Abstract
Sewage contains a mixed ecosystem of diverse sets of microorganisms, including human pathogenic viruses. Little is known about how conventional as well as advanced treatments of sewage, such as ozonation, reduce the environmental spread of viruses. Analyses for viruses were therefore conducted for three weeks in influent, after conventional treatment, after additional ozonation, and after passing an open dam system at a full-scale treatment plant in Knivsta, Sweden. Viruses were concentrated by adsorption to a positively charged filter, from which they were eluted and pelleted by ultracentrifugation, with a recovery of about 10%. Ion Torrent sequencing was used to analyze influent, leading to the identification of at least 327 viral species, most of which belonged to 25 families with some having unclear classification. Real-time PCR was used to test for 21 human-related viruses in inlet, conventionally treated, and ozone-treated sewage and outlet waters. The viruses identified in influent and further analyzed were adenovirus, norovirus, sapovirus, parechovirus, hepatitis E virus, astrovirus, pecovirus, picobirnavirus, parvovirus, and gokushovirus. Conventional treatment reduced viral concentrations by one to four log10, with the exception of adenovirus and parvovirus, for which the removal was less efficient. Ozone treatment led to a further reduction by one to two log10, but less for adenovirus. This study showed that the amount of all viruses was reduced by conventional sewage treatment. Further ozonation reduced the amounts of several viruses to undetectable levels, indicating that this is a promising technique for reducing the transmission of many pathogenic human viruses.
Keywords: Waste water, Next generation sequencing, Effluent, Adenovirus, Parvovirus
1. Introduction
Pathogenic human and animal viruses found in aquatic environments are usually shed from feces (enteric viruses), urine, and respiratory secretions from the infected host and enter into sewage water. The human viruses belong to different viral families. The most common viruses that are widely dispersed in sewage around the world include hepatitis A virus, hepatitis E virus, rotavirus, adenovirus, norovirus, astrovirus, parvovirus, coronavirus, poliovirus, and other enteroviruses (Hellmer et al., 2014; Laverick et al., 2004; Lodder and de Roda Husman, 2005). Not only human enteric viruses and animal pathogens, but also other viruses can be found in waters contaminated with sewage (Bosch, 1998; Cantalupo et al., 2011). If the pathogens are not removed in the treatment plants, they will be released into natural watersheds where many of them can persist for long periods (Fong and Lipp, 2005; Kotwal and Cannon, 2014). New hosts might be infected with these viruses through direct contact with contaminated water or by drinking it or by eating animals such as mollusks that have filtered and concentrated viruses from sewage-contaminated water (Nenonen et al., 2008).
In most western wastewater treatment plants, raw sewage is treated with combined mechanical, biological, and chemical processes such as screening, flocculation, sedimentation, and filtration. Gross pollutants and most organic and inorganic solids are removed during these steps. The effluent is thereafter either discharged into a receiving water system or reused for other purposes. Little is known, however, about the efficiency of removal of human viral pathogens from sewage by conventional treatment. Several studies have shown that such treatments are efficient for the reduction of the parasites Giardia and Cryptosporium and for bacteria but have little effect on adenoviruses and enteroviruses (Li et al., 2015; Ottoson et al., 2006a; Rodriguez-Manzano et al., 2012). Additional disinfection after conventional treatment is applied in some treatment plants to further remove pathogens, such as treatment with peracetic acid, chlorination, and ultraviolet irradiation (Das, 2001; Kitis, 2004). These treatments are efficient for inactivating and removing bacteria and protozoa, but not for most enteric viruses (Freese and Nozaic, 2004; Shannon et al., 2008).
Ozone treatment is an alternative for removing microcontaminants in sewage because ozone is an extremely reactive oxidant and thereby a powerful disinfectant. It has been used for disinfection of drinking water in Europe since 1906 (Rice et al., 1981) and has also been installed in some sewage treatment plants (Oh et al., 2007; Rakness et al., 1993). The disinfecting ability of ozone treatment has been shown to be efficient for bacteria and parasites in clean water (Kim et al., 1999; Peeters et al., 1989), and this treatment also has been shown to reduce the concentrations of enteric viruses and bacteriophages (Burleson et al., 1975; Kim et al., 1980). However, the ability of ozonation to inactivate pathogens in wastewater might be hampered due to the high contents of organic materials in sewage (Burleson et al., 1975). One mechanism for reducing viable viruses in water by ozone treatment is assumed to be due to a conformation change of the viral capsid proteins by oxidation that either destroys the capsid or suppresses the virus/host cell receptor binding by changing the viral capsid proteins (Shannon et al., 2008). Previous studies conducted in wastewater treatment plants have shown that ozone disinfection might be highly efficient in inactivating bacteria and bacteriophages after conventional sewage treatments (Kim et al., 1999; Tyrrell et al., 1995), but knowledge regarding its effect for reducing human enteric viruses is relatively scarce.
We used next-generation sequencing (NGS) technology and real-time PCR to investigate the efficiency of virus removal in sewage by conventional treatment and to evaluate the effect of additional ozone treatment at a full-scale pilot plant in Sweden.
2. Materials and methods
2.1. Ozone treatment of conventionally treated sewage
The investigated sewage treatment plant in Knivsta, which is situated 50 km north of Stockholm, Sweden, uses traditional activated sludge treatment and receives primarily household waste from up to 12,000 population equivalents with a hydraulic design flow of 300 m3/h. The initial treatment is mechanical with two parallel screens and an aerated grit chamber. The subsequent biological treatment includes activated sludge and reactors with carriers of active biofilms, and this is followed by a chemical treatment step where ferric chloride is added prior to the sewage entering the two final parallel sedimentation basins. Before release into the recipient river (Knivstaån), the effluent passes through a pond for the removal of phosphorus-containing fine particles. In 2015 an additional ozonation step treating the entire wastewater flow was added at the end of this process line. The full-scale ozonation step is divided into two parallel lines with a total maximum capacity of 560 m3 effluent wastewater per hour. The ozonation step includes lifting pumps, the production of ozone in generator units, the injection of ozone by static mixers, contact tanks, and final contact filters. Each line contains two lifting centrifugal pumps (APEX ISF C, Bristol, UK), one static mixer (NR Mixer, Statiflo International Ltd, UK), one 50 m3 stain-less steel contact tank with 5 m water depth and two compartments, one ozone destructor for off-gas (Primozone, Sweden), and two contact filters with a total area of 25 m2 filled with 1 m light-expanded clay aggregates (Leca, Saint-Gobain Linköping, Sweden) for potential stripping or quenching of ozone residues in ozonated wastewater. The ozone is produced from evaporated liquid oxygen with >99.5% O2 (YaraPraxair, Sweden) diluted to 98% O2 by addition of air in an ozone generator with a maximum production capacity of 2.4 kg O3/h (GM48, Primozone, Sweden). An ozone dose of around 6 mg/L is added to the effluent wastewater through static mixers that transfer more than 98% of the added ozone to the wastewater. Most of the ozone reacts or degrades rapidly after the addition to the wastewater. Analysis of the water samples in the inlets and outlets of the contact tanks show ozone concentrations of 1–3 mg O3/L and 0.1–0.3 mg O3/L respectively. To verify that adequate amounts of ozone are transferred, the removal of pharmaceutical residues was calculated based on frequent inlet and outlet samples, and the results showed a typical removal efficiency for an ozone dose of 6–7 mg O3/L, which is also reported in other studies and is related to total organic carbon (TOC) concentrations (Beijer et al., 2017). The hydraulic retention time in the contact tanks was on average 46 min and the minimum and maximum retention time was 15 and 180 min, respectively, during the period of ozonation that began in August 2015 and ended in February 2016.
The wastewater chemistry as well as effects on the recipient river were studied in parallel research projects before, during, and after the ozonation trial. For the present study, flow-proportional 24 h composite samples of influent (5 L per sample), effluent (10 L), effluent after ozonation (10 L), and effluent after the dam (outlet; 10 L) were collected on three occasions in 2015 (November 30 until December 4 (week 49); December 8 until December 12 (week 50); and December 19 until December 22 (week 51/52)), with time adjustment for the flow rate to represent the “same” water. All samples were cooled during sampling and then frozen at −20 °C until further processing.
2.2. Concentration of viruses in water
The water samples were first centrifuged at 8000 × g for 15 min before filtration twice through Nano-Ceram cartridge filters (Argonide, Sanford, Florida, USA) at an average flow rate of 2.5 L/min. The viruses were electrostatically attached to the filter from which they were eluted by 330 mL of 0.2 M phosphate buffer containing 0.05 M glycine (pH 9.5). The eluate was collected, and the pH was adjusted to 7.5 by the addition of 1 M HCl. The eluate was thereafter filtered through a 0.65/0.45 μm Sartobran Capsule filter (Sartorius, Göttingen, Germany) to remove remaining debris and most bacteria. The filtrate was then ultracentrifugated in eight tubes at 50,000 rpm for 4 h at 4 °C. The pellet in each tube was resuspended in 300 μL 10 mM Tris-HCl (pH 8.0) overnight, pooled, and stored at −80 °C until analysis.
2.3. Evaluation of the efficiency of the viral concentration
A fixed amount of human mastadenovirus 2 (HAdV-2) was added to 3.5 L raw sewage, and the sewage was concentrated by the method above. One milliliter of unconcentrated water and one mL from each concentration step was collected and analyzed for adenovirus by quantitative real-time PCR (qPCR) and isolation on cell culture.
Nucleic acids in the water samples were extracted from 200 μL concentrated sample using the QIAGEN DNA Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. qPCR for adenovirus was performed in a 20 μL reaction mix containing 2 μL extracted nucleic acids, 1 × universal DNA Master Mix (ThermoFisher, Waltham, MA, USA), 0.5 μM of forward and reverse primer, and 0.4 μM of probe (Table S1). The cycling conditions were 50 °C for 2 min and 95 °C for 10 min followed by two-step cycling 45 times at 95 °C for 15 s and 60 °C for 1 min on an ABI 7500 Fast Real-time PCR System (Applied Biosystems, Foster City, CA, USA).
For isolation of HAdV-2 on cell cultures, 100 μL of 10-fold serial dilutions (1/10 to 1/10,000) in Eaglés minimal essential medium (MEM, Gibco, Waltham, MA, USA) from each concentration step were inoculated in duplicate into wells in 48-well plates (ThermoFisher) containing confluent monolayers of A549 cells. The plates were incubated at 37 °C in an atmosphere of 5% CO2 for 2 h, after which the medium containing virus was removed from each well, followed by addition of 500 μL MEM containing 4% fetal calf serum and 1% L-glutamine (Gibco). The plates were incubated at 37 °C in 5% CO2 and were examined for cytopathogenic effects daily for 9 days.
2.4. Preparation of templates for Ion Torrent sequencing
2.4.1. Nuclease treatment of the dissolved pellets from the three concentrated incoming sewage samples
Before nucleic acid extraction, 400 μL of the dissolved pellet was treated with 50 U Benzonase nuclease (Sigma-Aldrich, St. Louis, MO, USA) and 1.25 mM MgCl2 (Applied Biosystems, Foster City, CA, USA) and incubated at 37 °C for 1 h. Thereafter EDTA was added to a final concentration of 50 mM to inhibit the nuclease activity. DNA was extracted from 200 μL of each treated sample using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and RNA was extracted from the remaining 200 μL of each sample using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany).
2.4.2. cDNA transcription
The RNA was reverse transcribed into cDNA in a 20 μL reaction mix containing 10.6 μL extracted RNA, 0.4 μg random hexamer primers (Roche Diagnostics, Indianapolis, IN, USA), 0.5 mM dNTP (Sigma-Aldrich), 1 × First Strand Buffer (Invitrogen, Waltham, MA USA), 5 mM dithiothreitol (Invitrogen), 40 U RNaseOUT (Invitrogen), and 200 U SuperScript III Reverse Transcriptase (Invitrogen). The synthesis was performed at 25 °C for 10 min followed by 50 °C for 90 min, and the cDNA was stored at −20 °C until further amplification.
2.4.3. PCR amplification with random primers
Extracted DNA and cDNA samples were amplified by nested PCR in triplicate. Briefly, touch-up gradient PCR using primers SISP3 and SIS3 (Table S1) was used as the first-round amplification in a 50 μL reaction mix containing 6.5 μL template, 1 × Taq buffer (Applied Biosystems), 2 mM MgCl2 (Applied Biosystems), 0.5 mM dNTP (Sigma-Aldrich), 1 U Taq DNA polymerase (Roche Diagnostics), and 0.8 μM of each primer. The PCR reaction was performed for one cycle at 94 °C for 3 min, followed by 12 cycles touch-up PCR with 94 °C for 30 s and 20 °C for 190 s (2 °C increase per cycle), followed by 30 cycles of 94 °C for 30 s, 48 °C for 30 s, and 68 °C for 2 min, and with a 5 min final extension at 68 °C. Five microliters of the first-round PCR product were further amplified with the primers SISP2 and SIS2 (Table S1) in a 50 μL reaction mix containing 1 × Taq buffer (Applied Biosystems), 2 mM MgCl2 (Applied Biosystems), 0.5 mM dNTP (Sigma-Aldrich), 1 U Taq DNA polymerase (Roche Diagnostics), and 0.8 μM of each primer. The PCR was performed with an initial denaturation at 94 °C for 3 min followed by 45 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 2 min, and with a 5 min final extension at 68 °C. The triplicates were pooled, and the PCR products were visualized by 1.5% agarose gel electrophoresis.
2.4.4. Sonication of PCR products
Before library building for next-generation sequencing (NGS), the majority of the amplified products were sheared into 200–500 bp fragments by sonication using a Bioruptor sonication device (Diagenode, Seraing, Belgium). The PCR products from each sample were pooled and thereafter aliquoted into five tubes each with 25 μL product. One aliquot was not sonicated, while the four other aliquots were sonicated for 3, 6, 12, and 18 cycles, respectively (each cycle was one minute with 30 s sonication and 30 s without sonication). After sonication, the five aliquots were pooled, and the products were visualized by 1.5% agarose gel electrophoresis.
2.4.5. Library construction and quantification of templates for ion torrent sequencing
Libraries were built from the fragmented PCR products by using the Ion Plus Fragment Library Kit on an AB Library Builder System (ThermoFisher, Waltham, MA USA) according to manufacturer's protocol. The newly built libraries were amplified for eight cycles and purified with AMPure XP beads (Beckman Coulter, Brea, CA, USA). Samples were then further size selected to about 370 bp by using a Pippin Prep (Sage Science, Beverly, MA, USA). The recovered materials were analyzed using the Agilent High Sensitivity D1000 ScreenTape System on a TapeStation 2200 (Agilent, Santa Clara, CA, USA). The DNA concentration was thereafter estimated using an Ion Quantitation Kit (ThermoFisher). The libraries were diluted and pooled to reach a final concentration of 50 pM.
Template preparation and chip loading was performed on the Ion Chef instrument using the Ion PGM™ Hi-Q™ View Chef Kit according to the manual (ThermoFisher). An Ion 318 chip (ThermoFisher) was used, and sequencing was conducted on an Ion Torrent PGM with an Ion PGM Hi-Q View Sequencing Kit (ThermoFisher).
2.5. NGS data analysis
The sequence data obtained was automatically trimmed by the Ion Torrent Suite software (ThermoFisher), and the resulting BAM files were imported into CLC Genomic Workbench 9.5.1 (Qiagen, Hilden, Germany) for analysis. The pipeline for viral identification is illustrated in Supplemental Figure A1 . Low-quality reads, reads below 30 bp, and the primer sequences were removed. The sequences were mapped to a human hg19 reference genome using stringent criteria, with length fraction set at 0.5 and similarity fraction set at 0.9. The mapped reads were discarded, while unmapped reads were assembled de novo using the built-in CLC de novo assembler with a word size of 20 and a minimal contig size of 80 bp.
The contigs obtained from the de novo assembly and singleton reads longer than 50 bp were blasted against the NCBI GenBank non-redundant nucleotide database (nr/nt) using BLASTn and a cut-off with an E-value of 10−3 for significant hits. After BLAST, contigs and singletons that satisfied an E-value <10−3 and an HSP length >80 bp were selected, and viral hits among them were used for further analysis.
2.6. Validation of NGS results by qPCR
Primers and probes for selected viruses related to human diseases were designed and used to verify the results obtained by NGS (Table A1) and to determine the prevalence of these viruses after conventional treatment and ozone treatment. The most common viruses identified were human feces pecovirus, picobirnavirus, parvovirus, parvovirus-like virus, adenovirus genotype 41, astrovirus 4, and gokushovirus. Three microliters of extracted nucleic acids from the 12 samples were analyzed by qPCR in a 25 μL reaction mixture containing 1 × Reaction Mix (Invitrogen), 20 U RNaseOUT™ (Invitrogen), 0.5 μL SuperScript® III/Platinum® Taq Mix (Invitrogen), and 0.6 μM of each primer, and 0.2 μM of probe. The qPCRs for astrovirus and picobirnavirus were performed with initial reverse transcription at 50 °C for 30 min and 95 °C for 15 min. The qPCR was performed for 45 cycles of 95 °C for 15 s and 55 °C for 1 min for most viruses, but with extension at 53 °C for adenovirus 41 and 54 °C for gokushovirus and astrovirus 4. The qPCRs were performed in a 7500 Fast Real-Time PCR system (Applied Biosystems), and all samples were analyzed in duplicate.
2.7. Detection of common enteric viruses by qPCR
The four concentrated water samples (incoming sewage, conventionally treated, ozone treated, and outlet water) from each of the three weeks were also analyzed by qPCR for 14 common enteric viruses (adenovirus, astrovirus, hepatitis A virus, hepatitis E virus, norovirus GI, norovirus GII, norovirus GIV, parechovirus, sapovirus, aichivirus, mengovirus, torovirus, enterovirus, and rotavirus). All primers and probes used for the detection are listed in Table A1. The method and conditions for the qPCR were as previously described (Hellmer et al., 2014). The qPCRs were performed on a 7500 Fast Real-Time PCR system (Applied Biosystems), and all samples were tested in duplicate.
A pUC57 plasmid containing all viral target regions was synthesized by GeneScript (GeneScript USA Inc. NJ, USA), and ten-fold serial dilutions were used as the positive control in all qPCR analyses. The Ct values of the samples were relative to the Ct values from the dilutions of the plasmid containing the virus sequences being analyzed, and from this the numbers of viral genomes per milliliter were estimated. These values were also compared to estimated amounts of viral genomes by assuming a perfect qPCR detecting one genome by using the formula: (Ct value of the sample) = −3.3 × log10 genomes/mL + 45. The number of estimated viral genomes was not adjusted to the estimated recovery in any of the analyzed waters, but was used directly to compare the reduction of viruses by the different treatments.
2.8. PCR amplification of adenovirus
Nested PCR was used to amplify the hexon protein-coding region of adenovirus. The 50 μL reaction mix contained 5 μL extracted DNA, 1 × Taq Buffer (Applied Biosystems), 2.5 mM MgCl2 (Applied Biosystems), 0.2 mM dNTP (Sigma-Aldrich), 0.5 μM of forward and reverse primers (AdvF 1 st and AdvR 1st; Table S1), and 1U Taq DNA polymerase (Roche Diagnostics). The PCR was initiated at 94 °C for 3 min, followed by 40 cycles at 94 °C for 20 s, 59 °C for 30 s, and 72 °C for 1 min, and a 5 min final extension at 72 °C. Five microliters of the PCR product from the first-round amplification were used as the template for nested PCR as described above but with different forward and reverse primers (AdvF 2nd, AdvR 2nd; Table S1). The fragments were purified using a QIAquick PCR purification kit (Qiagen) according to the manufacturer’s protocol, and the purified amplicons were sent for sequencing to GATC Biotech, Constance, Germany.
2.9. Sequence analysis
The sequences obtained were analyzed in the SeqMan Pro 13 program in the DNAStar Program package version 10.1.2 (DNAStar Inc., Madison, WI, USA). The sequences were aligned with the corresponding region of the hexon gene of 81 adenovirus sequences representing all human mastadenovirus types obtained from GenBank. Evolutionary distances were calculated using the Hasegawa–Kishino–Yano (HKY) algorithm in the DNADIST program in the PHYLIP package version 3.65 (Felsenstein, 1996a; Felsenstein, 1996b) with transition/transversion ratio of 9.14 and gamma correction with alpha 0.27. Phylogenetic trees were constructed using the unweighted pair-group method using arithmetic averages (UPGMA) and the neighbor-joining method in the NEIGHBOR program of the PHYLIP package. The trees were visualized with the program TreeView, version 1.6.6 (Page, 2002).
3. Results
3.1. Recovery of viruses after concentration of raw sewage
The added adenovirus could be detected by qPCR and isolation on cell cultures in all concentrations steps (Table 1 ). The raw sewage water was concentrated about 1450 times (from 3.5 L to 2.4 mL) with this technique, and the adenovirus was concentrated about 100 times, indicating a recovery of 7% (Table 1). The treatment of the pellet with nucleases before nucleic acid extraction did not significantly change the efficiency of the concentration procedure. By isolation on cell culture, it could be shown that most adenoviruses were viable, and the viruses from the final ultracentrifugation pellet could be detected up to a dilution of 10−5, indicating a 69% efficiency for the technique. However, when the pelleted samples treated with nuclease were isolated on cell culture, viruses could only be detected in a 10−4 dilution, which would indicate 7% recovery similar to the qPCR (Table 1).
Table 1.
Adenovirus type 2 concentration in raw sewage and after different concentration steps evaluated by real-time PCR and cell culture.
| Tested step | Ct Mean# | Viral genomes/mL | Fold concentration | Titer at cell culture | Fold concentration |
|---|---|---|---|---|---|
| Origin water (3.5 L) | 34.04 | 2100 | 1 | 10−2 | 1 |
| Elution from NanoCeram (350 mL, 100 × concentration) |
34.38 | 1660 | 0.8 | 10−3 | 10 |
| After satorious filtration (350 mL, 100 × concentration) |
32.92 | 4580 | 2.2 | 10−2 | 1 |
| After ultracentrifugation (2.4 mL, 1450 × concentration) |
27.58 | 190,000 | 90.4 | 10−5 | 1000 |
| After Benzonase treatment | 27.37 | 221,000 | 104.9 | 10−4 | 100 |
#: Concentrated from sewage water, with 100-fold dilution in the real-time PCR.
3.2. Viruses identified by NGS
By NGS sequencing, 309,881 to 444,559 reads were obtained after quality control for each of the three concentrated incoming sewages (Table 2 ). After de novo assembly, 5400 to 71,800 contigs and singletons were obtained. Between 350 and 2900 sequences were homologous to different viruses and could be classified into viral families, corresponding to about 4.1% to 6.4% of all contigs obtained (Table 2). In all, 327 viruses belonging to 25 different families were identified (Table A2).
Table 2.
Number of reads obtained by next-generation sequencing of PCR-amplified incoming sewage.
| Samples | Total reads after Quality control | Blast sequences (contigs + singletons) | Viral reads | Percentage viral reads |
|---|---|---|---|---|
| K399-DNA | 404,311 | 71,799 | 2934 | 4.09% |
| K399-RNA | 444,559 | 6535 | 368 | 5.63% |
| K416-DNA | 309,881 | 33,403 | 1387 | 4.15% |
| K416-RNA | 379,517 | 7241 | 461 | 6.37% |
| K462-DNA | 429,502 | 37,310 | 2039 | 5.47% |
| K462-RNA | 316 148 | 5 409 | 348 | 6.43% |
Bacteriophages from the families of Inoviridae, Microviridae, Myoviridae, Podoviridae, and Siphoviridae accounted for the largest proportion of the identified viruses. Sequences similar to gokushovirus, a member of Microviridae, were abundant and accounted for more than 70% of all viral sequences in one sample. Apart from bacteriophages, viruses infecting plants, vertebrates, invertebrates, and protists were also identified. Although there were fewer sequences similar to viruses that could infect vertebrates than bacteriophages, they had a high diversity and belonged to nine of the 25 viral families. Plant-related viruses, including Tombusviridae, Virgaviridae, Alphaflexiviridae, Betaflexiviridae, Partitiviridae, and Tymoviridae, were also widely distributed in the samples.
Sequences similar to a number of viruses related to human diseases were also detected. As shown in Table 3 , sequences representing some common viral families, such as Adenoviridae, Astroviridae, Papillomaviridae, Parvoviridae, and Picobirnaviridae were found. Contigs longer than 400 bp identified human adenovirus F, serotype 41 in samples from weeks 49 and 50. Shorter contigs had sequences similar to human astrovirus 1 and 4 in weeks 50 and 51/52. Sequences similar to three strains of human papillomavirus (HPV) were identified in samples from week 49 and week 51/52. Two could be classified into genus Betapapillomavirus and the other to Mupapillomavirus. In addition, sequences similar to those for other viruses that might cause human enteric disease were also identified, such as human picobirnavirus, parvovirus, and human feces pecovirus.
Table 3.
Next-generation sequencing data revealed sequences homologous to viruses belonging to 25 different viral families, of which the 11 most common are listed.
| Virus family | Genus/species | Week 49 | Week 50 | Week 51/52 | Longest contigs or reads |
|---|---|---|---|---|---|
| Number of different viral families identified | 19 | 18 | 20 | ||
| Adenoviridae | Human adenovirus F/HAdV41 Human adenovirus type A | + | + | -# | 425 bp |
| Astroviridae | Mamastrovirus/ Human astrovirus 1/4 | – | + | + | 292 bp |
| Papillomaviridae | Human papillomavirus | + | – | + | 315 bp |
| Picobirnaviridae | Human picobirnavirus | + | + | + | 284 bp |
| Parvoviridae | Parvovirus-like virus | + | + | + | 647 bp |
| unclassified | Human feces pecovirus | + | – | + | 343 bp |
|
Microviridae/ Gokushovirinae |
Gokushovirus | + | + | + | 971 bp |
| Mimiviridae | Mimivirus | + | + | + | 266 bp |
| Siphoviridae | Several phages | + | + | + | 796 bp |
| Genomoviridae | Gemycircularvirus | + | + | + | 1070 bp |
| Virgaviridae | Pepper mild mottle virus | + | + | + | 333 bp |
+ = identified.
# Not identified.
3.3. Identification of common enteric viruses and NGS-selected viruses before and after each treatment step by qPCR
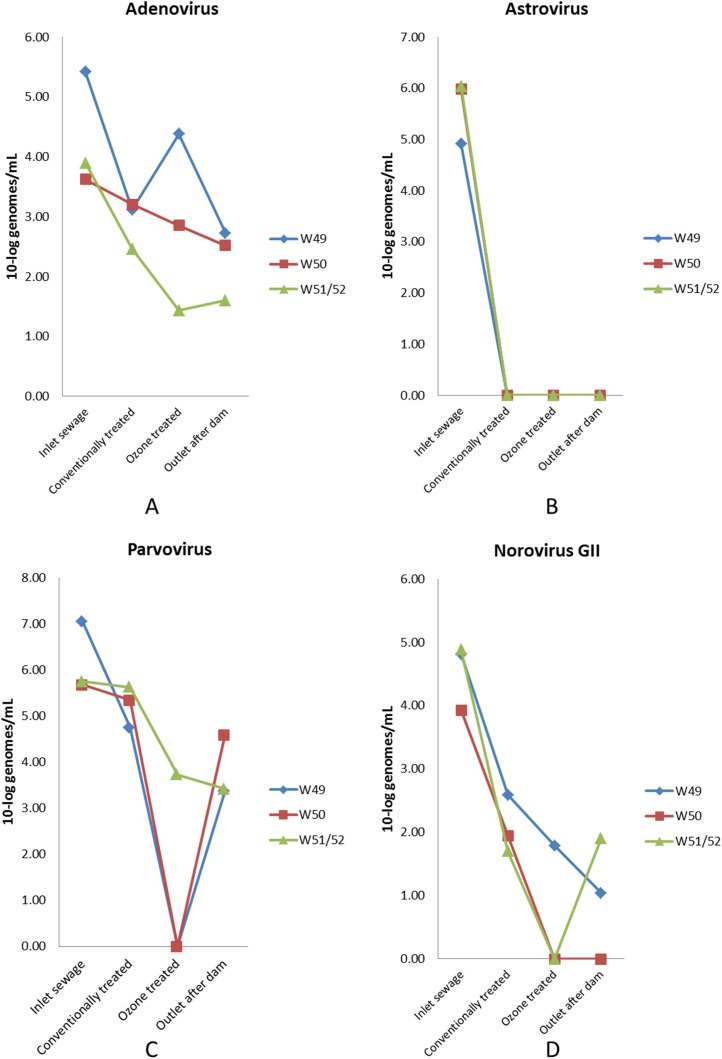
qPCR was used for identification of 14 common human enteric viruses in influent, treated sewage, ozone-treated effluents, and after passing an open dam system. Three of these viruses (adenovirus, norovirus GII, and astrovirus) were detected in samples from the influent and after treatment at 62% to 99% reduced concentrations, with adenovirus being the least affected (Table 4 , Fig. 1 ). An additional four viruses (norovirus GI, sapovirus, parechovirus, and hepatitis E virus) were only detected in the inlet samples with concentrations reduced to undetectable levels after conventional treatment.
Table 4.
Ct values and estimated numbers of genomes of enteric viruses per milliliter of water in untreated and treated sewage samples from three weeks in 2015, where K numbers indicate the internal sample numbers.
| Virus | Week 49 |
Week 50 |
Week 51/52 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K399 | K400 | K401 | K402 | K416 | K417 | K418 | K419 | K462 | K463 | K464 | K465 | ||
| Inlet sewage |
Conven-tionally treated | Ozone treated |
Outlet after dam | Inlet sewage |
Conven-tionally treated | Ozone treated |
Outlet after dam | Inlet sewage | Conven-tionally treated | Ozone treated |
Outlet after dam | ||
| Adenovirus | Ct value | 27.1# | 34.7 | 30.6 | 36.0 | 33.0 | 34.4 | 35.6 | 36.7 | 32.1 | 36.9 | 40.3 | 39.7 |
| Viral genomes/mL | 260,000 | 1300 | 24,000 | 530 | 4200 | 1600 | 720 | 330 | 7900 | 290 | 27 | 40 | |
| Norovirus GI | Ct value | 30.0 | -* | – | – | 35.6 | – | – | – | 29.7 | – | – | – |
| Viral genomes/mL | 36,000 | – | – | – | 730 | – | – | – | 43,000 | – | – | – | |
| Norovirus GII | Ct value | 29.1 | 36.5 | 39.1 | 38.3 | 32.1 | 38.6 | – | – | 28.9 | 36.1 | – | 38.7 |
| Viral genomes/mL | 65,000 | 390 | 61 | 11 | 8400 | 88 | – | – | 76,000 | 50 | – | 80 | |
| Sapovirus | Ct value | 26.6 | – | – | – | 31.4 | – | – | – | 27.1 | – | – | – |
| Viral genomes/mL | 380,000 | – | – | – | 14,000 | – | – | – | 270,000 | – | – | – | |
| Parechovirus | Ct value | 33.6 | – | – | – | 35.3 | – | – | – | 35.4 | – | – | – |
| Viral genomes/mL | 2800 | – | – | – | 850 | – | – | – | 780 | – | – | – | |
| HEV | Ct value | – | – | – | – | – | – | – | – | 39.6 | – | – | – |
| Viral genomes/mL | – | – | – | – | – | – | – | – | 45 | – | – | – | |
| Astrovirus | Ct value | 28.8 | – | – | – | 25.3 | – | – | – | 25.0 | 37.5 | – | – |
| Viral genomes/mL | 81,000 | – | – | – | 960,000 | – | – | – | 1,100,000 | 180 | – | – | |
#: mean Ct value (Ct = cycle threshold in the quantitative PCR).
*: Undetected.
Fig. 1.
Reduction of adenovirus, astrovirus, parvovirus and norovirus GII by the different treatments during the three weeks of investigation.
Based on the obtained NGS sequences, new qPCR primers and probes were designed to detect seven selected viruses related to humans in the incoming sewage, including human feces pecovirus, picobirnavirus, parvovirus, parvovirus-like virus, adenovirus type 41, astrovirus type 4, and gokushovirus (Table 5 ). For these viruses the reduction of viral genomes after the conventional treatment ranged from 99.9% for gokushovirus to 0–53% for parvovirus and parvovirus-like virus during weeks 50 and 51/52 (Table 5). The reduction was higher, 99%, for parvovirus during week 49.
Table 5.
Real-time PCR for seven of the viruses identified by NGS and the estimated numbers of genomes of these viruses per milliliter of water in untreated and treated sewage samples from three weeks in 2015, where K numbers indicate internal sample numbers.
| Virus | Week 49 |
Week 50 |
Week 51/52 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K399 | K400 | K401 | K402 | K416 | K417 | K418 | K419 | K462 | K463 | K464 | K465 | ||
| Inlet sewage | Conven-tionally treated | Ozone treated |
Outlet after dam | Inlet sewage | Conven-tionally treated | Ozone treated | Outlet after dam | Inlet sewage | Conven-tionally treated | Ozone treated | Outlet after dam | ||
| Pecovirus | Ct value | 21.03# | 31.2 | -* | – | 24.32 | 31.02 | – | 32.08 | 24.22 | 29 | – | – |
| Viral genomes/mL | 18,000,000 | 15,000 | – | – | 1,900,000 | 17,000 | – | 8200 | 2,000,000 | 71,000 | – | – | |
| Picobirnavirus | Ct value | 27.73 | 31.16 | – | – | 28.05 | 32.87 | – | – | 27.17 | 30.94 | – | – |
| Viral genomes/mL | 170,000 | 16,000 | – | – | 140,000 | 4700 | – | – | 250,000 | 18,000 | – | – | |
| Parvovirus | Ct value | 21.8 | 29.34 | – | 33.91 | 26.29 | 27.38 | – | 29.9 | 26.02 | 26.46 | 32.72 | 33.74 |
| Viral genomes/mL | 11,000,000 | 56,000 | – | 2300 | 470,000 | 220,000 | – | 38,000 | 560,000 | 420,000 | 5300 | 2600 | |
| Parvovirus-like virus | Ct value | 20.3 | 26.92 | – | – | 25.11 | 26 | – | 28.6 | 25.04 | 25.02 | – | – |
| Viral genomes/mL | 31,000,000 | 300,000 | – | – | 1,100,000 | 570,000 | – | 93,000 | 1,100,000 | 1,100,000 | – | – | |
| Gokushovirus | Ct value | 23.29 | 34.89 | – | – | 20.51 | – | – | – | 20.11 | 31.78 | – | – |
| Viral genomes/mL | 3,800,000 | 1200 | – | – | 26,000,000 | – | – | – | 35,000,000 | 10,000 | – | – | |
| Astrovirus 4 | Ct value | 22.27 | 27.66 | – | – | 24.11 | 27.98 | – | 29.70 | 21.84 | 25.74 | 33.20 | 35.50 |
| Viral genomes/mL | 7,700,000 | 180,000 | – | – | 2,100,000 | 140,000 | – | 43,000 | 10,000,000 | 680,000 | 3800 | 760 | |
| Adenovirus 41 | Ct value | 29.49 | 35.56 | – | – | – | 37.94 | – | 37.26 | 35.1 | 37.68 | 37.23 | – |
| Viral genomes/mL | 50,000 | 730 | – | – | – | 140 | – | 220 | 1000 | 170 | 230 | – | |
#: mean Ct value.
*: Undetected.
Ozone treatment of conventionally treated effluent lowered the amount of viruses further to undetectable levels for pecovirus, picobirnavirus, parvovirus-like virus, and gokushovirus for all three weeks. This treatment reduced the adenovirus concentration by 55% to 91%, norovirus GII by 85% to 100%, and astrovirus 4 and parvovirus by 99% to 100% (Table 4, Table 5). Despite not being detected after ozone treatment, several of these viruses were found in the outlet water after the dams, as were parvovirus during weeks 49 and 50 and parvovirus-like virus, astrovirus 4, pecovirus, and adenovirus 41 during week 50. For these viruses, the amount of virus identified in the outlet water was between 4% and 100% of that identified in the conventionally treated sewage samples (Table 4, Table 5).
3.4. Phylogenetic analysis of adenoviruses identified in inlet sewage and in treated water
It was possible to amplify and sequence a partial region of the adenovirus hexon gene in all 12 water samples except for the outlet samples from weeks 49 and 51/52. The sequence obtained for the ozone-treated sample in week 50 could not be analyzed because it contained multiple sequences. Phylogenetic analysis was performed for the remaining nine sequences and revealed that six belonged to species F and two to species A, and for the ozone-treated sample in week 49 the species could not be determined, but it was found on the same branch as species F strains in the phylogenetic tree (Fig. 2 ). The strains were divergent from each other, and the most similar strains were from conventionally treated sewage from weeks 49 and 50 within species F and were found on the same branch as the strains in the incoming and ozone-treated samples from week 51/52 (Fig. 2).
Fig. 2.
Phylogenetic tree based on 185 nucleotides of the hexon protein-coding region of the adenovirus genome. Nine sequences from water samples and 81 adenovirus sequences representing all human mastadenovirus types from GenBank are included, and water samples are labeled in bold. The tree shows the dominant strains during the sewage treatment.
4. Discussion
In this study we demonstrated a high diversity of viruses in incoming sewage. Human-related viruses were strongly reduced by conventional treatment of the sewage, and the elimination of several of these was achieved by additional ozone treatment. Other studies have previously shown reduction of parasitic protozoa, bacteria, and phages by conventional treatment of sewage (Ottoson et al., 2006a; Tyrrell et al., 1995), but little was known regarding the reduction of human enteric viruses (Montazeri et al., 2015; Ottoson et al., 2006b, Ottoson et al., 2006c) or their sensitivity to ozonation of sewage. While ozonation is already recognized as a promising technology for removing microcontaminants, such as pharmaceuticals, in sewage (Beijer et al., 2017), the additional benefit of reducing risks for the spread of human pathogenic viruses via contaminated waters should be considered as well.
The conventional treatment of sewage eliminated or reduced the amount of most viruses detected in inlet sewage in this study, but it was less efficient for parvovirus and adenovirus. Additional ozone treatment lowered the amount further, for some viruses to undetectable levels. This result is in line with other studies that demonstrated a reduction of enteroviruses and F+ coliphages by more than 2.9 and 2.2 log with ozone disinfection of wastewater effluents (Xu et al., 2002).
Because there is a relatively high quantity of suspended solids and organic materials in secondary effluent after conventional treatment that could affect the ozone treatment efficiency, we do not know if an increased amount of ozone or longer ozone treatment time would have reduced the amounts of viruses even further.
However, in this study some viruses that were undetectable in the ozone-treated samples reoccurred in the outlet water, including parvovirus, norovirus GII, human feces pecovirus, parvovirus-like virus, gokushovirus, and HAdV-F41, although the amounts were significantly lower compared with raw sewage. The reason for the identification of viruses in the outlet after the dam system despite the fact that they were not detected by qPCR after ozonation is unclear, but two circumstances identified during the evaluation of the operation of the treatment plant might explain these findings. During week 49, the protective sieves for the lifting pumps in the ozonation step were partly clogged with large particles released by high hydraulic flow in the treatment plant leading to by-pass of 4% of the conventionally treated wastewater directly to the dam. In week 50/51, the concentration of suspended solids in the conventionally treated wastewater was 13 mg SS/L, which was 60% higher than in samples from week 49 and week 50 that contained 8 mg SS/L. This might have allowed some of these viruses to escape the ozone treatment and to reappear in the dams. Additional explanations could be that our attempts to follow the “same” water based on the flow through the treatment plant had been disrupted and that the waters analyzed from the outlet of the dams were not the same as the ozonized waters. Another reason might be that the viruses were present in amounts too low to be detected in the ozonized water by qPCR. Larger amounts of water might be needed to determine if these viruses were also present in the waters after ozone treatment, and further studies are also needed to determine the impact of the ozone treatment on these viruses.
Adenoviruses are resistant to UV disinfection (Baxter Carole et al., 2007; Baxter et al., 2007) and were also rather insensitive to the ozone treatment used in this study. In clean water, human mastadenovirus F40 (HAdV-F40) has been shown to be more resistant to ozone treatment than feline calicivirus, which is often used as a surrogate for noroviruses (Thurston-Enriquez et al., 2005). In the present study, the dominant strains of adenovirus varied, both between the differently treated waters and within the sample groups. There were different HAdV-F40 and HAdV-F41 strains in all inlet sewage samples, while in one ozone-treated sample a divergent type F strain dominated and in some of the treated waters there were type A strains. A somewhat similar finding was also obtained by Fernandez-Cassi and co-workers (Fernandez-Cassi et al., 2017). The pattern observed in this study suggests that there was a complex composition comprising multiple adenovirus types in the raw sewage, and these might have varying degrees of sensitivity to the conventional processing and ozone treatment. However, we do not know if these viruses are viable after the ozone treatment, and the oxidization might have changed the conformation of the viral head protein and thereby inhibited viral-host binding (Shannon et al., 2008). Because the amount of viral genomes decreased significantly after the ozonation, the treatment might not only change the conformation of the viral capsids, but might also destroy or open the capsid of some viruses and thereby release the viral genomes into the water where nucleases are present. Further studies are needed on ozonation of water containing adenovirus types that are easy to isolate on cell cultures in order to identify the viability of the viruses after treatment.
The high diversity of viruses identified in the inlet sewage in this study is in agreement with a previous study on sewage where 237 known viruses representing 26 taxonomic families, as well as a large number of novel viruses, were identified (Cantalupo et al., 2011), confirming that raw sewage contains a vast and diverse viral pool. The identified viruses could infect bacteria, plants, vertebrates, invertebrates, and protists, with bacteriophages contributing the largest proportion. The bacteriophage gokushovirus, which primarily targets Chlamydia, Bdellovibrio, and Spiroplasma (Labonte et al., 2015), accounted for most viral reads in all samples. This virus has been shown to be widespread in global environmental samples, especially in aquatic ecosystems (Hopkins et al., 2014; Labonte et al., 2015), and can be used as a potential viral indicator for treatment efficacy in different waters. Human disease-related viruses, which can cause diarrhea, respiratory diseases, conjunctivitis, meningoencephalitis, and infantile gastroenteritis (Fong et al., 2010; Nguyen et al., 2007) were also identified. Some of these that might cause gastroenteritis, such as human picobirnavirus and human feces pecovirus (Giordano et al., 1999; Phan et al., 2016; Phan et al., 2012), were widely detected in the sewage samples but are usually not tested for at hospital laboratories. To understand their importance for public health, more enteric viruses identified in environmental samples should be incorporated into the routine monitoring of patients with gastroenteritis.
There were also other human pathogenic viruses detected in the sewage during the whole study period, including HPV, which might cause warts, papilloma, and malignant tumors. Other recent studies have also identified multiple types of HPV, including the oncogenic high-risk HPV16, in raw sewage (Bibby and Peccia, 2013; Cantalupo et al., 2011; La Rosa et al., 2013). The presence of this virus in large amounts in aquatic environments has caused concern about its potential for waterborne transmissions, which might cause anogenic or oral HPV infections even among people who have not been sexually active (Fratini et al., 2014); however, further research into its transmission by the waterborne route is still needed.
When comparing the results obtained with real-time PCR and NGS, only adenovirus and astrovirus could be identified by NGS data, and sequences corresponding to sapovirus, norovirus, and parechovirus were not found even though these viruses could be identified with real-time PCR. This might be due to their low quantity in sewage. Most of the viruses detected by NGS had about one to two log10 higher viral genome concentrations than the overlooked viruses, indicating that NGS has some limitations when dealing with complex environmental samples and might underestimate the number of different viruses if the number reads are not high enough to exhaustively probe the sample. The concentration efficiency of about 10% used in this study might also have influenced the amount of viruses identified. However, this efficiency is comparable to that obtained by other concentration methods (Cashdollar et al., 2013; Fout et al., 2015; Hellmer et al., 2014; Ikner et al., 2011). By using adsorption to charged filters, HAdV-2, which was used for the evaluation of this technique, has previously been used in another study with a recovery of about 14%, while the recovery was 77% to 83% for enteroviruses (Ikner et al., 2011). The low recovery of adenovirus might be due to its surface capsid structure and electrostatic charge, which might facilitate the entrapment of these viruses in the charged filters and hamper their elution from the filters (Gibbons et al., 2010; Shi et al., 2016). HAdv-2 was used in this study as an indicator of the effectiveness of the method because it could easily be detected by both qPCR and isolation on cell cultures and was shown to be viable in all concentration steps used. This method might therefore have a higher concentration efficiency for other viruses than adenovirus and thus should also be analyzed for other viruses that can be isolated on cell culture.
This work expands our knowledge of the possible use of ozone treatment to eliminate viruses from water. The ability of ozonation to reduce the transmission risks of human pathogens might therefore be considered when decisions are taken as to whether it is worthwhile to install advanced sewage treatments to remove microcontaminants and bacterial pathogens. It should be acknowledged that in addition to the apparent advantages, ozonation also come with a cost, and high ozone doses in particular can sometimes lead to the formation of compounds that can negatively affect aquatic life (Samuelsson et al., 2011). Further studies are needed to determine the most efficient methods, including different ozonation procedures, for viral elimination in sewage and whether these pathogens in treated waters are viable and can spread. The elimination of human pathogens is imperative in order to prevent waterborne viruses from re-entering into the environment and thus reduce the potential risk they pose to public health.
Conflict of interest
There is no conflict of interest.
Acknowledgment
This work was financially supported by The Swedish Research Council for Sustainable Development, Formas, grant number 942-2015-306.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.ijheh.2018.01.012.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Baxter Carole S., Hofmann R., Templeton Michael R., Brown M., Andrews Robert C. Inactivation of adenovirus types 2, 5, and 41 in drinking water by UV light, free chlorine, and monochloramine. J. Environ. Eng. 2007;133:95–103. doi: 10.1061/(ASCE)0733-9372(2007)133:1(95). [DOI] [Google Scholar]
- Baxter C.S., Hofmann R., Templeton M.R., Brown M., Andrews R.C. Inactivation of adenovirus types 2, 5, and 41 in drinking water by UV light free chlorine, and monochloramine. J. Environ. Eng. 2007;133:95–103. [Google Scholar]
- Beijer K., Bjorlenius B., Shaik S., Lindberg R.H., Brunstrom B., Brandt I. Removal of pharmaceuticals and unspecified contaminants in sewage treatment effluents by activated carbon filtration and ozonation: evaluation using biomarker responses and chemical analysis. Chemosphere. 2017;176:342–351. doi: 10.1016/j.chemosphere.2017.02.127. [DOI] [PubMed] [Google Scholar]
- Bibby K., Peccia J. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ. Sci. Technol. 2013;47:1945–1951. doi: 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A. Human enteric viruses in the water environment: a minireview. Int. Microbiol. 1998;1:191–196. [PubMed] [Google Scholar]
- Burleson G.R., Murray T.M., Pollard M. Inactivation of Viruses and Bacteria by Ozone: with and without Sonication. Appl. Microbiol. 1975;29:340–344. doi: 10.1128/am.29.3.340-344.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo P.G., Calgua B., Zhao G., Hundesa A., Wier A.D., Katz J.P., Grabe M., Hendrix R.W., Girones R., Wang D., Pipas J.M. Raw sewage harbors diverse viral populations. mBio. 2011:2. doi: 10.1128/mBio.00180-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashdollar J.L., Brinkman N.E., Griffin S.M., McMinn B.R., Rhodes E.R., Varughese E.A., Grimm A.C., Parshionikar S.U., Wymer L., Fout G.S. Development and evaluation of EPA method 1615 for detection of enterovirus and norovirus in water. Appl. Environ. Microbiol. 2013;79:215–223. doi: 10.1128/AEM.02270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T.K. Ultraviolet disinfection application to a wastewater treatment plant. Clean Technol. Environ. Policy. 2001;3:69–80. [Google Scholar]
- Felsenstein J. Inferring phylogeneies from protein sequences by parsimony, distance and likelyhood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- Fernandez-Cassi X., Timoneda N., Martinez-Puchol S., Rusinol M., Rodriguez-Manzano J., Figuerola N., Bofill-Mas S., Abril J.F., Girones R. Metagenomics for the study of viruses in urban sewage as a tool for public health surveillance. Sci. Total Environ. 2017;618:870–880. doi: 10.1016/j.scitotenv.2017.08.249. [DOI] [PubMed] [Google Scholar]
- Fong T.T., Lipp E.K. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 2005;69:357–371. doi: 10.1128/MMBR.69.2.357-371.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fout G.S., Cashdollar J.L., Varughese E.A., Parshionikar S.U., Grimm A.C. EPA Method 1615: measurement of enterovirus and norovirus occurrence in water by culture and RT-qPCR. I. Collection of virus samples. J. Vis. Exp. 2015 doi: 10.3791/52067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratini M., Di Bonito P., La Rosa G. Oncogenic papillomavirus and polyomavirus in water environments: is there a potential for waterborne transmission? Food Environ. Virol. 2014;6:1–12. doi: 10.1007/s12560-013-9134-0. [DOI] [PubMed] [Google Scholar]
- Freese S.D., Nozaic D.J. Chlorine: is it really so bad and what are the alternatives? Water Sa. 2004;30:566–572. [Google Scholar]
- Gibbons C.D., Rodriguez R.A., Tallon L., Sobsey M.D. Evaluation of positively charged alumina nanofibre cartridge filters for the primary concentration of noroviruses, adenoviruses and male-specific coliphages from seawater. J. Appl. Microbiol. 2010;109:635–641. doi: 10.1111/j.1365-2672.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- Giordano M.O., Martinez L.C., Rinaldi D., Espul C., Martinez N., Isa M.B., Depetris A.R., Medeot S.I., Nates S.V. Diarrhea and enteric emerging viruses in HIV-infected patients. Aids Res. Hum. Retrov. 1999;15:1427–1432. doi: 10.1089/088922299309937. [DOI] [PubMed] [Google Scholar]
- Hellmer M., Paxeus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergstrom T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/aem.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M., Kailasan S., Cohen A., Roux S., Tucker K.P., Shevenell A., Agbandje-McKenna M., Breitbart M. Diversity of environmental single-stranded DNA phages revealed by PCR amplification of the partial major capsid protein. ISME J. 2014;8:2093–2103. doi: 10.1038/ismej.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikner L.A., Soto-Beltran M., Bright K.R. New method using a positively charged microporous filter and ultrafiltration for concentration of viruses from tap water. Appl. Environ. Microbiol. 2011;77:3500–3506. doi: 10.1128/AEM.02705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.K., Gentile D.M., Sproul O.J. Mechanism of ozone inactivation of bacteriophage f2. Appl. Environ. Microbiol. 1980;39:210–218. doi: 10.1128/aem.39.1.210-218.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.G., Yousef A.E., Dave S. Application of ozone for enhancing the microbiological safety and quality of foods: a review. J. Food Prot. 1999;62:1071–1087. doi: 10.4315/0362-028X-62.9.1071. [DOI] [PubMed] [Google Scholar]
- Kitis M. Disinfection of wastewater with peracetic acid: a review. Environ. Int. 2004;30:47–55. doi: 10.1016/S0160-4120(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Kotwal G., Cannon J.L. Environmental persistence and transfer of enteric viruses. Curr. Opin. Virol. 2014;4:37–43. doi: 10.1016/j.coviro.2013.12.003. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Fratini M., Accardi L., D'Oro G., Della Libera S., Muscillo M., Di Bonito P. Mucosal and cutaneous human papillomaviruses detected in raw sewages. PLoS One. 2013:8. doi: 10.1371/journal.pone.0052391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte J.M., Hallam S.J., Suttle C.A. Previously unknown evolutionary groups dominate the ssDNA gokushoviruses in oxic and anoxic waters of a coastal marine environment. Front. Microbiol. 2015:6. doi: 10.3389/fmicb.2015.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverick M.A., Wyn-Jones A.P., Carter M.J. Quantitative RT-PCR for the enumeration of noroviruses (Norwalk-like viruses) in water and sewage. Lett. Appl. Microbiol. 2004;39:127–136. doi: 10.1111/j.1472-765X.2004.01534.x. [DOI] [PubMed] [Google Scholar]
- Li B., Ju F., Cai L., Zhang T. Profile and fate of bacterial pathogens in sewage treatment plants revealed by high-Throughput metagenomic approach. Environ. Sci. Technol. 2015;49:10492–10502. doi: 10.1021/acs.est.5b02345. [DOI] [PubMed] [Google Scholar]
- Lodder W.J., de Roda Husman A.M. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl. Environ. Microbiol. 2005;71:1453–1461. doi: 10.1128/AEM.71.3.1453-1461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montazeri N., Goettert D., Achberger E.C., Johnson C.N., Prinyawiwatkul W., Janes M.E. Pathogenic enteric viruses and microbial indicators during secondary treatment of municipal wastewater. Appl. Environ. Microbiol. 2015;81:6436–6445. doi: 10.1128/AEM.01218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenonen N.P., Hannoun C., Horal P., Hernroth B., Bergstrom T. Tracing of norovirus outbreak strains in mussels collected near sewage effluents. Appl. Environ. Microbiol. 2008;74:2544–2549. doi: 10.1128/AEM.02477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.A., Yagyu F., Okame M., Phan T.G., Trinh Q.D., Yan H., Hoang K.T., Cao A.T.H., Le Hoang P., Okitsu S., Ushijima H. Diversity of viruses associated with acute gastroenteritis in children hospitalized with diarrhea in Ho Chi Minh City, Vietnam. J. Med. Virol. 2007;79:582–590. doi: 10.1002/jmv.20857. [DOI] [PubMed] [Google Scholar]
- Oh B., Park S., Jung Y., Park S., Kang J. Disinfection and oxidation of sewage effluent water using ozone and UV technologies. Water Sci. Technol. 2007;55:299–306. doi: 10.2166/wst.2007.036. [DOI] [PubMed] [Google Scholar]
- Ottoson J., Hansen A., Bjorlenius B., Norder H., Stenstrom T.A. Removal of viruses, parasitic protozoa and microbial indicators in conventional and membrane processes in a wastewater pilot plant. Water Res. 2006;40:1449–1457. doi: 10.1016/j.watres.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Ottoson J., Hansen A., Westrell T., Johansen K., Norder H., Stenstrom T.A. Removal of noro- and enteroviruses, Giardia cysts, Cryptosporidium oocysts, and fecal indicators at four secondary wastewater treatment plants in Sweden. Water Environ. Res. 2006;78:828–834. doi: 10.2175/106143006X101719. [DOI] [PubMed] [Google Scholar]
- Ottoson J., Hansen A., Westrell T., Johansen K., Norder H., Stenstrom T.A. Removal of noro- and enteroviruses Giardia cysts, Cryptosporidium oocysts, and fecal indicators at four secondary wastewater treatment plants in Sweden. Water Environ. Res. 2006;78:828–834. doi: 10.2175/106143006x101719. [DOI] [PubMed] [Google Scholar]
- Page R.D. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinf. 2002;6.2 doi: 10.1002/0471250953.bi0602s01. 1–62. 15. [DOI] [PubMed] [Google Scholar]
- Peeters J.E., Mazas E.A., Masschelein W.J., Dematurana I.V.M., Debacker E. Effect of disinfection of drinking-Water with ozone or chlorine dioxide on survival of cryptosporidium-parvum oocysts. Appl. Environ. Microbiol. 1989;55:1519–1522. doi: 10.1128/aem.55.6.1519-1522.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., Vo N.P., Bonkoungou I.J.O., Kapoor A., Barro N., O'Ryan M., Kapusinszky B., Wang C.L., Delwart E. Acute diarrhea in West African children: diverse enteric viruses and a novel parvovirus genus. J. Virol. 2012;86:11024–11030. doi: 10.1128/JVI.01427-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.G., da Costa A.C., Mendoza J.D., Bucardo-Rivera F., Nordgren J., O'Ryan M., Deng X.T., Delwart E. The fecal virome of South and Central American children with diarrhea includes small circular DNA viral genomes of unknown origin. Arch. Virol. 2016;161:959–966. doi: 10.1007/s00705-016-2756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakness K.L., Corsaro K.M., Hale G., Blank B.D. Wastewater disinfection with ozone – process control and operating results. Ozone: Sci. Eng. 1993;15:497–513. doi: 10.1080/01919512.1993.10555741. [DOI] [Google Scholar]
- Rice R.G., Robson C.M., Miller G.W., Hill A.G. American Water Works Association; 1981. Uses of Ozone in Drinking Water Treatment; pp. 44–57. [Google Scholar]
- Rodriguez-Manzano J., Alonso J.L., Ferrus M.A., Moreno Y., Amoros I., Calgua B., Hundesa A., Guerrero-Latorre L., Carratala A., Rusinol M., Girones R. Standard and new faecal indicators and pathogens in sewage treatment plants, microbiological parameters for improving the control of reclaimed water. Water Sci. Technol. 2012;66:2517–2523. doi: 10.2166/wst.2012.233. [DOI] [PubMed] [Google Scholar]
- Samuelsson L.M., Bjorlenius B., Forlin L., Larsson D.G. Reproducible (1)H NMR-based metabolomic responses in fish exposed to different sewage effluents in two separate studies. Environ. Sci. Technol. 2011;45:1703–1710. doi: 10.1021/es104111x. [DOI] [PubMed] [Google Scholar]
- Shannon M.A., Bohn P.W., Elimelech M., Georgiadis J.G., Marinas B.J., Mayes A.M. Science and technology for water purification in the coming decades. Nature. 2008;452:301–310. doi: 10.1038/nature06599. [DOI] [PubMed] [Google Scholar]
- Shi H., Xagoraraki I., Parent K.N., Bruening M.L., Tarabara V.V. Elution is a critical step for recovering human adenovirus 40 from tap water and surface water by cross-Flow ultrafiltration. Appl. Environ. Microbiol. 2016;82:4982–4993. doi: 10.1128/AEM.00870-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston-Enriquez J.A., Haas C.N., Jacangelo J., Gerba C.P. Inactivation of enteric adenovirus and feline calicivirus by ozone. Water Res. 2005;39:3650–3656. doi: 10.1016/j.watres.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Tyrrell S.A., Rippey S.R., Watkins W.D. Inactivation of bacterial and viral indicators in secondary sewage effluents, using chlorine and ozone. Water Res. 1995;29:2483–2490. doi: 10.1016/0043-1354(95)00103-R. [DOI] [Google Scholar]
- Xu P., Janex M.L., Savoye P., Cockx A., Lazarova V. Wastewater disinfection by ozone: main parameters for process design. Water Res. 2002;36:1043–1055. doi: 10.1016/S0043-1354(01)00298-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.