM |
CpG DNA (for KIR2DL2) | 49, 50, 51, 52, 53 |
| CD300c CD300a |
NK cells, mast cells, eosinophils, basophils and neutrophils | PE and PS | Apoptotic cells | ND | Enveloped viruses containing PE and PS, such as dengue, vaccinia, West Nile, Sindbis and Ebola viruses | 7, 9, 25, 32, 36, 37, 45, 54, 55, 56, 57, 58, 59 |
| CEACAM1-S, CEACAM3 CEACAM1-L |
T cells, NK cells, mast cell and eosinophils | CEACAM1 | Most abundant in glandular epithelia of the gastrointestinal tract, bile canaliculi of the liver and intercalated ducts of the salivary gland. Also, activated immune cells (mainly T cells), resting and activated mast cells and eosinophils | ND | Opa (Neisseria spp.) P5 (Haemophilus influenzae) UspA1 (Moraxella catarrhalis) Carbohydrates on Escherichia coli and Salmonella spp. HopQ (Helicobacter pylori) |
12, 34, 60 |
| DNAM1 TIGIT |
NK cells, T cells and mast cells (DNAM1 only) | PVR | Wide tissue distribution, notable expression in the alimentary canal and nervous system | DNAM1: 114 nM TIGIT: 3.15 nM |
Fap2 of Fusobacterium nucleatum (for TIGIT only) | 8, 20, 38, 43 |
| DNAM1 CD112R |
NK cells and T cells | Nectin2 | Wide cellular distribution, including eosinophils (part of the AEU) | DNAM1: 8.97 mM CD112R: 88.4 nM |
ND | [13] |
| Siglec-14 Siglec-5 |
Monocytes | Sialylated glycans HSP70 |
Almost all cell types | ND | Group B Streptococcus | [14] |
| NKG2C NKG2A |
NK cells | HLA-E | All nucleated cells | NKG2C: 30 μM NKG2A: 1 μM |
UL-40 Leader peptide of HCMV bound to HLA-E | [15] |
| CD200RLa-e CD200R |
Myeloid cells, and B and T cells | CD200 | Cells of the hematopoietic lineage, specifically those of the myeloid lineage such as macrophages, DCs, neutrophils, mast cells, eosinophils and basophils | ND | K14 of HHV8 U85 of HHV6 and HHV7 (for CD200R) 1 vOX-2 of myxoma virus M141R 2 vCD200 for rhesus rhadinovirus R15 |
16, 33, 61, 62, 63 |
| Siglec-16 Siglec-11 |
Human brain microglia | α2–8-linked sialic acids | Widely distributed in mammalian tissues | ND | Polysialic acids on Escherichia coli | [64] |
HHV, human herpesvirus; ND, not determined; PE, phosphatidylethanolamine; PS, phosphatidylserine; SEB, Staphylococcus aureus enterotoxin B.
There is sometimes great variability in reported affinities; measurements representative of the range of values are shown.
NK cells, which express numerous activating and inhibitory receptors, with no single dominant activating receptor, thus function mainly as balanced effectors, indeed express the widest variety of these paired receptors (Table 1) [19]. Interestingly, the paired receptors, identified so far, do not include independently functioning, bona fide activating receptors that function independently [such as CD16 on naïve NK cells, NKG2D and the natural cytotoxicity receptors (NCRs) on activated NK cells, or FcεRI on mast cells]. Instead, it is coactivating receptors (that do not function independently) that are paired with the inhibitory receptors.
For the paired receptors, the inhibitory receptor seems to bind the shared ligand with greater affinity than does the coactivating receptor, as determined by in vitro assays (Table 1) [19]. Although in vitro affinities may not accurately reflect the cell membrane environment in vivo, these observations suggest that under healthy, steady-state conditions, when the inhibitory receptor and coactivating receptor are expressed on the same cell bind their shared ligand, the dominant signal is most likely inhibition. This high-affinity binding of the inhibitory receptor also results in competitive binding, whereby the inhibitory receptors physically interrupt the binding of the coactivating receptors to the ligand (Figure 1B). In addition, some of the inhibitory receptors can also exert their function in a receptor-extrinsic manner. For example, the inhibitory receptor TIGIT has been shown to disrupt the homodimerisation of its binding partner DNAM1, thus interfering with the coactivating receptor function of DNAM1 (Figure 1B) [20].
Activating and coactivating receptors signal mainly through immunoreceptor tyrosine-based activation motifs (ITAMs), whereas inhibitory receptors signal through immunoreceptor tyrosine-based inhibitory motifs (ITIMs). Several of the coactivating paired receptors have an ITIM sequence in the 3′ untranslated region, which suggests that they may have evolved from the inhibitory receptors [18]. Indeed, because inhibitory receptors are often used by pathogens for immune evasion (as discussed below), immune cells seem to have acquired coactivating receptors later in their evolution than inhibitory receptors, to counterattack these pathogens. This model is well represented by the Ly49 paired receptors expressed on NK cells. Inhibitory Ly49 receptors recognise host MHC class I molecules, which can be downregulated on the surface of virus-infected cells, leading to NK cell activation. Mouse cytomegalovirus (MCMV) therefore evades the immune response by producing viral MHC class I-like ligand m157 for inhibitory Ly49I. However, some MCMV-resistant mouse strains also express activating Ly49 receptors, which recognise m157 to directly kill MCMV-infected cells [21]. Still, even in this case it seems that the inhibitory interaction is dominant. The binding constants determined by Adams et al. using surface plasmon resonance (SPR) were KD = 166 nM for Ly49I/m157 interaction and KD = 1 μM for Ly49H/m157 interaction [22].
Independent Activity of Members of the Paired Receptors
If activating and inhibitory receptors of a pair interact with the same ligand and inhibition is dominant, when does the coactivating receptor of the pair have an effect? Some paired receptors (such as DNAM1 and TIGIT on NK cells [8]) are expressed constitutively and on the same cell; however, other immune cells only express the coactivating receptor of a pair. For example, mast cells and eosinophils express the coactivating receptor DNAM1 [23] but not its inhibitory receptor counterpart TIGIT. Interestingly, these effector cells of allergy also express Nectin 2, which is a ligand of TIGIT and DNAM1 [23]. The expression by mast cells and eosinophils of only the coactivating receptor and its ligand means that when these cells come together there is enhanced activation, which can explain the self-fuelling, proinflammatory allergic effector unit (AEU) 23, 24. Thus, tissue-resident mast cells start the allergic response and when eosinophils penetrate into the tissue, they interact via soluble mediators and physical binding with mast cells to form the AEU. This synergistic overactivation of mast cells and eosinophils is an example of the pathology that can be ‘lying in wait’ when the paired receptors become unbalanced in certain situations.
The paired receptors can be expressed on the same type of immune cell, but not necessarily on the same clone, which enables clonal-dependent activity of the coactivating receptors. The human killer cell immunoglobulin-like receptors (KIRs) have short or long cytoplasmic domains, with the long forms (such as KIR2DL and KIR3DL) being inhibitory and the short forms (such as KIR2DS and KIR3DS) being activating or coactivating. They are stochastically expressed on NK cells (some KIR haplotypes have few activating receptors) and thus may function independently on certain NK cell clones. Similarly, in the case of CD300a and CD300c, which bind phosphatidylserine and phosphatidylethanolamine with opposing effects [25], NK cells express the members of this receptor pair stochastically [7].
In cases where the paired receptors are expressed on the same cell, the spatial expression of the receptors may determine their ability to function. For example, differential expression of paired receptors in different zones of the NK cell inhibitory and activating synapse [26] can influence their activity. The concept of the immune synapse is an important reminder that cells exist in three dimensions and that location is crucial. Thus, even if inhibitory and coactivating paired receptors are present on the same cell, they might be segregated into different cellular domains following their gathering at intercellular contacts on the immune synapse. These domains are sometimes associated with different signalling molecules and, in such situations, the coactivating receptor might function.
The level of expression (number and density) of inhibitory and coactivating members of the paired receptors might also affect their activities. Indeed, although inhibitory receptors have overall a higher affinity for their ligand, they may be present at lower densities at the cell surface than their coactivating counterpart. Therefore, in the context of target cells expressing a low level of the common ligand, the inhibitory receptors will engage ligand first, thus preventing engagement of the activating counterpart, which will lead to a dominant inhibition. By contrast, in the context of a target cell expressing high levels of the ligand, the activating receptors will also be engaged and owing to their greater density at the cell surface of the responding cells compared with the inhibitory receptors, the dominant response will be activation.
Finally, paired receptors can be expressed on the same cells, but at different time points. A well-characterised example is that of CD28 and cytotoxic T lymphocyte antigen (CTLA)4; both of which bind CD80 and CD86 [6]. The activity of immature T cells is well controlled by a rigorous education process in the thymus, but the later stages of T cell activity in the periphery must be checked as well, and this is achieved by paired receptors. The coactivating receptor CD28 is expressed on naïve T cells, which allows them to respond to signals presented by a dendritic cell (DC) that has sensed a pathogen. These naïve T cells, however, do not yet express the inhibitory receptor CTLA4. Expression of CTLA4 by conventional T cells occurs only after TCR activation [6]. Once it is expressed, CTLA4 can inhibit T cell activity by various mechanisms, including direct signalling, competing for ligands and even ‘grabbing’ and degrading ligands from target cells [27].
Keeping Immune Cells in a Responsive State
As a result of the higher affinity of inhibitory receptors for their ligand, paired receptors seem to provide a balanced form of inhibition under homeostatic conditions, by which inactivity is favoured by this group of receptors and thereby by a large part of the immune system as a whole. This enables T and NK cells harbouring cytolytic granules – and mast cells containing granules with proinflammatory mediators – not to harm normal tissues, but still respond quickly through the already present coactivating receptors when there is danger. The simultaneous presence of inhibitory and coactivating receptors also keeps immune cells in a responsive state that allows them to detect small variations within their environment. For example, NK cells, in the absence of MHC class I are hyporesponsive, which indicates that inhibitory receptor signalling might be involved in the education and responsiveness of NK cells [28]. It has also been proposed that chronic activating receptor signals, that are delivered in the absence of inhibition, desensitize NK cells, leading to their hyporesponsiveness, by a process referred to as disarming [29]. Thus, paired coactivating and inhibitory receptors might coexist to fine tune the response of immune cells to their environment. This ensures that under homeostatic conditions, cells remain quiescent, but still functional, and upon subtle but rapid changes in the environment (e.g., an increase in ligand density, providing additional ligands for activating receptors) these cells are able to respond efficiently. This theory is directly related to the discontinuity theory that has been postulated by Vivier and colleagues, which proposes that immune cells generate an effector response when there is a discontinuity, that is, sudden changes in antigenic stimulation, whereas they tend to become tolerant to continuously (slow or continuous stimulation) [30].
Ligands for the Paired Receptors
The variety of shared ligands for the paired receptors is impressive. Paired receptors recognise everything from proteins to sugars, to lipids, to nucleic acids ([31] and Table 1). This vast diversity of potential ligands indicates that they might be involved in a large number of immune responses and clinical situations, many of which remain to be discovered.
Many of the shared ligands are ubiquitously expressed under normal conditions (Table 1). This may represent the role of paired receptors as a basic method of immune homeostasis, with healthy tissues constantly being sensed by the paired receptors so that immune responses are dampened.
Pathogens are also sensed by the paired receptors; for example, the carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) short (CEACAM1-S) and long (CEACAM1-L) isoforms recognise a particularly large set of pathogen ligands of different viruses and bacteria (Table 1). In many cases, virus recognition by paired receptors is beneficial to the virus. For example, CD300a facilitates the infection of several mosquito-borne viruses including Dengue, West Nile and Chikungunya viruses [32]. In addition, the K14 protein of human herpesvirus 8 mimics CD200 in downregulating macrophage activation through the CD200 receptor [33]. Interestingly, the coactivating CD200 receptors were mainly described in mice and do not show substantial binding to CD200.
Expression of the cellular ligands for the paired receptors can also be modulated by pathogens. For example, the cell surface expression of CEACAM1, which binds homophilically to the paired receptors CEACAM1-L and CEACAM1-S expressed on effector cells, is upregulated on the infected target cells following infection with several viruses [34]. Cellular transformation can also lead to the upregulation of ligands for the paired receptors; for example, expression of PVR (a TIGIT and DNAM1 ligand) is upregulated in many solid tumours [35]. Finally, some ligands such as phosphatidylserine and phosphatidylethanolamine that are normally expressed on the cell surface following apoptosis and are recognised by CD300a and CD300c, can be upregulated, even in the absence of apoptosis, following viral infection, malignant transformation [25], and in allergic inflammation 36, 37. The underlying mechanism is not well understood. As discussed below, as inhibition is the dominant response of paired receptors, pathogens and tumours will benefit from upregulating the cellular ligands of the paired receptors to protect themselves from immune cell attack.
Pathogen Manipulation of Paired Receptors
A wide variety of pathogens use the shared ligands of the paired receptors as their cellular adhesion or entry receptors (see Table 2 for virus examples).
Table 2.
Paired Receptor Ligands That Function as Entry Receptors for Viruses
| Cellular ligands for paired receptors | Viral opportunist | Refs |
|---|---|---|
| CD80 and CD86 | Adenovirus subgroup B | [65] |
| Phosphatidylethanolamine and phosphatidylserine | Dengue virus | [32] |
| CEACAM1 | Mouse hepatitis virus | [66] |
| PVR | Bovine herpesvirus, pseudorabies virus, poliovirus | 67, 68 |
| Nectin2 | Herpes simplex virus type 2 | [69] |
| Sialylated glycans |
Human JC polyomavirus, BK virus, mouse polyomavirus mPy, bovine parvovirus, minute virus of mice, porcine rotavirus, bovine coronavirus, human coronavirus OC43, enterovirus 70, influenza A, B, and C viruses, human parainfluenza 1 virus, human parainfluenza 3 virus, Newcastle disease virus, Sendai virus | 70, 71, 72, 73, 74, 75, 76, 77 |
| HLA-E | ND | |
| CD200 | ND |
In particular, sialic acids are used widely as adhesion and/or entry receptors for viruses. It is possible that the ubiquitous expression of sialic acids makes them an attractive target as viruses can adhere to and thereafter invade many types of cell. Other paired receptor ligands that are used as entry receptors for viruses are not as widely expressed. A classic example is PVR, which was discovered for its role as the entry receptor for poliovirus. Years later, it was shown that PVR binds the paired receptors DNAM1 and TIGIT 8, 38, 39. PVR is also a ligand for CD96 [40], although it is unclear whether this receptor has an inhibitory or activating function.
Bacteria can also use the paired receptor ligands as entry receptors. For example, Helicobacter pylori binds CEACAM1 using its HopQ protein to facilitate translocation of its virulent protein CagA into host cells [41]. This is of particular clinical importance as CagA has been implicated in the promotion of gastric cancer and other pathologies [42].
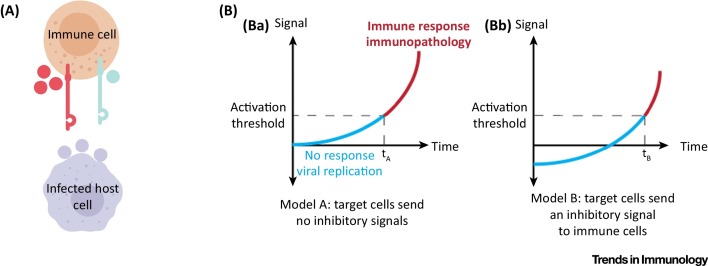
Altogether, given what we know about the functions of paired receptors in the immune system, there is a great deal of evolutionary sense behind the use of shared ligands as entry receptors, especially since is seems as if the inhibitory receptors of the paired receptors were developed prior to the development of the activating ones (Figure 2 A). The expression of shared ligands may shield the invading pathogens during the early stages of cellular infection, where they are generally more vulnerable to elimination by immune cells. In addition, the baseline immune-inhibitory property of a cell expressing shared ligands also means that there is a lag period before the cell can be tipped towards an immune-activating state (Figure 2B). This would allow the pathogen to proliferate more, reaching higher numbers and thus increasing its chances of surviving immune attack when it starts (Figure 2B). Using a shared ligand as an entry receptor ensures these benefits, and provides the pathogen with a distinct advantage in outperforming immune pressure.
Figure 2.
Using a Shared Ligand as a Cellular Entry Receptor Is Advantageous for Pathogens. (A) Evolutionary pressure favours viruses that use the shared ligands of paired receptors as entry and/or adhesion receptors. Host cells that express a shared ligand (shown in purple) are relatively protected from immune system attack because the shared ligand sends a dominant inhibitory signal to immune cells expressing the paired inhibitory (marked in red) and coactivating (marked in black) receptors. Therefore, viruses that enter and reproduce in cells expressing a shared ligand can avoid an immune response. Virus-infected host cells that do not express a shared ligand can be killed by the immune system. (B) Invading a host cell that sends an inhibitory signal to immune cells allows pathogens more time to replicate safely. (Ba) A host cell sends no net signal to the immune system at the initiation of the pathological process (t = 0). This is true of several cell types, such as beta cells in the pancreas as well as some cells in immune privileged sites throughout the body. The time that the host cell takes to produce a signal that is sufficient to activate immune cells is termed tA. (Bb) If the host cell sends an inhibitory signal to the immune system at baseline (through its expression of a shared ligand), then the same processes that induce immune-activating signals will take longer to cross the threshold required to activate the immune system (tB > tA). This lag period (tB − tA) allows for greater viral replication and increased pathology to develop before immune intervention (blue portion of the graph). In turn, the larger viral population is more likely to survive immune attack. Onset of immunopathology is indicated by the red portion of the graph.
However, that pathogens use ligands for paired receptors as their entry receptors might be viewed as risky because the ligand can also be recognised by a coactivating receptor. Thus, we hypothesise that some pathogens will target the inhibitory receptor of the pair to further inhibit and escape immune recognition. Indeed, as mentioned above some viruses and bacteria directly target the paired receptors themselves.
An example of a bacterial pathogen that targets paired receptors is Fusobacterium nucleatum, which uses its Fap2 protein to bind TIGIT only (and not its coactivating partner DNAM1) and inhibits immune cell activities [43].
Using Paired Receptors for Therapy and Implication for Education
Given that viruses use ligands of the paired receptors as entry proteins and that tumours can upregulate the expression of paired receptor ligands to inhibit immune cell activity, it is possible that viruses can be used for tumour therapeutic purposes. If a specific type of cancer overexpresses a shared ligand – for example, PVR in pancreatic cancer [35] – then a virus that uses the same shared ligand as an entry receptor and that can lyse the cells could be used therapeutically. In the case of pancreatic cancer, a poliovirus strain that is engineered to be oncolytic [44] could prove useful in combating this otherwise lethal disease. This idea is particularly attractive given the fact that patients with pancreatic cancer who have higher expression levels of PVR (and would therefore potentially be more responsive to oncolytic viral therapy) currently have poorer prognoses [35]. Using this strategy, immune evasion, which is critical for cancer cell survival, may turn out to be a targetable weak spot.
Antibodies against checkpoint receptors such as anti-PD1 (Keytruda) are currently very successfully used for the treatment of various tumours. Similarly, using an array of agonistic, antagonistic, and bispecific antibodies against the paired receptors (which are difficult to obtain due to the high resemblance of their extracellular portions), it is possible to tip the balance of the immune response in any direction. Agonism of a coactivating receptor or antagonism of an inhibitory receptor of the paired receptors could encourage the immune system to fight off pathogens or cancer; the converse response could be used to treat a range of autoimmune disorders and allergy.
Phase I trials are already underway examining the use of a TIGIT-blocking antibody in the treatment of solid tumours. Bispecific antibodies could be used to help their homing to the correct cell types, and to avoid some of the side effects that are associated with biological therapies. For example, we previously generated a bispecific antibody targeting the CC chemokine receptor 3, that preferentially recognises this receptor on mouse eosinophils, and the inhibitory paired receptor CD300a [45]. This antibody was used in a mouse model of asthma and showed efficacy in reducing eosinophilia and lung inflammation [45]. Similarly, we developed a bispecific antibody that targets IgE and CD300a on mast cells [37]. Given the extensive involvement of paired receptors in different processes, similar antibody-based strategies could be tested in various diseases.
MHC molecules are involved in education processes of both T cells and NK cells [46]. MHC molecules are important in regulating the strength of T cell reactivity. A too strong or a too weak signal will lead to deletion of a T cell clone during the course of the education process. As the paired receptors are also balanced, immune cells might be educated via the interaction of the shared receptors with their shared ligands, as was recently shown for NK cells [47].
Concluding Remarks
In this review, we examine the paired receptors from several perspectives. The curious phenomenon of two receptors sharing a ligand but having opposite effects on immune cells has exceptionally far-reaching consequences. We describe that this is essentially a form of carefully balanced inhibition because inhibition is dominant. We propose that the paired receptors are sometimes not truly paired and that they can be separated temporally and/or clonally, or sometimes being uncoupled in the even when expressed on the same cell, in the immunological synapse. This enables the co-stimulating partner of the pair to be active in some situations.
The biochemically diverse shared ligands of the paired receptors are no less fascinating. Some ligands are upregulated in various pathologies, or are used by pathogens to infect cells. We hypothesise that using the shared ligands as entry or adhesion receptors is beneficial for pathogens because it better protects them from immune attack (as inhibition is the dominant response). We suggest that we can use this property to generate oncolytic viruses that could be used to treat tumours that upregulate shared ligands. Furthermore, we hypothesise that some pathogens target the inhibitory receptor of the pair to further inhibit and escape immune recognition. We also propose that the paired receptors might be involved in education of various immune cells.
Looking at the paired receptors as a whole, and understanding the intricacy of this system in both health and disease may prove useful for clinical applications. These receptors have already been linked to many immunological processes, and many more roles await discovery (see Outstanding Questions). Immunotherapy has undergone great improvements in the past few years, and we hope that understanding of the function of paired receptors will continue to contribute to this success.
Outstanding Questions.
Do additional paired receptors and paired ligands exist?
What are the mechanisms controlling the expression of the paired ligands? Better understanding of the regulation of the paired ligand expression is also critical to understand the biology of paired receptors.
Are paired receptors involved in the education of additional immune cell subsets, other than NK cells, for example of mast cells or eosinophils?
Can pathogens affect the expression of the paired receptors and enhance the expression of the inhibitory partner to escape immune attack?
Can the expression of the paired receptors and their ligands be manipulated for therapy against cancer and pathogens?
Will paired receptors be found and function in pathogens and in nonimmune cells and if so, what is their purpose?
Acknowledgements
We thank N. Stein for excellent ideas and discussions and for the careful editing of the manuscript. F.L.-S. is supported by grants from the United States–Israel Binational Science Foundation (BSF) and the Israel Science Foundation (ISF). O.M. is supported by a European Research Council (ERC) advanced grant and by a grant from the ISF.
References
- 1.Ni L., Dong C. New checkpoints in cancer immunotherapy. Immunol. Rev. 2017;276:52–65. doi: 10.1111/imr.12524. [DOI] [PubMed] [Google Scholar]
- 2.Bulfone-Paus S. Positive and negative signals in mast cell activation. Trends Immunol. 2017;38:657–667. doi: 10.1016/j.it.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Guillerey C. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 4.Barclay A.N., Hatherley D. The counterbalance theory for evolution and function of paired receptors. Immunity. 2008;29:675–678. doi: 10.1016/j.immuni.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colonna M., Samaridis J. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 6.Bour-Jordan H. Intrinsic and extrinsic control of peripheral T-cell tolerance by costimulatory molecules of the CD28/B7 family. Immunol. Rev. 2011;241:180–205. doi: 10.1111/j.1600-065X.2011.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lankry D. Expression and function of CD300 in NK cells. J. Immunol. 2010;185:2877–2886. doi: 10.4049/jimmunol.0903347. [DOI] [PubMed] [Google Scholar]
- 8.Stanietsky N. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munitz A. The inhibitory receptor IRp60 (CD300a) suppresses the effects of IL-5, GM-CSF, and eotaxin on human peripheral blood eosinophils. Blood. 2006;107:1996–2003. doi: 10.1182/blood-2005-07-2926. [DOI] [PubMed] [Google Scholar]
- 10.Campbell K.S. Signaling through human killer cell activating receptors triggers tyrosine phosphorylation of an associated protein complex. Eur. J. Immunol. 1998;28:599–609. doi: 10.1002/(SICI)1521-4141(199802)28:02<599::AID-IMMU599>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 11.Markel G. CD66a interactions between human melanoma and NK cells: a novel class I MHC-independent inhibitory mechanism of cytotoxicity. J. Immunol. 2002;168:2803–2810. doi: 10.4049/jimmunol.168.6.2803. [DOI] [PubMed] [Google Scholar]
- 12.Muller M.M. Homophilic adhesion and CEACAM1-S regulate dimerization of CEACAM1-L and recruitment of SHP-2 and c-Src. J. Cell Biol. 2009;187:569–581. doi: 10.1083/jcb.200904150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y. Identification of CD112R as a novel checkpoint for human T cells. J. Exp. Med. 2016;213:167–176. doi: 10.1084/jem.20150785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ali S.R. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J. Exp. Med. 2014;211:1231–1242. doi: 10.1084/jem.20131853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braud V.M. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 16.Akkaya M., Barclay A.N. Heterogeneity in the CD200R paired receptor family. Immunogenetics. 2010;62:15–22. doi: 10.1007/s00251-009-0415-6. [DOI] [PubMed] [Google Scholar]
- 17.Akkaya M., Barclay A.N. How do pathogens drive the evolution of paired receptors? Eur. J. Immunol. 2013;43:303–313. doi: 10.1002/eji.201242896. [DOI] [PubMed] [Google Scholar]
- 18.Arase H., Lanier L.L. Specific recognition of virus-infected cells by paired NK receptors. Rev. Med. Virol. 2004;14:83–93. doi: 10.1002/rmv.422. [DOI] [PubMed] [Google Scholar]
- 19.Martinet L., Smyth M.J. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- 20.Johnston R.J. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Arase H. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 22.Adams E.J. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:10128–10133. doi: 10.1073/pnas.0703735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachelet I. Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J. Biol. Chem. 2006;281:27190–27196. doi: 10.1074/jbc.M602359200. [DOI] [PubMed] [Google Scholar]
- 24.Gangwar R.S., Levi-Schaffer F. Eosinophils interaction with mast cells: the allergic effector unit. Methods Mol. Biol. 2014;1178:231–249. doi: 10.1007/978-1-4939-1016-8_20. [DOI] [PubMed] [Google Scholar]
- 25.Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood. 2013;121:1951–1960. doi: 10.1182/blood-2012-09-435057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCann F.E. The size of the synaptic cleft and distinct distributions of filamentous actin, ezrin, CD43, and CD45 at activating and inhibitory human NK cell immune synapses. J. Immunol. 2003;170:2862–2870. doi: 10.4049/jimmunol.170.6.2862. [DOI] [PubMed] [Google Scholar]
- 27.Rudd C.E. CD28 and CTLA-4 coreceptor expression and signal transduction. Immunol. Rev. 2009;229:12–26. doi: 10.1111/j.1600-065X.2009.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viant C. SHP-1-mediated inhibitory signals promote responsiveness and anti-tumour functions of natural killer cells. Nat. Commun. 2014;5 doi: 10.1038/ncomms6108. [DOI] [PubMed] [Google Scholar]
- 29.Elliott J.M., Yokoyama W.M. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 2011;32:364–372. doi: 10.1016/j.it.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pradeu T. The speed of change: towards a discontinuity theory of immunity? Nat. Rev. Immunol. 2013;13:764–769. doi: 10.1038/nri3521. [DOI] [PubMed] [Google Scholar]
- 31.Burshtyn D.N., Morcos C. The expanding spectrum of ligands for leukocyte Ig-like receptors. J. Immunol. 2016;196:947–955. doi: 10.4049/jimmunol.1501937. [DOI] [PubMed] [Google Scholar]
- 32.Carnec X. The phosphatidylserine and phosphatidylethanolamine receptor CD300a binds dengue virus and enhances infection. J. Virol. 2015;90:92–102. doi: 10.1128/JVI.01849-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster-Cuevas M. Human herpesvirus 8 K14 protein mimics CD200 in down-regulating macrophage activation through CD200 receptor. J. Virol. 2004;78:7667–7676. doi: 10.1128/JVI.78.14.7667-7676.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitenshtein A. CEACAM1-mediated inhibition of virus production. Cell Rep. 2016;15:2331–2339. doi: 10.1016/j.celrep.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiwada S. Clinical significance of CD155 expression in human pancreatic cancer. Anticancer Res. 2015;35:2287–2297. [PubMed] [Google Scholar]
- 36.Sabato V. CD300a is expressed on human basophils and seems to inhibit IgE/FcepsilonRI-dependent anaphylactic degranulation. Cytom. B Clin. Cytom. 2012;82:132–138. doi: 10.1002/cyto.b.21003. [DOI] [PubMed] [Google Scholar]
- 37.Bachelet I. Abrogation of allergic reactions by a bispecific antibody fragment linking IgE to CD300a. J. Allergy Clin. Immunol. 2006;117:1314–1320. doi: 10.1016/j.jaci.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 38.Yu X. The surface protein TIGIT suppresses T cell activation by promoting the generation of mature immunoregulatory dendritic cells. Nat. Immunol. 2009;10:48–57. doi: 10.1038/ni.1674. [DOI] [PubMed] [Google Scholar]
- 39.Boles K.S. A novel molecular interaction for the adhesion of follicular CD4 T cells to follicular DC. Eur. J. Immunol. 2009;39:695–703. doi: 10.1002/eji.200839116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan C.J. The receptors CD96 and CD226 oppose each other in the regulation of natural killer cell functions. Nat. Immunol. 2014;15:431–438. doi: 10.1038/ni.2850. [DOI] [PubMed] [Google Scholar]
- 41.Javaheri A. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat. Microbiol. 2016;2 doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- 42.Tohidpour A. CagA-mediated pathogenesis of Helicobacter pylori. Microb. Pathog. 2016;93:44–55. doi: 10.1016/j.micpath.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Gur C. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown M.C., Gromeier M. Oncolytic immunotherapy through tumor-specific translation and cytotoxicity of poliovirus. Discov. Med. 2015;19:359–365. [PMC free article] [PubMed] [Google Scholar]
- 45.Munitz A. Reversal of airway inflammation and remodeling in asthma by a bispecific antibody fragment linking CCR3 to CD300a. J. Allergy Clin. Immunol. 2006;118:1082–1089. doi: 10.1016/j.jaci.2006.07.041. [DOI] [PubMed] [Google Scholar]
- 46.He Y., Tian Z. NK cell education via nonclassical MHC and non-MHC ligands. Cell. Mol. Immunol. 2016;14:321–330. doi: 10.1038/cmi.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y. Contribution of inhibitory receptor TIGIT to NK cell education. J. Autoimmun. 2017;81:1–12. doi: 10.1016/j.jaut.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Arad G. Binding of superantigen toxins into the CD28 homodimer interface is essential for induction of cytokine genes that mediate lethal shock. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spits H. NK cells and type 1 innate lymphoid cells: partners in host defense. Nat. Immunol. 2016;17:758–764. doi: 10.1038/ni.3482. [DOI] [PubMed] [Google Scholar]
- 50.Tu M.M. Licensed and unlicensed NK cells: differential roles in cancer and viral control. Front. Immunol. 2016;7:166. doi: 10.3389/fimmu.2016.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burian A. HLA-F and MHC-I open conformers bind natural killer cell Ig-like receptor KIR3DS1. PLoS One. 2016;11 doi: 10.1371/journal.pone.0163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Beltran W.F. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016;17:1067–1074. doi: 10.1038/ni.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sivori S. A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood. 2010;116:1637–1647. doi: 10.1182/blood-2009-12-256586. [DOI] [PubMed] [Google Scholar]
- 54.Alvarez Y. The CD300a (IRp60) inhibitory receptor is rapidly up-regulated on human neutrophils in response to inflammatory stimuli and modulates CD32a (FcgammaRIIa) mediated signaling. Mol. Immunol. 2008;45:253–258. doi: 10.1016/j.molimm.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bachelet I. Suppression of normal and malignant kit signaling by a bispecific antibody linking kit with CD300a. J. Immunol. 2008;180:6064–6069. doi: 10.4049/jimmunol.180.9.6064. [DOI] [PubMed] [Google Scholar]
- 56.Bachelet I. The inhibitory receptor IRp60 (CD300a) is expressed and functional on human mast cells. J. Immunol. 2005;175:7989–7995. doi: 10.4049/jimmunol.175.12.7989. [DOI] [PubMed] [Google Scholar]
- 57.Gibbs B.F. Expressions and inhibitory functions of CD300a receptors on purified human basophils. Exp. Dermatol. 2012;21:884–886. doi: 10.1111/exd.12018. [DOI] [PubMed] [Google Scholar]
- 58.Nissim Ben Efraim A.H. The inhibitory receptor CD300a is up-regulated by hypoxia and GM-CSF in human peripheral blood eosinophils. Allergy. 2013;68:397–401. doi: 10.1111/all.12092. [DOI] [PubMed] [Google Scholar]
- 59.Takahashi M. Human CD300C delivers an Fc receptor-gamma-dependent activating signal in mast cells and monocytes and differs from CD300A in ligand recognition. J. Biol. Chem. 2013;288:7662–7675. doi: 10.1074/jbc.M112.434746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gray-Owen S.D., Blumberg R.S. CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 61.Shiratori I. Down-regulation of basophil function by human CD200 and human herpesvirus-8 CD200. J. Immunol. 2005;175:4441–4449. doi: 10.4049/jimmunol.175.7.4441. [DOI] [PubMed] [Google Scholar]
- 62.Cameron C.M. Myxoma virus M141R expresses a viral CD200 (vOX-2) that is responsible for down-regulation of macrophage and T-cell activation in vivo. J. Virol. 2005;79:6052–6067. doi: 10.1128/JVI.79.10.6052-6067.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langlais C.L. Rhesus rhadinovirus R15 encodes a functional homologue of human CD200. J. Virol. 2006;80:3098–3103. doi: 10.1128/JVI.80.6.3098-3103.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwarz F. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. 2017;36:751–760. doi: 10.15252/embj.201695581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van de Ven R. Selective transduction of mature DC in human skin and lymph nodes by CD80/CD86-targeted fiber-modified adenovirus-5/3. J. Immunother. 2009;32:895–906. doi: 10.1097/CJI.0b013e3181b56deb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taguchi F., Hirai-Yuki A. Mouse hepatitis virus receptor as a determinant of the mouse susceptibility to MHV infection. Front. Microbiol. 2012;3:68. doi: 10.3389/fmicb.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan G. Neutralization of poliovirus by cell receptors expressed in insect cells. J. Virol. 1990;64:4697–4702. doi: 10.1128/jvi.64.10.4697-4702.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geraghty R.J. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 69.Lopez M. Nectin2alpha (PRR2alpha or HveB) and nectin2delta are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 2000;74:1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maginnis M.S. JC polyomavirus attachment, entry, and trafficking: unlocking the keys to a fatal infection. J. Neurovirol. 2015;21:601–613. doi: 10.1007/s13365-014-0272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krempl C. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 1995;380:371–374. doi: 10.1007/978-1-4615-1899-0_60. [DOI] [PubMed] [Google Scholar]
- 72.Nokhbeh M.R. Enterovirus 70 binds to different glycoconjugates containing alpha2,3-linked sialic acid on different cell lines. J. Virol. 2005;79:7087–7094. doi: 10.1128/JVI.79.11.7087-7094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson F.B. Attachment of bovine parvovirus to sialic acids on bovine cell membranes. J. Gen. Virol. 2004;85:2199–2207. doi: 10.1099/vir.0.79899-0. [DOI] [PubMed] [Google Scholar]
- 74.Nam H.J. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 2006;281:25670–25677. doi: 10.1074/jbc.M604421200. [DOI] [PubMed] [Google Scholar]
- 75.Haselhorst T. Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 2009;5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 76.Korownyk C. Antiviral medications for influenza. Can. Fam. Physician. 2015;61:351. [PMC free article] [PubMed] [Google Scholar]
- 77.Matrosovich M. Sialic acid receptors of viruses. Top. Curr. Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]