Highlights
-
•
During July 2014–June 2017, 854,575 human coronavirus tests were reported to NREVSS.
-
•
The percent of positive human coronavirus tests peaked during December–March.
-
•
Different human coronavirus species predominated in different years.
-
•
Human coronavirus OC43 was the most commonly detected species.
Keywords: Coronavirus, Epidemiology, Respiratory tract infection, Respiratory virus
Abstract
Background
Human coronaviruses (HCoVs) -OC43, -229E, -NL63 and -HKU1 cause upper and lower respiratory tract infections. HCoVs are globally distributed and the predominant species may vary by region or year. Prior studies have shown seasonal patterns of HCoV species and annual variation in species prevalence but national circulation patterns in the US have not yet been described.
Objectives
To describe circulation patterns of HCoVs -OC43, -229E, -NL63 and -HKU1 in the US.
Study design
We reviewed real-time reverse transcription polymerase chain reaction (rRT-PCR) test results for HCoV-OC43, -229E, -NL63 and -HKU1 reported to The National Respiratory and Enteric Virus Surveillance System (NREVSS) by U.S. laboratories from July 2014–June 2017. We calculated the total number of tests and percent positive by week. For a subset of HCoV positive submissions with age and sex of the patient available, we tested for differences in age and sex across the four HCoV species using Chi Square and Kruskal Wallace tests.
Results
117 laboratories reported 854,575 HCoV tests; 2.2% were positive for HCoV-OC43, 1.0% for HCoV-NL63, 0.8% for HCoV-229E, and 0.6% for HCoV-HKU1. The percentage of positive tests peaked during December – March each year. No significant differences in sex were seen across species, although a significant difference in age distribution was noted.
Conclusions
Common HCoVs may have annual peaks of circulation in winter months in the US, and individual HCoVs may show variable circulation from year to year. Different HCoV species may be detected more frequently in different age groups. Further years of data are needed to better understand patterns of activity for HCoVs.
1. Background
Human coronaviruses (HCoVs) HCoV-NL63, HCoV-HKU1, HCoV-229E, and HCoV-OC43 circulate worldwide and cause a range of respiratory symptoms [1]. Infections are often asymptomatic or associated with mild to moderate upper respiratory tract illness in immunocompetent children and adults; HCoVs are considered the second most common cause of the common cold [2]. Infections can also result in lower respiratory tract illness including bronchiolitis and pneumonia, especially in immunocompromised individuals, infants, and older adults [1]. Increased availability of molecular test methods and more frequent testing for multiple respiratory pathogens have allowed for opportunities to characterize circulation patterns of individual HCoVs.
Although HCoVs are globally distributed, the predominant species may vary by region or year [[3], [4], [5]]. Previous studies have shown seasonal patterns of HCoV species and annual variation in species prevalence [[3], [4], [6]]. However, national circulation patterns across the United States have not been described and few studies have described circulation of all four HCoVs across multiple years [6].
2. Objectives
Our objective was to describe laboratory detections of HCoVs -NL63, -HKU1, -229E, and -OC43 in the United States during 2014–2017, using data collected by The National Respiratory and Enteric Virus Surveillance System (NREVSS) [7].
3. Study design
NREVSS is a passive surveillance network established by the U.S. Centers for Disease Control and Prevention (CDC) in the 1980s that collects specimen test results for several respiratory and enteric viruses from multiple laboratories across the United States [7]. NREVSS currently collects data from three different sources: directly from clinical, state, and local laboratories; indirectly from state or local partners on behalf of laboratories within their jurisdictions; and indirectly through the Public Health Laboratory Interoperability Project (PHLIP).
PHLIP is a collaborative partnership between CDC, state and local public health labs (PHLs), and the Association of Public Health Laboratories (APHL) to strengthen the submission of automated specimen-level surveillance laboratory test results directly to CDC [8]. Laboratories submitting to NREVSS via PHLIP submit specimen level results for respiratory virus tests along with patient demographics, such as sex and age. The remaining majority of NREVSS participants (i.e. non-PHLIP submitters) submit weekly aggregates of positive detections for the four HCoV species by RT-PCR along with the aggregate number of RT-PCR HCoV tests performed.
To better understand HCoV circulation in the US, we assessed reports of specimens tested for HCoVs, and submitted to NREVSS during July 1, 2014–June 30, 2017. Reporting laboratories were excluded if they did not report HCoV results at the species level. We summarized the total number of HCoV tests submitted during the study period and calculated the overall percent positive for each HCoV species by week. Test numbers and positive HCoV results were also summarized separately for individual US census regions.
To understand demographics of patients with HCoV detections we further analyzed the subset of NREVSS data reported through PHLIP. We selected reports of specimens tested for all four HCoV species and calculated the percent positive for each HCoV species. We used all PHLIP reports with a single positive HCoV detection to test differences in age distribution among the four HCoV species using the Kruskal Wallis Test. Differences in sex distribution among the four HCoV species were tested using the Chi Square Test. We then summarized viral co-detections for each HCoV species using reports of specimens tested for all four HCoVs and which were also tested for the following viruses: parainfluenza viruses 1–4, respiratory syncytial virus (RSV), human metapneumovirus, human adenovirus, rhinovirus/enterovirus, influenza A and influenza B. Analysis was performed using R version 3.3.1.
4. Results
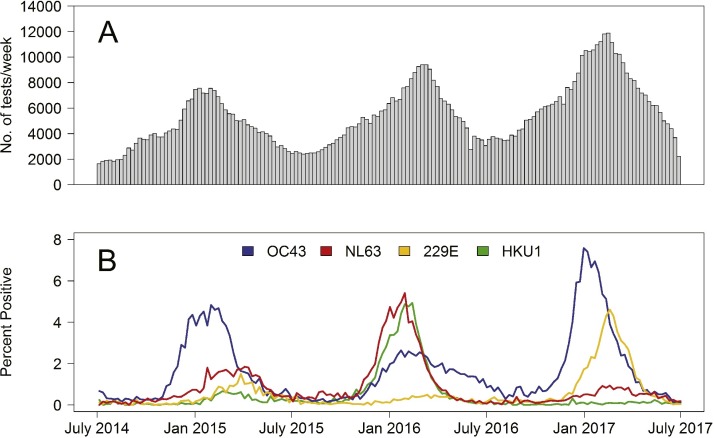
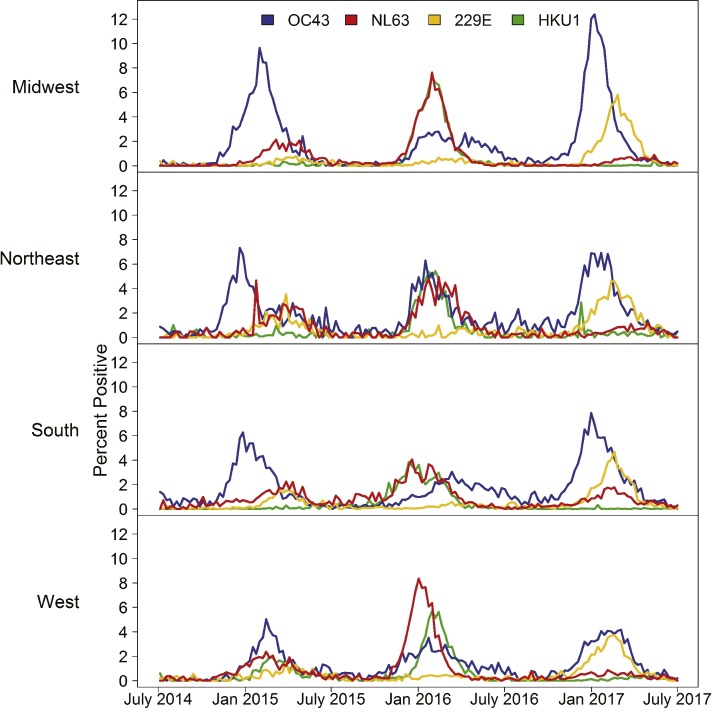
During July 1, 2014–June 30, 2017, 854,575 HCoV tests were reported by 117 laboratories in 42 states submitting to NREVSS. Overall, 18,804 (2.2%) were positive for HCoV-OC43, 8558 (1.0%) for HCoV-NL63, 7001 (0.8%) for HCoV-229E and 5225 (0.6%) for HCoV-HKU1. The number of HCoV tests submitted to NREVSS per week varied seasonally, with the testing peak occurring each year in winter, generally between December and March (Fig. 1 A). Overall HCoV testing increased during the three years (Fig. 1A). The percent of HCoV positive tests varied throughout each year, and also peaked each year between December and March. The percent positive varied annually by HCoV species (Fig. 1B). HCoV-OC43 demonstrated a distinct peak each of the three years, with a less pronounced peak in 2016. HCoV-NL63 and HCoV-HKU1 demonstrated similar patterns to one another; both had a small peak in 2015 and larger peaks in 2016, although only HCoV-NL63 had a small peak in 2017. HCoV-229E showed a slight peak in 2015, no peak in 2016 and a relatively large peak in 2017 (Fig. 1B). The highest percent positive for any single species was 7.6% of tests positive for HCoV-OC43 in the week beginning December 31, 2016. Across each census region, minimal differences in seasonal and annual patterns of percentage of tests positive for each HCoV species were seen compared to national data (Fig. 2 ). The most notable difference was in the percentage of positive HCoV-OC43 tests during the 2016–2017 season, with the West region showing a peak percent positive of 5.0% and the Midwest region showing a peak percent positive of 12.4%.
Fig. 1.
A) The number of tests performed to detect any of the four human coronaviruses (HCoVs) -OC43, -NL63, -229E, and -HKU1 reported to the National Respiratory and Enteric Viruses Surveillance System (NREVSS) by week, July 2014–June 2017, B) The percentage of tests positive for HCoVs -OC43, -NL63, -229E, and -HKU1 reported to NREVSS by week, July 2014–June 2017.
Fig. 2.
The percentage of tests positive for human coronaviruses (HCoVs) -OC43, -NL63, -229E, and -HKU1 reported to the National Respiratory and Enteric Viruses Surveillance System (NREVSS) by week, stratified by US Census Region, July 2014–June 2017.
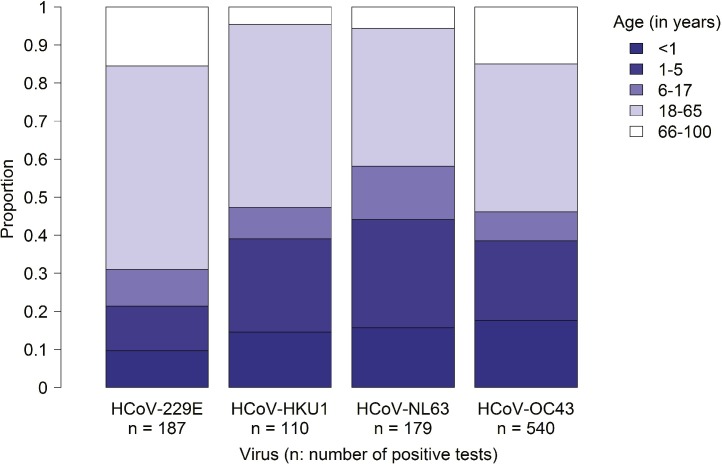
Data reported to NREVSS through PHLIP was further analyzed to understand sex and age characteristics. During the study period, 20,806 specimens tested for all four HCoVs were submitted via PHLIP from six laboratories. Overall 1569 tests (7.5%) were positive for any HCoV; 852 (4.1%) were positive for HCoV-OC43, 255 (1.2%) for HCoV-NL63, 335 (1.6%) for HCoV-229E and 154 (0.7%) for HCoV-HKU1. The majority of specimens with a single HCoV detection (92.2% of 1543 specimens) included the sex of the patient, and approximately half (50.6%) of all these HCoV detections were reported as male. No significant difference was seen in the sex distribution between the four HCoVs (p = 0.19). Age was available for 1016 (67%) of specimens with a single HCoV detection and the median patient age of these specimens was 23 years (range 0–96 years). The patient age distribution of specimens with a single HCoV detection was significantly different between species (p < 0.01) (Fig. 3 ). The median ages of patients with specimens testing positive for a single HCoV species were as follows: HCoV-OC43, 24 years; HCoV-NL63, 11 years; HCoV-229E, 30 years; and HCoV-HKU1, 19 years. For HCoV-OC43, HCoV-NL63, and HCoV-HKU1 >45% of detections were in children <18 years old (Fig. 3). By contrast, 31% of HCoV-229E detections were in children <18 years old (Fig. 3).
Fig. 3.
Age distribution of human coronaviruses (HCoVs) -229E, -HKU1, -NL63 and -OC43 positive tests with age available reported to NREVSS via the Public Health Laboratory Interoperability Project (PHLIP), from July 2014–June 2017. Note, specimens with more than one coronavirus detected were excluded (n = 18).
Among the 1569 HCoV positive detections reported via PHLIP, 1538 (98%) were also tested for parainfluenza viruses 1–4, respiratory syncytial virus (RSV), human metapneumovirus, human adenovirus, rhinovirus/enterovirus, influenza A and influenza B. Among these, 68.6% reported a single HCoV species detection only, 1.7% reported two or more HCoV species, and 30.2% detected another respiratory virus. The most common HCoV co-detections were HCoV-OC43 with HCoV-NL63 (8 specimens, 0.5%), and HCoV-OC43 with HCoV-229E (8 specimens, 0.5%). The most common co-detected non-HCoV viruses were RSV (11% of HCoV positive specimens), rhinovirus/enterovirus (6.6%), and influenza A (5.7%); 51 (3.3%) specimens had ≥2 viral species detected in addition to HCoV. Co-detection patterns were broadly similar among the four HCoVs (Table 1 ).
Table 1.
Frequency and percentage (in parentheses) of viral co-detections for by individual human coronavirus (HCoV) detection and all HCoV detections, reported to NREVSS via PHLIP from July 2014–June 2017.
| HCoV-OC43 | HCoV-NL63 | HCoV-229E | HCoV-HKU1 | PIV | RSV | HMPV | HAdV | RV/EV | Flu A | Flu B | Any non-HCoV co-detection | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HCoV-OC43 n = 836 | 836 (100) | 8 (1.0) | 8 (1.0) | 4 (0.5) | 28 (3.3) | 100 (12.0) | 23 (2.8) | 54 (6.5) | 59 (7.1) | 44 (5.3) | 10 (1.2) | 259 (31.0) |
| HCoV-NL63 n = 253 | 8 (3.2) | 253 (100) | 3 (1.2) | 5 (2.0) | 5 (2.0) | 26 (10.3) | 10 (4.0) | 13 (5.1) | 19 (7.5) | 11 (4.3) | 0 (0.0) | 74 (29.2) |
| HCoV-229E n = 325 | 8 (2.5) | 3 (0.9) | 325 (100) | 0 (0.0) | 16 (4.9) | 21 (6.5) | 9 (2.8) | 11 (3.4) | 11 (3.4) | 27 (8.3) | 5 (1.5) | 89 (27.4) |
| HCoV-HKU1 n = 151 | 4 (2.6) | 5 (3.3) | 0 (0.0) | 151 (100) | 5 (3.3) | 21 (13.9) | 5 (3.3) | 10 (6.6) | 13 (8.6) | 8 (5.3) | 2 (1.3) | 53 (35.1) |
| All HCoVs | 836 (54.4) | 253 (16.4) | 325 (21.1) | 151 (9.8) | 54 (3.5) | 164 (10.7) | 47 (3.1) | 86 (5.6) | 101 (6.6) | 87 (5.7) | 17 (1.1) | 465 (30.2) |
PIV = parainfluenza virus, RSV = respiratory syncytial virus, HMPV = human metapneumovirus, HAdV = human adenovirus, RV/EV = rhinovirus/enterovirus, Flu A = influenza A, Flu B = influenza B
5. Discussion
This report is the first to describe the national patterns of circulation of the four common HCoV species in the United States during a multi-year period. During the study period, HCoVs showed a peak prevalence during December– March each year, which coincides with the winter respiratory virus season [[7], [9]]. HCoV-OC43 was the most commonly detected HCoV with 2.2% of all tests positive. Different HCoV species predominated in different years; HCoV-OC43 appeared to peak annually, while HCoV-NL63, HCoV-HKU1, and HCoV-229E showed more variability, with distinct peaks in one or two of the three years studied. This is consistent with previously published site-specific data indicating that individual species may only demonstrate peak activity every 2–3 years [[10], [11]].
Factors associated with annual differences in activity for HCoV species are currently unknown. Individual HCoV species activity could fluctuate independently, or cross-immunity within or between Alphacoronavirinae (HCoV-229E and HCoV-NL63) and Betacoronavirinae (HCoV-HKU1 and HCoV-OC43) might affect annual activity of the four HCoVs [[4], [12]]. During the study period, HCoV-229E and HCoV-NL63 did not show large contemporaneous peaks of activity, although both showed smaller peaks of activity in 2014–2015. When data was visualized by census region, annual and seasonal patterns were similar to those seen nationally (Fig. 2).
The age distribution of patients with reported HCoV infections differed between HCoV species (Fig. 3). HCoV-229E detections were more common in adults >18 years old compared to HCoV-HKU1, -NL63, and -OC43. HCoV-229E has previously been reported as disproportionately affecting immunocompromised individuals relative to the other HCoV species [4], possibly affecting the median reported age at infection. HCoV-NL63 showed the lowest median age of infection, and has been shown to be associated with croup in young children [13], which may lower the average age of infection. Non-HCoV viral co-detections were seen in 30% of specimens positive for HCoV, and 3.3% of specimens had two or more co-detected viral species. The clinical impact of coronaviruses in co-detections is not fully understood, with prior studies reporting both increased and unchanged morbidity and mortality with respiratory viral co-detections [14]. Single infections with HCoVs have been associated with morbidity due to lower and upper respiratory tract infections [4], however Prill et al demonstrated that HCoVs were not found more frequently in children hospitalized for acute respiratory illness and/or fever than asymptomatic controls [15].
There were limitations to this report. NREVSS is a passive, voluntary surveillance system, collecting results from specimens submitted to U.S. laboratories. Many HCoV infections are subclinical or mild, and do not require clinical care; therefore these infections are unlikely to require laboratory testing and would not be captured by NREVSS. The relative proportions of HCoV species reported here may not be representative of all HCoV infections. Within NREVSS all reporters of HCoV surveillance data at the species level were included, including those inconsistently reporting over time. This may result in certain laboratories or regions being overrepresented at certain times e.g. during winter, when the majority of respiratory virus tests are conducted.
Data reported to NREVSS through PHLIP represent a smaller subset of six laboratories that also report additional data including sex and age; data reported through PHLIP may not be representative of the entire NREVSS population. The overall percent of HCoV positive tests varied between PHLIP and NREVSS; the percent of HCoV positive tests submitted to NREVSS via PHLIP was 7.5%, vs 4.6% for NREVSS as a whole. This may result from differences in reporting laboratories; reporting through PHLIP is limited to military, state and public health laboratories, whereas data submitted directly to NREVSS through non-PHLIP sources primarily includes clinical laboratories. State and local public health laboratories test for a variety of reasons including surveillance and public health response to outbreaks and clusters of cases, whereas clinical laboratories often test in the course of managing individual cases.
Within the peak in detections noted during December-March, we were not able to define precise seasonal onset and offset periods for any individual HCoV species. Additionally, we assessed HCoV percent positivity over time based on the date of submission of a report to NREVSS rather than the date of specimen collection, and we anticipate that our findings may reflect this slight delay. Finally, aggregate data reported to NREVSS via PHLIP might include multiple specimens from the same patient, potentially impacting the demographic characteristics of reported cases.
Surveillance of HCoVs is important to determine seasonality and annual circulation patterns. Continued use of respiratory virus multiplex assay panels could facilitate further definition of HCoV circulation through public health surveillance in the future. Further years of data are needed to better understand patterns of activity for HCoVs, and studies with additional epidemiologic data will be useful to better characterize HCoV burden and spectrum of illness.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
Funding
National Center for Immunization and Respiratory Diseases, Centers for Disease Control.
Ethical approval
Collection of data for the National Respiratory and Enteric Virus Surveillance System has been approved by Centers for Disease Control and Prevention as routine public health surveillance.
Competing interests
None declared.
Acknowledgements
Dean Erdman, Xiaoyan Lu, National Respiratory and Enteric Virus Surveillance System laboratories.
Footnotes
This information was presented at IDWeek November 2017, San Diego, CA, USA (abstract number: 63424).
References
- 1.Su S., Wong G., Shi W. Epidemiology, genetic recombination, and pathogenesis of Coronaviruses. Trends Microbiol. 2016;24(June(6)):490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mäkelä M.J., Puhakka T., Ruuskanen O. Viruses and bacteria in the etiology of the common cold. J. Clin. Microbiol. 1998;36(February(2)):539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sipulwa L.A., Ongus J.R., Coldren R.L., Bulimo W.D. Molecular characterization of human coronaviruses and their circulation dynamics in Kenya, 2009–2012. Virol. J. 2016;13(Febraury(18)) doi: 10.1186/s12985-016-0474-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 2010;48(August(8)):2940–2947. doi: 10.1128/JCM.00636-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez S.R., Robinson C.C., Holmes K.V. Detection of four human coronaviruses in respiratory infections in children: a one-year study in Colorado. J. Med. Virol. 2009;81(September(9)):1597–1604. doi: 10.1002/jmv.21541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talbot H.K., Shepherd B.E., Crowe J.E., Jr The pediatric burden of human coronaviruses evaluated for twenty years. Pediatr. Infect. Dis. J. 2009;28(August(8)):682–687. doi: 10.1097/INF.0b013e31819d0d27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes A.K., Prill M.M., Iwane M.K., Gerber S.I. Centers for disease control and prevention (CDC). respiratory syncytial virus–United states, july 2012–June 2014. MMWR Morb. Mortal Wkly. Rep. 2014;63(December(48)):1133–1136. [PMC free article] [PubMed] [Google Scholar]
- 8.2016. Association of Public Health Laboratories: Informatics Initiatives; PHLIP Electronic Laboratory Surveillance Message for Influenza. Available at https://www.aphl.org/programs/informatics/Pages/Technical-Assistance-for-Data-Exchange.aspx. (Accessed 25 August 2017) [Google Scholar]
- 9.2016. Centers for Disease Control and Prevention: Influenza (Flu) The Flu Season. Available at https://www.cdc.gov/flu/about/season/flu-season.htm. (Accessed 25 August 2017) [Google Scholar]
- 10.van der Hoek L., Ihorst G., Sure K. Burden of disease due to human coronavirus NL63 infections and periodicity of infection. J. Clin. Virol. 2010;48(June(2)):104–108. doi: 10.1016/j.jcv.2010.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monto A.S., Lim S.K. The Tecumseh study of respiratory illness: VI. Frequency of and relationship between outbreaks of coronavirus infection. J. Infect. Dis. 1974;129(March(3)):271–276. doi: 10.1093/infdis/129.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkman R., Jebbink M.F., Gaunt E., Rossen J.W., Templeton K.E., Kuijpers T.W., van der Hoek L. The dominance of human coronavirus OC43 and NL63 infections in infants. J. Clin. Virol. 2012;53(February(2)):135–139. doi: 10.1016/j.jcv.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Hoek L., Sure K., Ihorst G. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005;2(August(8)):e240. doi: 10.1371/journal.pmed.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asner S.A., Science M.E., Tran D., Smieja M., Merglen A., Mertz D. Clinical disease severity of respiratory viral co-infection versus single viral infection: a systematic review and meta-analysis. PLoS One. 2014;9(June(6)):e99392. doi: 10.1371/journal.pone.0099392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prill M.M., Iwane M.K., Edwards K.M. Human coronavirus in young children hospitalized for acute respiratory illness and asymptomatic controls. Pediatr. Infect. Dis. J. 2012;31(March(3)):235–240. doi: 10.1097/INF.0b013e31823e07fe. [DOI] [PMC free article] [PubMed] [Google Scholar]