Summary
Molecular diagnostics are revolutionising the clinical practice of infectious disease. Their effects will be significant in acute-care settings where timely and accurate diagnostic tools are critical for patient treatment decisions and outcomes. PCR is the most well-developed molecular technique up to now, and has a wide range of already fulfilled, and potential, clinical applications, including specific or broad-spectrum pathogen detection, evaluation of emerging novel infections, surveillance, early detection of biothreat agents, and antimicrobial resistance profiling. PCR-based methods may also be cost effective relative to traditional testing procedures. Further advancement of technology is needed to improve automation, optimise detection sensitivity and specificity, and expand the capacity to detect multiple targets simultaneously (multiplexing). This review provides an up-to-date look at the general principles, diagnostic value, and limitations of the most current PCR-based platforms as they evolve from bench to bedside.
Pathogen identification: scope of the problem
In the USA, hospitals report well over 5 million cases of recognised infectious-disease-related illnesses annually.1 Significantly greater numbers remain unrecognised, both in the inpatient and community settings, resulting in substantial morbidity and mortality.2 Critical and timely intervention for infectious disease relies on rapid and accurate detection of the pathogen in the acute-care setting and beyond. The recent anthrax-related bioterrorist events and the outbreak of severe acute respiratory syndrome (SARS) further underscore the importance of rapid diagnostics for early, informed decision-making related to patient triage, infection control, treatment, and vaccination with life-and-death consequences for patients, health providers, and the public.3, 4, 5 Unfortunately, despite the recognition that outcomes from infectious illnesses are directly associated with time to pathogen identification, conventional hospital laboratories remain encumbered by traditional, slow multistep culture-based assays, which preclude application of diagnostic test results in the acute and critical-care settings. Other limitations of the conventional laboratory include extremely prolonged assay times for fastidious pathogens (up to several weeks); requirements for additional testing and wait times for characterising detected pathogens (ie, discernment of species, strain, virulence factors, and antimicrobial resistance); diminished test sensitivity for patients who have received antibiotics; and inability to culture certain pathogens in disease states associated with microbial infection.2, 6
The failure of either clinical judgment or diagnostic technology to provide quick and accurate data for identifying the pathogen infecting patients leads most clinicians to adopt a conservative management approach. Empiric intravenous antibiotic therapy (most common in acute-care settings such as emergency departments and intensive care units) offers the advantages of maximum patient safety and improved outcomes. The benefits of conservative management may be offset, however, by added costs and potential iatrogenic complications associated with unnecessary treatment and hospitalisations, as well as increased rates of antimicrobial resistance.7, 8, 9 A rapid reliable diagnostic assay, which allows for accurate identification of infected patients and informed early therapeutic intervention, would thus be invaluable for emergency and critical care physicians.
For more than a decade, molecular testing has been heralded as the “diagnostic tool for the new millennium”, whose ultimate potential could render traditional hospital laboratories obsolete.10, 11, 12 However, with the evolution of novel diagnostics tools, difficult questions have arisen regarding the role of such testing in the assessment of clinical infectious diseases. As molecular diagnostics continue to flow from bench to bedside, clinicians must acquire a working knowledge of the principles, diagnostic value, and limitations of varied assays.13 Here we discuss the most promising molecular diagnostic techniques for infectious diseases in hospital-based settings: the emphasis is on PCR-based methods since they have reached greatest maturity; existing assays, current, and future applications are described. Further, a framework for describing limitations that have been encountered, as well as speculation regarding the potential effect of these developments from the patient, physician, hospital, and societal perspective is provided.
Nucleic-acid-based amplification: historical perspective
The first nucleic-acid-based assays used DNA probe technology.14, 15, 16 DNA probes are short, labelled, single-strand segments of DNA that are designed and synthesised to hybridise targeted complementary sequences of microbial DNA. By contrast with traditional culture-based methods of microbial identification, which rely on phenotypic characteristics, this molecular fingerprinting technique relies on sequence-based hybridisation chemistry, which confers greater specificity to pathogen identification. Direct detection of target microbial DNA in clinical samples also eliminates the need for cultivation, drastically reducing the time required for reporting of results. In 1980, the description of DNA hybridising probes for detecting enterotoxigenic Escherichia coli in stool samples raised hopes that nucleic-acid-based technologies would eventually replace traditional culture techniques.17 Since that time, however, a more restrained approach has been adopted due to recognition of technical limitations of the methodology; most notably, the large amount of starting target DNA required for analysis, which results in poor detection sensitivity.18
To attain optimum sensitivity, critical for most clinical applications, researchers sought to directly amplify target microbial DNA. The development of the PCR technique in 1985 answered this need, and provided what is now the best-developed and most widely used method for target DNA amplification. Other approaches, including amplification of the hybridising probes (eg, ligase chain reaction and Q-beta replicase amplification) and amplification of the signals generated from hybridising probes (eg, branched DNA and hybrid capture), and transcription-based amplification (eg, nucleic-acid-sequence-based amplification and transcription-mediated amplification) have also been incorporated into various detection systems.19 Detailed descriptions of these technologies are beyond the scope of this review, but are well summarised elsewhere.20
PCR: basic principles and overview
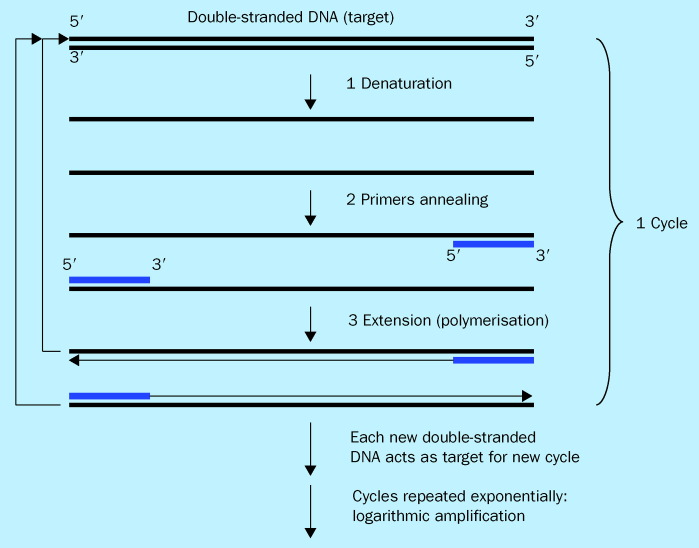
PCR is an enzyme-driven process for amplifying short regions of DNA in vitro. The method relies on knowing at least partial sequences of the target DNA a priori and using them to design oligonucleotide primers that hybridise specifically to the target sequences. In PCR, the target DNA is copied by a thermostable DNA polymerase enzyme, in the presence of nucleotides and primers. Through multiple cycles of heating and cooling in a thermocycler to produce rounds of target DNA denaturation, primer hybridisation, and primer extension, the target DNA is amplified exponentially (figure 1 ). Theoretically, this method has the potential to generate billions of copies of target DNA from a single copy in less than 1 h. For more detailed discussion of the basic principles of PCR see 21, 22, 23, 24, 25.
Figure 1.
Schematic of PCR. The PCR reaction takes place in a thermocycler. Each PCR cycle consists of three major steps: (1) denaturation of template DNA into single-stranded DNA; (2) primers annealing to their complementary target sequences; and (3) extension of primers via DNA polymerisation to generate new copy of the target DNA. At the end of each cycle the newly synthesised DNA act as new targets for the next cycle. Subsequently, by repeating the cycle multiple times, logarithmic amplification of the target DNA occurs.
Over the past two decades, PCR has been extensively modified to expand its utility and versatility. Multiplex PCR enables the simultaneous detection of several target sequences by incorporation of multiple sets of primers.26 To increase sensitivity and specificity, a double amplification step can be done with appropriately designed “nested” primers.27 Amplification may be made less specific to detect divergent genomes by randomising portions of the primer sets.28 Finally, RNA (rather than DNA) can be detected by converting RNA into a complementary DNA copy, and then amplifying (so-called reverse transcriptase PCR, or RT-PCR), enabling evaluation of RNA viruses or viable organisms.27
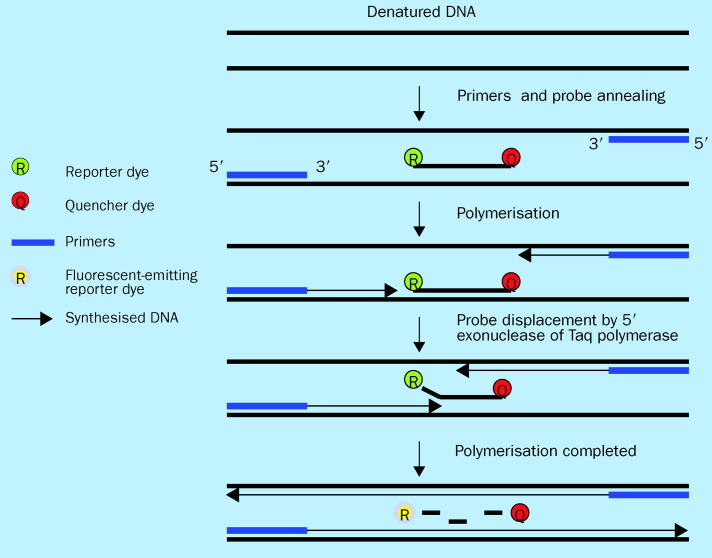
A significant advancement in PCR technology is quantitative real-time PCR, in which amplification and detection of amplified products are coupled in a single reaction vessel. For purposes of clinical applicability, this process represents a major breakthrough since it eliminates the need for laborious post-amplification processing (ie, gel electrophoresis) conventionally needed for amplicon detection, and allows for measurement of product simultaneous with DNA synthesis. One approach for real-time monitoring of amplicon production is to use fluorescent DNA intercalating dyes, such as SYBR-Green I, which bind non-specifically to double-stranded DNA generated during amplification.29 A more popular alternative approach is to use a fluorescent-labelled internal DNA probe which specifically anneals within the target amplification region. The choice of probe format depends on the compatibility of its hybridisation chemistry with the experimental design. Variations in probe format include TaqMan (Applied Biosystems; figure 2 ), fluorescence resonance energy transfer (FRET), and molecular beacon probes.30, 31, 32 Regardless of the format chosen, the internal probe emits a fluorescent signal during each amplification cycle only in the presence of target sequences, with signal intensity increasing in proportion to the amount of amplified products generated. The amount of starting templates in a specimen can be quantified by comparing the exact cycle number at which amplified products accumulate significantly over baseline with a pre-derived quantitative standard. Development of automated instrumentation with quantitative capacity insures reproducibility. Practical advantages of real-time PCR over conventional PCR are thus myriad and include speed, simplicity, reproducibility, and quantitative capacity.
Figure 2.
Real-time PCR using Taqman probe. Taqman probe is a single-stranded oligonucleotide that is labelled with two different fluorescent dyes. On the 5′ terminus is a reporter dye and on the 3′ terminus is a quenching dye. This oligonucleotide probe sequence is homologous to an internal target sequence present in the PCR amplified product. When the probe is intact, the proximity of the two fluorescent dyes results in quenching of the reporter dye emission by the quencher dye. During the extension phase of PCR the probe is cleaved by 5′ exonuclease activity of Taq polymerase thereby releasing the reporter from the quencher and producing an increase in reporter emission intensity which can detected and quantified. As amplification continues, the amount of reporter dye signal measured is proportional to the amount of PCR product made.
PCR-based diagnostics have been effectively developed for a wide range of microbes. Due to its incredible sensitivity, specificity, and speed of amplification, PCR has been championed by infectious disease experts for identifying organisms that cannot be grown in vitro, or in instances where existing culture techniques are insensitive and/or need prolonged incubation times.33, 34 Application to the clinical arena, however, has met with variable success so far. Only a limited number of assays have been approved by the US Food and Drug Administration (FDA; table 1 ) and fewer still have achieved universal acceptance in clinical practice.47 Furthermore, surprisingly limited effort has been focused on harnessing these time-saving diagnostics for emergency department and other acute critical-care settings where time is of the essence. A discussion of the progress made and the obstacles remaining to be addressed follows.
Table 1.
FDA-approved nucleic-acid-based assays for detection of microbial pathogens
| Organism detected | Trade name | Company/institution | Method | Clinical sensitivity | Clinical specificity |
|---|---|---|---|---|---|
| Chlamydia trachomatis | Amplicor | Roche | PCR | 93·219 | 98·4119 |
| LCX | Abbott | LCR | >9519 | >9919 | |
| AMP | Gen-Probe | TMA | 86·7–99·219 | >9919 | |
| PACE 2 | Gen-Probe | Hybridisation | 60·8–78·135 | >9935 | |
| BDProbeTec | Becton Dickinson | SDA | 94·036 | >9936 | |
| Hybrid capture II CT-ID | Digene | Hybrid capture | 95·437 | 9937 | |
| Cytomegalovirus | CMV pp67 mRNA | Organon Teknika | NASBA | 9538 | 9838 |
| Hybrid capture CMV DNA test | Digene | Hybrid capture | 9539 | 9539 | |
| Gardnerella vaginalis | Affirm VIP III | Becton Dickinson | Hybridisation | 9440 | 8140 |
| Group A streptococcus | GP-ST test | Gen-Probe | Hybridisation | 88·619 | 97·819 |
| Group B streptococcus | IDI-StrepB | Infectio Diagnostics | Real-time PCR | 9719 | 10019 |
| HCV | Amplicor HCV | Roche | PCR | 9819 | NA |
| Versant HCV RNA qualitative assay | Bayer | TMA | NA | 9841 | |
| Versant HCV RNA 3·0 | Bayer | BDNA | NA | 98·242 | |
| HIV | Amplicor HIV-1 Monitor Test | Roche | RT-PCR | NA | >9919 |
| Trugene HIV drug resistance and OpenGene DNA sequencing | Visible Genetics | DNA sequencing | NA | NA | |
| NucliSens EasyQ HIV-1 | bioMerieux | NASBA | NA | >9919 | |
| Procleix HIV-1/HCV | Chiron | TMA | >9943 | >9943 | |
| Versant HIV-1 RNA 3·0 | Bayer | BDNA | 97·644 | ||
| ViroSeq | Applied Biosystems | DNA sequencing | NA | NA | |
| HPV | Hybrid capture II HPV DNA | Digene | Hybrid capture | >9945 | 85–9045 |
| M tuberculosis | TB Amplicor | Roche | PCR | 79·4–91·919 | >9919 |
| E-MTD | Gen-Probe | TMA | 90·9–95·219 | >9919 | |
| Neisseria gonorrhoeae | Amplicor | Roche | PCR | NA | NA |
| LCX | Abbott | LCR | >9519 | >9919 | |
| Hybrid capture II CT/GC | Digene | Hybrid capture | 9346 | 98·546 | |
| BDProbeTec | Becton Dickinson | SDA | 88·936 | >9936 | |
| PACE-2 | Gen-Probe | Hybridisation | 9735 | 9935 | |
| Trichomonas vaginalis | Affirm VIP III | Becton Dickinson | Hybridisation | 88–91 938 | 10038 |
BDNA=branched DNA; LCR=ligase chain reaction; NASBA=nucleic-acid-sequence-based amplification; PCR=polymerase chain reaction; RT-PCR=reverse transcriptase PCR; SDA=strand displacement amplification; TMA=transcription mediated amplification; NA=not applicable. Adapted from reference 19.
Specific PCR diagnostics: development and clinical applications
With the increasing number of genomes of infectious pathogens being sequenced, catalogues of genes can be exploited to serve as amplification targets fundamental to the design of clinically useful diagnostic tests. As a result, over the past decade the number of PCR assays developed commercially and in hospital-based laboratories (“inhouse”) has continued to expand. Among the assays that have been developed for detection of specific microbes, three are described in more detail below, along with a discussion of their pros and cons relative to conventional diagnostic methodologies (table 2 ).
Table 2.
The pros and cons of PCR-based versus conventional diagnostic methods for detection of three target organisms
| Target organism | Conventional diagnostic method | PCR-based method | ||||
|---|---|---|---|---|---|---|
| Method | Pros | Cons | Method | Pros | Cons | |
| M tuberculosis | Culture | Allows susceptibility testing | Inadequate sensitivity | PCR (TB Aamplicor) | High sensitivity (93%)/ specificity (98–100%)19 | Potential contamination |
| High specificity with nucleic-acid-based identification | Prolonged time to result (>2 weeks) | Rapid detection time | Unable to assess viability | |||
| Requires further identification after positive culture | No transport requirement | |||||
| High cost (direct and indirect) associated with delayed diagnosis | Allows detection from non-invasive specimens | Limited ability for genotype and susceptibility testing | ||||
| Best for diagnosis and screening | ||||||
| Acid-fast stain | Rapid detection | Inadequate sensitivity/specificity | ||||
| C trachomatis | Culture | High specificity (100%)48 | Low sensitivity (70–80%)49 | PCR (Amplicor) | Rapid detection time | Limited ability for testing multidrug-resistance |
| Invasive specimen collection | Moderate to high sensitivity (80–90%)19 with high specificity (95–100%)19 | |||||
| Detects only viable organisms | Prolonged time to result | Currently used as an adjunctive test | ||||
| Allows further genotype or susceptibility testing | Cold transport | |||||
| High cost | Probably cost effective51 | |||||
| Antigen detection method (dfa) | High specificity (98–99%)50 | Requires technical expertise | ||||
| Allows assessment of specimen adequacy | Moderate sensitivity (80–90%)50 | |||||
| Rapid detection time | Requires high expertise | |||||
| No transport requirement | ||||||
| Enterovirus | Viral isolation | High specificity (100%)52 | Prolonged time to result (10–14 days) | RT-PCR (Amplicor EV) | Higher sensitivity (98%)53 | |
| High specificity (94%)53 | ||||||
| Low sensitivity (65–75%)53 | Rapid detection time | |||||
| Multiple cell lines needed for isolation | More adaptable for serotyping | |||||
| Serotyping time-consuming, labour intensive, and costly | Cost effective54 | |||||
One of the earliest recognised applications of PCR for clinical practice was for detection of Mycobacterium tuberculosis.54, 55 Disease characteristics favouring the development of a non-culture-based test for tuberculosis included week-long to month-long delays associated with standard testing, and the public-health imperative associated with early recognition, isolation, and treatment of infected patients. Two PCR-based assays are approved by the FDA for direct detection of M tuberculosis from clinical specimens (table 1). Although these assays result in significant improvements in time to diagnosis, the only FDA approved use at this time is as a diagnostic adjunct to the conventional smear and culture. Nonetheless, recent studies suggest that more widespread use of these assays may significantly affect patient management, clinical outcomes, and cost efficacy.56, 57 Potential yet unrealised applications of the assay for use in acute-care settings include earlier informed decision-making for appropriate use of isolation beds in high prevalence sites, regional outbreaks, or where isolation beds are scarce. The use of this and other PCR-based diagnostics in these settings, however, will have to be balanced against costs, expertise, and time associated with routine around-the-clock availability of testing, as discussed in more detail below.
Another PCR-based diagnostic assay, which has gained widespread acceptance, is that for Chlamydia trachomatis. Conventional detection systems for this organism (ie, culture and direct antigen testing by immunoassays), have been limited by the requirement for specialised facilities to culture this fastidious microbe, as well as inadequate sensitivity and specificity of immunoassays relative to the gold standard culture results.58 Laboratory development and subsequent clinical validation testing have indicated excellent sensitivity and specificity of the PCR assay leading to its commercial development (table 1). Proven efficacy of the PCR assay for both genital and urine samples has resulted in its application to a range of clinical settings, most recently routine screening of emergency department patients considered at risk for sexually transmitted diseases.59, 60 Although studies in the acute-care settings have not yet used PCR assays for C trachomatis on site and in real time (thus not taking full advantage of the speed of PCR), routine use of this assay in the aforementioned studies has resulted in nearly three-fold greater rates of disease detection and treatment relative to standard care.
Distinguishing life-threatening causes of fever from more benign causes in children is a fundamental clinical dilemma faced by clinicians, especially when infections of the central nervous system are being considered. Bacterial causes of meningitis can be highly aggressive but generally cannot be differentiated on a clinical basis from aseptic meningitis, a benign condition generally appropriate for outpatient management.61 Culture methods often take several days to show positive results and are confounded by poor sensitivity or false-negative findings in patients receiving empiric antimicrobials.62 One well developed assay, which has the potential to influence the management of patients in the acute-care setting, allows early and rapid diagnosis of diseases of viral cause. Testing and application of a PCR assay for enteroviral meningitis has been seen to be highly sensitive.63, 64 With reporting of results within 1 day, preliminary clinical trials have shown significant decreases in hospital costs due to decreased duration of hospital stays and courses of antibiotic therapy.65, 66 Other viral PCR assays, now routinely available, include those for herpes simplex virus, cytomegalovirus, Epstein-Barr virus, hepatitis viruses, and HIV.67 Each has a proven cost-saving role in clinical practice, including detection of otherwise difficult to diagnose infections, and a newly realised capacity to monitor progression of disease and response to therapy, vital to the management of chronic infectious diseases.68
With the increasing number of genomes of infectious pathogens being sequenced, catalogues of genes can be exploited to serve as amplification targets. As a result, the number of PCR assays developed both commercially and in-house continues to expand.
Broad-ranged PCR
The notion of a universal detection system has been proposed for the identification of classes of pathogens and speaks most directly to the future potential effect of PCR-based assays for clinical practice in emergency and other acute critical-care settings.69 Experimental work has focused on using sequences of the 16S rRNA gene, an evolutionarily conserved gene seen exclusively in bacterial species.70, 71 By designing primers that are complementary to these regions, investigators can, in theory, establish the presence of any bacteria in an otherwise sterile clinical specimen (such as cerebrospinal fluid or whole blood). Clinical applications are profound. Acute-care physicians could rapidly identify the presence of bacteraemia. Previous empiric decision-making could be abandoned in favour of educated practice, allowing appropriate, expeditious decision-making about the need for antibiotic therapy and hospitalisation. In principle, this approach could be applied to other taxonomic groups of pathogens (eg, genus of species, families of viruses, or fungi) by exploiting common features of classes of organisms for broad-range PCR assay design.72
Validation of this technique for eubacterial detection has focused on “high yield” clinical settings where expeditious identification of the presence of systemic bacterial infection has immediate high morbidity and mortality consequences. Notable clinical trials have included assessment of patients at risk for infective endocarditis,73, 74, 75 febrile infants at risk for sepsis,76, 77 febrile neutropenic cancer patients,78 and critically ill patients in the intensive care unit.79 While several of these studies have reported promising results (with sensitivity and specificity for bacteraemia well above 90%), significant technical difficulties remain, preventing general acceptance of these assays in clinics and hospitals (see Limitations below).
One significant investigational role for broad-range PCR has been its use as a “molecular petri dish” to identify emerging or existing infectious causes for diseases previously described as idiopathic. The DNA amplified using this broad-range approach may contain intervening sequence information that is phylogenetically specific to a unique microbe when compared with existing microbial genetic databases. For example, sequencing of the 16S rRNA gene amplified via highly conserved primer sets has led to the identification of Bartonella henselae in bacillary angiomatosis, and Tropheryma whipplei as the uncultured bacillus associated with Whipple's disease.80 Further, recent epidemiological studies that suggest a strong association between Chlamydia pneumoniae and coronary artery disease serve as an example of the possible widespread, yet undiscovered, links between pathogen and host which may ultimately lead to new insights into pathogenesis and development of novel life sustaining or saving therapeutics.81
The most recent, high-profile investigational use of broad-range PCR was in the molecular identification of a coronavirus as the causative agent in SARS. In a variant approach to PCR assay development, broad-based primers with degenerate sequences designed to detect unknown viruses were used to randomly amplify the genetic contents of infected clinical isolates. A subset of the amplified sequences showed homologies to the genus of coronavirus,82, 83 consistent with other confirmatory laboratory test results. Soon afterwards, a coronavirus-specific PCR assay was developed for rapid laboratory diagnosis of SARS.84 Notably, these advancements came only weeks after the first reports of the disease surfaced—a veritable tour de force bespeaking the power of broad-range PCR.85
Antimicrobial resistance profiling
With multidrug-resistant pathogens on the rise, early antimicrobial resistance profiling is crucial both for timely, objective treatment of infected patients, as well as for broader public-health surveillance. Conventional tests of this type are limited by prolonged culturing time (48–72 h) and poor accuracy due to variability in inoculum size and culturing conditions. To address these shortcomings, nucleic-acid-based assays are being advanced as genetic mechanisms of drug resistance are elucidated. Three examples of the clinical applicability of resistance profiling follow.
Although the presence of a resistance gene does not necessarily imply its expression and conferment of phenotypic resistance, its absence does establish a lack of resistance through that particular genetic mechanism: for example, meticillin resistance is mediated by the mecA gene. A distinctive feature of meticillin resistance is its heterogenous expression. As such, when typical phenotypic susceptibility testing is used to assess resistance, meticillin-resistant strains may seem falsely susceptible to some β-lactam antibiotics in vitro.86 For this reason, direct detection of the mecA gene by PCR is more desirable. With its high detection sensitivity and specificity, mecA PCR has gained wide acceptance and is becoming the most reliable method of identifying meticillin-resistant Staphylococcus aureus (MRSA).87
PCR-based resistance testing in M tuberculosis has also been developed for the detection of rifampicin resistance. Rifampicin resistance is well characterised and conferred by mutations within a short sequence of the rpoB gene of M tuberculosis, which result in aminoacid substitutions in the rpoB subunit of RNA polymerase.88 The Line Probe assay (LiPA; Inno-Genetics) is a commercially available PCR-based assay that targets the mutation-prone segment of the rpoB gene.89 Correlation with standard resistance-detection methods has been more than 90% and is shown to provide clinicians with a drastic reduction in detection time, critical for treatment decisions.90 Genotypic analysis of other M tuberculosis drug resistance is more challenging due to the number of mutations and genetic loci involved. Technical innovations (ie, multiplex PCR or DNA microarray) that allow simultaneous amplification and analysis of multiple target sequences will likely provide the means to surmount this later limitation.91, 92
With ever-increasing evidence supporting the prognostic value of identifying drug-resistant mutations, routine genotypic resistance testing is now standard care in the treatment of HIV-infected patients.93, 94, 95 PCR followed by nucleotide sequencing is the most commonly used method. Although genotypic tests are more complex than typical antimicrobial susceptibility tests, their ability to detect mutations at concentrations too low to affect drug susceptibility in a phenotypic assay provides insight into the potential for resistance to emerge. They also have the advantage of detecting transitional mutations that do not themselves cause drug resistance but indicate the presence of selective drug pressure, with potential importance for individual patient treatment decisions.
Applications in bioterrorism
The increasing threat of bioterrorism has gained considerable attention in light of the anthrax outbreak that came after the September 11, 2001 terrorist attacks. It has become increasingly apparent that responsibility for the rapid recognition and accurate diagnosis of real or suspected bioterrorism events will fall principally to front-line acute-care physicians who will be critical in initiating appropriate response measures.96 Unfortunately, as was seen with the 2001 anthrax episode, the clinical presentation of bioterrorism victims may be non-specific and difficult to distinguish from commonly encountered disease processes.96
The previously described limitations of conventional culture-based assays make such tests wholly inadequate for detection of bioterrorism agents in suspected clinical outbreaks. Furthermore, traditional microbiological methods, which require prolonged incubation, increase biohazard risk at the hospital laboratory due to unnecessary propagation of bioterrorism pathogens in culture-based systems. Wide recognition of these limitations has led to recent developments and refinements of PCR-based assays for a number of category A bioterrorism agents, including variola major, Bacillus anthracis, Yersinia pestis, and Francisella tularensis. 97, 98, 99, 100 PCR diagnostics for bioterrorism agents will likely be used both for diagnosis of symptomatic individuals, as well as larger scale screening of exposed victims (preclinical phase), who would be candidates for early prophylactic therapy.
Although most bioterrorism-induced illnesses resemble natural outbreaks, there is the possibility that causative bioterrorism agents are genetically engineered to increase virulence, acquire resistance to antibiotics or vaccines, or produce phenotypic characteristics that resemble multiple, simultaneous infections, so-called binary agents (via insertion of recombinant genes).101 In such cases, it is likely that nucleic-acid-based approaches will be more invaluable than conventional detection methods since they are the more easily adaptable and capable of uncovering detailed information embedded in genetic sequences.
Cost effectiveness
PCR is more expensive than conventional approaches. The direct costs of PCR reagents, equipment, dedicated space, personnel training, and labour have been reported to be as high as US$125 per reaction.102 Even among PCR methods, there is variability in cost with the most expensive being fluorogenic-based systems. Moreover, the labour intensity needed for most assays as well as technical limitations of most thermocyclers to do multiple runs of PCR simultaneously have prevented routine around-the-clock testing in the clinical setting. On the other hand, continued refinement in PCR technology, as well as improvements in automation and reproducibility via high throughput robotics, will probably lead to increasing demand and marked cost reductions to rates competitive with traditional methods. Already, this development has been reported for Neisseria gonorrhoeae and C trachomatis PCR tests which now cost around $9 per reaction.103
In assessing the overall benefit of PCR, however, direct monetary costs should not be the only consideration since the assay has several significant advantages over traditional methods. One study, which took a global methodological approach to cost, involved assessment of perinatal screening for Group B streptococcus using PCR versus culture techniques.104 Considered variables of the assays in addition to the direct monetary cost included infections averted, mortality, infant disabilities, hospital stays, and the societal benefits of healthy infants. Overall, the authors concluded that the benefits of PCR outweighed its cost. Notably, this result was reached even without inclusion of important but difficult-to-measure parameters, such as the societal benefit of decreased drug resistance due to targeted therapy made possible by the PCR assay.
Limitations of PCR and emerging innovations
The principal shortcomings in applying PCR assays to the clinical setting include false-positive results from background DNA contamination; the potential for false-negative test results; detection sensitivity exceeding clinical significance; and limited detection space of the assay or platform for simultaneous identification of multiple species, virulence factors, or drug resistance.
False positives
The widespread use of PCR in clinical settings has been hampered largely by background contamination from exogenous sources of DNA.105 In most pathogen-specific assays, the predominant source of contamination is derived from “carryover” products from earlier PCR reactions, which can be harboured and transmitted through PCR reagents, tubes, pipettes, and laboratory surfaces. Coupled with the robust amplification power of PCR, even very minor amounts of carry-over contamination may serve as substrates for amplification and lead to false-positive results. Meticulous control measures such as good laboratory practices and physical separation of preamplification and postamplification areas can reduce contamination risks but are not foolproof. The use of enzymatic inactivation of carry-over DNA (ie, uracil N-glycosylase) can further reduce contamination risk.106
Contamination issues are most pronounced in assays that use universal primers, such as those targeting conserved regions of the eubacterial 16S rRNA gene. Here, the ubiquitous presence of eubacterial DNA in either the environment or working reagents may lead to false-positive findings. Attempts to decontaminate PCR materials have involved nearly all known methods of destroying DNA including ultraviolet irradiation, chemical treatment, and enzymatic digestion.107, 108 None of these methods has been shown to be entirely effective without significant diminution of assay sensitivity. We have recently reported an alternative method that uses a size-based ultrafiltration step for reducing contaminating DNA from PCR reagents, primers, and DNA polymerase before amplification. Although this method of decontamination has been shown to be effective without compromising detection sensitivity in vitro,109 validation, and optimisation of the method in clinical samples needs further study. More importantly, effective and reliable methods of decontamination have not yet been developed for steps outside the assay proper such as sample collection and preparation. Towards this end, one promising area of investigation involves development of methods to integrate sample preparation, amplification and detection on a single platform, the so-called “lab-on-a-chip”. Self-contained microchip platforms thus hold promise for the best means of decontamination and overall assay efficiency.110
False negatives
PCR assays for microbial detection may give false-negative results for two principal reasons: the relatively small sample volume permissible for PCR reactions; and problems associated with PCR processing. The sample volume most PCR assays can accommodate is quite small relative to the volume used in conventional culture methods; as such, in cases in which the concentration of infectious organisms is low, the assay may yield false-negative findings. To account for this, DNA extraction and purification steps are usually performed before PCR amplification as a means of concentrating total DNA from a larger sample volume. Additional methods to optimise starting concentration of target DNA for the PCR reaction include: selecting specimen sources (eg, cerebrospinal fluid) or specimen fractions (eg, buffy coat instead of whole blood) with the highest abundance of microbial DNA for the DNA extraction; briefly cultivating samples to increase microbial load before DNA extraction;20 or introducing specific capture probes to concentrate only microbial DNA in a given sample.111 Several sample processing obstacles may also lead to false-negative findings. Three of the most commonly encountered problems are (1) inadequate removal of PCR inhibitors in the sample, such as haemoglobin, blood culture media, urine, and sputum; (2) ineffective release of microbial DNA content from the cells; or (3) poor DNA recovery after extraction and purification steps. Methods to ensure best sample processing include: incorporating internal amplification controls (eg, the human β-globin gene) to the PCR assay to monitor for presence of both purified sample DNA as well as potential PCR inhibitors;112 and inducing various chaotropic, enzymatic, or thermal methods of cell lysis to effectively liberate microbial DNA content.20 Because of the varying effectiveness of each of these measures, efforts to improve an assay's detection sensitivity may need to be individually adjusted based on the assay's clinical application and the microbial pathogen of interest.
Clinical significance of positive PCR
PCR assays may detect microbial pathogens at concentrations below those of previously established gold standard reference methods. Distinguishing whether this result represents a false-positive finding and establishing the clinical significance of these findings is challenging. In the past, discrepant analysis based on the results of additional ancillary tests was used to provide estimates of sensitivity and specificity in the presence of an imperfect gold standard.113 One example can be seen in assessments of novel nucleic acid-based assays in detecting C trachomatis. 114, 115 In these studies, “false positives” (DNA-amplification positive and tissue culture negative) were adjudicated by either antigen detection methods or another well-established DNA-amplification test. Despite its popularity, recent concerns have been raised regarding the potential bias incurred by discrepant analysis in favour of the new tests.116, 117 Up to now, the issue has not been completely resolved.
The complexity of the clinical interpretation of positive PCR findings is further underscored by one study that reported that a universal PCR assay (using primers from conserved regions of the 16S rRNA gene) amplified eubacterial DNA in blood samples from healthy people.118 It is unknown whether such findings are indicative of latent disease processes or sub-clinical colonisation. Moreover, the finding that microbial DNA can be detected even after successful antimicrobial treatment suggests that the assays detect both viable and non-viable organisms.119, 120 Clearly, interpretive guidelines based on the correlation of test results with clinical presentation and existing standards will be required before these assays can be used for definitive diagnosis and/or treatment decisions.
One breakthrough in establishing the meaning of positive PCR results involves the development of reliable quantitative measures of pathogen load. While traditional PCR assays are used primarily for dichotomous outcome, innovative real-time PCR methods allow for quantitative measurement of starting template in the sample, which will probably be useful in differentiating benign colonisation from either latent or active disease. Other non-PCR amplification methods with quantitative capacities include branched DNA and nucleic-acid-sequence-based amplification. Quantification of pathogen load is already well established in clinical virology (eg, HIV-1, cytomegalovirus, hepatitis B virus, hepatitis C virus, and Epstein-Barr virus), where it has proven useful in assessing disease severity or monitoring treatment efficacy.121, 122, 123, 124, 125, 126 The value and importance of PCR-based pathogen quantifications in clinical bacteriology remains under investigation.127
Alternative innovations regarding PCR technologies may help in differentiating viable from non-viable organisms, important for clinical practice decisions. RNA is known to be rapidly degraded with a typical half-life of minutes after cell death; thus, it has been proposed as a more accurate indicator of viable microorganisms.128, 129 In some clinical situations, detection of RNA species by RT-PCR has been shown to correlate well with the presence of viable organisms and has been effectively used to monitor antibiotic therapy.130, 131, 132, 133, 134 Clinical application of RNA-based approaches will need further improvement, however, because they have been hampered in development by difficulties in extracting detectable concentrations of intact RNA from small numbers of bacteria.
Limited detection space for characterising the detected pathogen
Conventional methods for pathogen detection will not be supplanted by PCR-based assays if the latter cannot be elaborated to further characterise detected pathogens. As described previously, genetic sequences contain rich sources of information that can be analysed to ascertain pathogens′ species or strains, virulence factors, and antimicrobial susceptibilities. However, to do so in a single reaction, simultaneous amplification of several target genes is needed. Repeating amplifications with different primer pairs, so-called multiplexing, is notoriously difficult since often one or more of the target sequences do not amplify.91
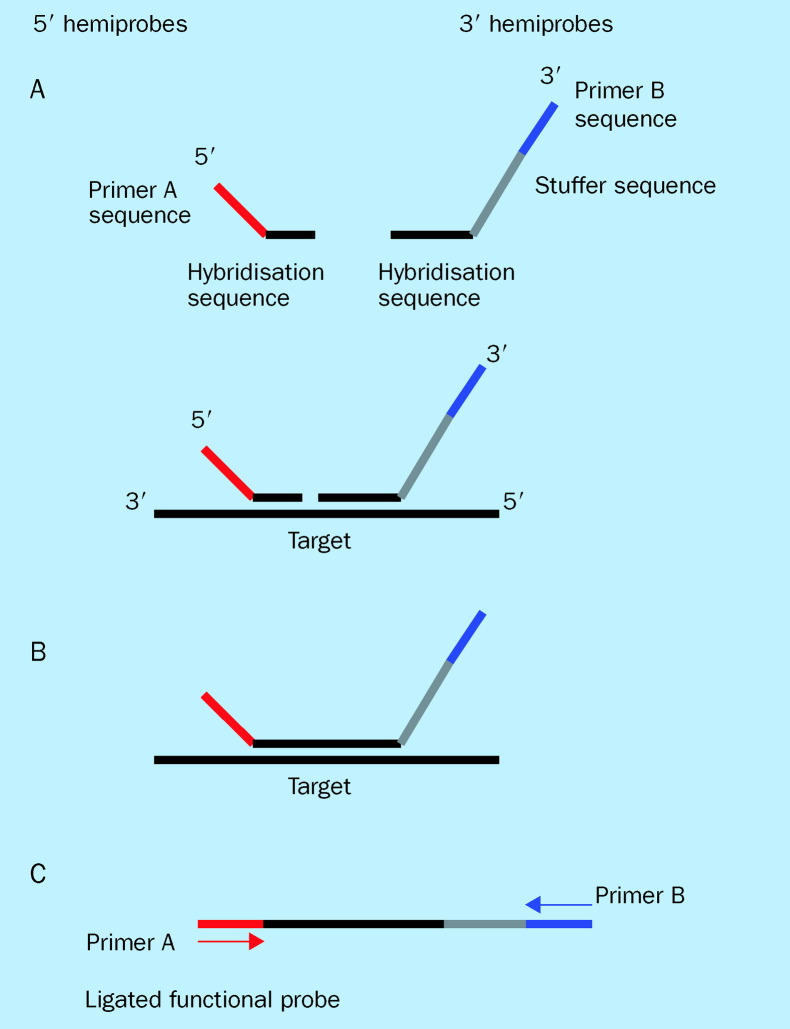
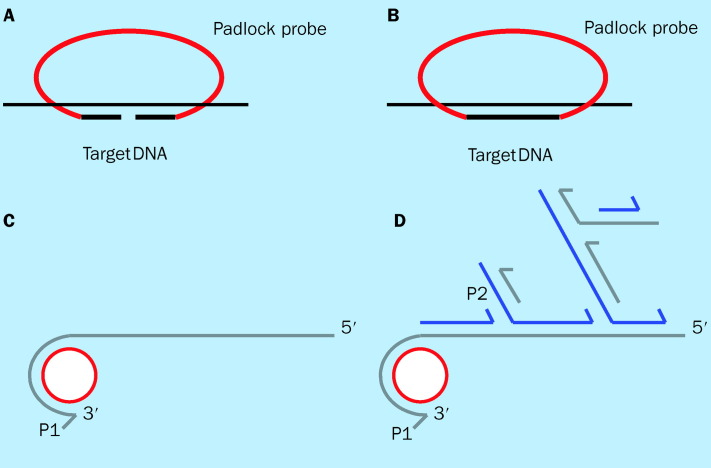
Recent studies have shown that PCR can be used for simultaneous reproducible amplification of multiple DNA fragments in a single reaction, provided that only a single primer set is used for amplification of all these fragments. Repetitive-sequence-based PCR (rep-PCR), which uses consensus PCR primers to amplify DNA sequences located between successive repetitive elements in eubacterial genomes, has been shown to simultaneously amplify fragments of different sizes, allowing discrimination of bacteria at the subspecies level.135, 136 This conceptual breakthrough has led various investigators to develop and explore various technical approaches which harness the same idea. By exploiting the conserved and variable sequences on the 16S rRNA gene, we have shown through use of quantitative PCR that a single consensus primer set can multiplex amplify multiple species of the 16S rRNA gene with equal efficiencies.109 Similarly, ligation-dependent PCR (figure 3 )137 and padlock probes with rolling circle amplification (figure 4 ),138 which are both probe amplification methods, have also shown that multiple genetic targets can be queried simultaneously by using a single primer pair for amplification. Both of these assays rely on multiple oligonucleotide probes, each containing a unique target sequence and a consensus primer sequence, that are amplifiable in the presence of their targets. Progress in these approaches could greatly enhance throughput in genotyping pathogens detected, and may represent the next generation of PCR-based assays that hold tremendous promise with regard to their clinical applications.
Figure 3.
Ligation-dependent PCR (LD-PCR). LD-PCR is a process in which non-amplifiable hemiprobes for each target will be constructed as follows: (A) For the 5′ hemiprobe the 5′ end will be a generic primer sequence shared by all 5′ hemiprobes, and the 3′ end will be target-specific. The 3′ hemiprobe will have a mirror symmetrical arrangement with an intervening stuffer sequence of variable length. (B) In the presence of target sequences the hemiprobes are juxtaposed to each other as they hybridise to their targets. (C) A single PCR primer set based on the generic sequences on the hemiprobes will be used for amplification of any ligated functional probes. The amplified product of each ligated functional probe has a unique length that can be separated by electrophoresis.
Figure 4.
Padlock probe with rolling circle amplification. (A) Padlock probe hybridises to the target sequence. (B) The padlock probe can be converted to a circle by ligation only if ends of the probe are matched to their target. (C) Rolling circle amplification begins when primer 1 (P1) anneals to circularised probe and polymerase copies probe sequence, eventually copyin the entire circle. The polymerase begins displacing the previously synthesised product, opening up single-stranded primer 2 (P2) binding site. (D) Each turn of displacement synthesis around the circle exposes another P2 binding site, and the resulting synthesis from the annealed P2 primers results in the displacement of downstream primers and products. Displacement of the downstream primers and products also opens up additional P1 binding sites, and the process continues in an exponential cascade. The amplification process occurs in an isothermal reaction. Adapted from reference 138.
Even if this problem is resolved, the best way in which to analyse the resultant PCR products remains unclear. While PCR product detection and analysis have typically been achieved using gel-electrophoresis and sequencing techniques, these approaches are laborious and time-consuming, which detracts from clinical applicability.139 The introduction of real-time PCR technology with the potential use of differentially labelled fluorescent probes for simultaneous identification of multiple amplified products in a single assay holds promise.140 Unfortunately, current ability to spectrally differentiate multiple fluorescent signals is quite limited.
Another possible approach that can be used to analyse multiple amplified sequences is to incorporate microarray technology. DNA microarrays are constructed by spatially isolating specific genome sequences to prearranged areas on a microchip.141, 142, 143 Fluorescently labelled amplification products are then allowed to anneal to complementary sequences on the chip, and the resultant pattern is spectrally analysed. The main advantage of using microarrays for pathogen detection is the potentially large number of target sequences the system can discriminate simultaneously. The use of microarray technology for pathogen detection is still in the development phase however. Efforts to improve sensitivity, reproducibility, and to streamline approaches to complex data analysis are still needed before these platforms can be used clinically.
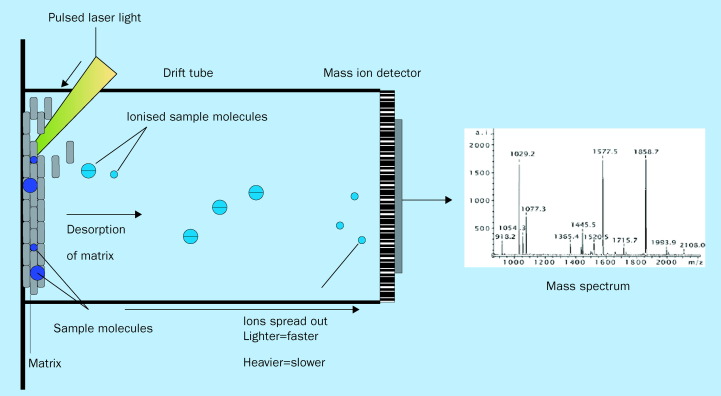
A final novel approach for analysing amplified products is mass spectrometry. The recent advent of matrix assisted laser description/ionisation (MALDI) technology coupled with time of flight mass spectrometry (TOF-MS) has created a robust means to characterise mostly proteins, but increasingly, nucleic acid. In MALDI-TOF-MS, the organic molecule—for example an amplified product—is ionised and subsequently identified based on its mass-to-charge ratio (figure 5 ).144, 145, 146, 147, 148, 149 The advantages of MALDI-TOF-MS lie in the inherent accuracy and the high-speed (1 second) of signal acquisition, making this technology an attractive candidate for high-throughput DNA analysis.
Figure 5.
MALDI-TOF mass spectrometry. Sample molecules (ie, amplified DNA products) are co-crystallised with matrix and then subjected to desorption and ionisation by an incident laser pulse. An applied electric field accelerates the resultant ionised sample molecules across the time-of-flight (TOF) drift tube in vacuum, and a detector at the end of the tube accurately measures the flight time from the ion source to the detector. Typically, ions with larger mass-to-charge (M/z) ratios travel more slowly than those with smaller m/z. The data are recorded as a “spectra” that displays ion intensity vs m/z value.
Practical aspects of rapid and point-of-care testing
The real-time PCR-based platform holds great promise in replacing conventional laboratory-based testing for future point-of-care testing. With advancements in automation, integration of specimen preparation with target identification, and miniaturisation, it will become much easier to bring analyses near bedside to be done by less-trained personnel. The ability to interface with high throughput PCR systems is already seen in many new automated extraction instruments. Technical limitations of most PCR instruments to run overlapping reactions in parallel have restricted analyses to batches, thereby compromising the assay's overall turnaround time. However, with the new generation of thermocycler (eg, SmartCycler, Cepheid), separate PCR reactions, each with a unique set of cycling protocols and data analysis, can now be done simultaneously. Furthermore, the recent introduction of hand-held battery-operated real-time PCR instruments (BioSeeq, Smiths Detection-Edgewood BioSeeq) is the latest iteration of the moving trend from laboratory to near patient testing.150 Ultimately, as “lab-on-a-chip” technologies mature, routine point-of-care testing will be realised.
The true effect of PCR development for true point-ofcare testing remains to be seen. Significant practical and regulatory requirements slow and often halt the transition of laboratory developments for bedside applications. Compliance with federal, state, and local regulations must be met when operating point-of-care testing devices. Novel tests must go through the complex and time-consuming process of FDA approval. For in-house assays, strict clinical laboratory improvement amendment requirements must be met to define the operational characteristics of the assay relative to current gold standards. Institutional resources, manpower issues, and cost effectiveness also have to be carefully considered when making decisions about the practicalities of replacing traditional diagnostic methodologies. Additional programmatic steps for true point-of-care testing must be developed to insure effectiveness, including (1) operational turn-around time (vs speed of the test); (2) education of practitioners in interpretation of results; (3) development of protocols for optimal treatment and decision-making based on results of novel tests; and (4) establishment of quality assurance and quality improvement programmes. As PCR-based technologies continue to mature, each of these issues will need to be systematically addressed in order to realise their benefit for routine patient care.151
Continuing clinical need: how PCR diagnostics may revolutionise clinical care
PCR technology offers a great potential in the arena of infectious disease. A universally reliable infectious disease diagnostic system will certainly become a fundamental tool in the evolving diagnostic armamentarium of the 21st century clinician. For front-line acute care physicians, or physicians working in disaster settings, a quick universal PCR assay, or panels of PCR assays targeting categories of pathogens involved in specific syndromes such as meningitis, pneumonia, or sepsis, would allow for rapid triage and early aggressive targeted therapy. Resources could thus be appropriately applied, and patients with suspected infections rapidly risk-stratified to the different treatment settings, depending on the pathogen and virulence. The ability to discern species and subtype would allow for more precise decision-making regarding antimicrobial agents. Patients who are colonised with highly contagious pathogens could be appropriately isolated on entry into the medical setting without delay. Targeted therapy would diminish development of antibiotic resistance, because the identification of antibiotic-resistant strains would permit precise pharmacological intervention. Both physicians and patients would benefit from less repetitive testing and elimination of wait times for traditional laboratory results. Furthermore, links with data management systems, locally, regionally, and nationally, would allow for effective epidemiological surveillance with obvious benefits for antibiotic selection and control of disease outbreaks.
It is certain that the individual patient will benefit directly from this approach. Patients with unrecognised or difficult-to-diagnose infections could be identified and treated promptly. Inpatient stays would be reduced with a concomitant decrease in iatrogenic events. Societal benefits will need to be carefully explored with attention to relative costs of the novel diagnostics in relation to existing standards.
Search strategy and selection criteria
Data for this review were identified by a search of Medline and from the references of relevant articles. Search terms used were: “PCR”, “molecular diagnostics”, “emerging infectious disease”, “bioterrorism agents”, “antimicrobial resistance”, “DNA microarrays”, and “mass spectrometry”. Only English language papers were considered.
Conflicts of interest
RER has served as an expert consultant to Ibis Therapeutics, a division of Isis Pharmaceuticals (Carlsbad, CA, USA), which develops diagnostic assays using both PCR and mass spectrometry techniques.
References
- 1.Centers for Disease Control and Prevention Increase in national hospital discharge survey rates for septicemia—United States. MMWR Morb Mortal Wkly Rep. 1990;39:31–34. [PubMed] [Google Scholar]
- 2.Sands KE, Bates DW, Lanken PN. Epidemiology of sepsis syndrome in 8 academic medical centers. JAMA. 1997;278:234–240. [PubMed] [Google Scholar]
- 3.Pavlin JA, Gilchrist MJR, Osweiler GD. Diagnostic analyses of biological agent-caused syndromes: laboratory and technical assistance. Emerg Med Clin North Am. 2002;20:331–350. doi: 10.1016/s0733-8627(01)00004-9. [DOI] [PubMed] [Google Scholar]
- 4.Klietmann WF, Ruoff KL. Bioterrorism: Implications for the Clinical Microbiologist. Clin Micro Rev. 2001;14:364–381. doi: 10.1128/CMR.14.2.364-381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerberding JL. Faster…but fast enough? Responding to the epidemic of severe acute respiratory syndrome. N Engl J Med. 2003;348:2030–2031. doi: 10.1056/NEJMe030067. [DOI] [PubMed] [Google Scholar]
- 6.Jungkind D. Tech.Sight. Molecular testing for infectious disease. Science. 2001;294:1553–1555. doi: 10.1126/science.294.5546.1553. [DOI] [PubMed] [Google Scholar]
- 7.Marantz PR, Linzer MM, Feiner CJ. Inability to predict diagnosis in febrile intravenous drug abusers. Ann Int Med. 1987;106:823828. doi: 10.7326/0003-4819-106-6-823. [DOI] [PubMed] [Google Scholar]
- 8.Samet JH, Shevitz A, Fowle J. Hospitalization decision in febrile intravenous drug users. Am J Med. 1990;89:5356. doi: 10.1016/0002-9343(90)90098-x. [DOI] [PubMed] [Google Scholar]
- 9.Cosgrove SE, Carmeli Y. The impact of antimicrobial resistance on health and economic outcomes. Clin Infect Dis. 2003;36:1433–1437. doi: 10.1086/375081. [DOI] [PubMed] [Google Scholar]
- 10.Pitt TL, Saunders NA. Molecular bacteriology: a diagnostic tool for the millennium. J Clin Pathol. 2000;53:71–75. doi: 10.1136/jcp.53.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich HA, Gelfand D, Sninsky JJ. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- 12.Naber SP. Molecular pathology—diagnosis of infectious disease. N Engl J Med. 1994;331:1212–1215. doi: 10.1056/NEJM199411033311808. [DOI] [PubMed] [Google Scholar]
- 13.Santis G, Evans TW. Molecular biology for the critical care physician. Crit Care Med. 1999;27:997–1004. doi: 10.1097/00003246-199905000-00043. [DOI] [PubMed] [Google Scholar]
- 14.Drake TA, Hindler JA, Berlin OG. Rapid identification of Mycobacterium avium complex in culture using DNA probes. J Clin Microbiol. 1987;25:1442–1445. doi: 10.1128/jcm.25.8.1442-1445.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Musial CE, Tice LS, Stockman L. Identification of mycobacteria from culture by using the Gen-Probe rapid diagnostic system for Mycobacterium avium complex and Mycobacterium tuberculosis complex. J Clin Microbiol. 1988;26:2120–2123. doi: 10.1128/jcm.26.10.2120-2123.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis JS, Fakil O, Foss E. Direct DNA probe assay for Neisseria gonorrhoeae in pharyngeal and rectal specimens. J Clin Microbiol. 1993;31:27838–27845. doi: 10.1128/jcm.31.10.2783-2785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moseley SL, Huq I, Alim AR, So M, Samadpour-Motalebi M, Falkow S. Detection of enterotoxigenic Escherichia coli by DNA colony hybridization. J Infect Dis. 1980;142:892–898. doi: 10.1093/infdis/142.6.892. [DOI] [PubMed] [Google Scholar]
- 18.Engleberg NC, Eisenstein BI. Detection of microbial nucleic acids for diagnostic purposes. Annu Rev Med. 1992;43:147–155. doi: 10.1146/annurev.me.43.020192.001051. [DOI] [PubMed] [Google Scholar]
- 19.Tang YW, Procop GW, Persing DH. Molecular diagnositics of infectious diseases. Clin Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- 20.Wolk D, Mitchell S, Patel R. Principles of molecular microbiology testing methods. Inf Dis Clin of North Am. 2001;15:1157–1204. doi: 10.1016/s0891-5520(05)70190-2. [DOI] [PubMed] [Google Scholar]
- 21.Mullis KB, Faloona FA. Specific synthesis of DNA in vitro via a polymerase-catalyzed reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 22.Mullis KB. The unusual origin of the polymerase chain reaction. Sci Am. 1990;262:56–65. doi: 10.1038/scientificamerican0490-56. [DOI] [PubMed] [Google Scholar]
- 23.Eisenstein BI. The polymerase chain reaction: a new method of using molecular genetics for medical diagnosis. N Engl J Med. 1990;332:178–183. doi: 10.1056/NEJM199001183220307. [DOI] [PubMed] [Google Scholar]
- 24.White TJ, Madej R, Persing DH. The polymerase chain reaction: clinical applications. Adv Clin Chem. 1992;29:161–196. doi: 10.1016/s0065-2423(08)60224-3. [DOI] [PubMed] [Google Scholar]
- 25.Persing DH. Diagnostic molecular microbiology: current challenges and future directions. Diagn Microbiol Infect Dis. 1993;16:159–163. doi: 10.1016/0732-8893(93)90015-y. [DOI] [PubMed] [Google Scholar]
- 26.Chamberlain JS, Gibbs R, Rainier JE. Detection screening of Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erlich HA, Gelfand D. Sninsky JJ. Recent advances in the polymerase chain reaction. Science. 1991;252:1643–1651. doi: 10.1126/science.2047872. [DOI] [PubMed] [Google Scholar]
- 28.Power EG. RAPD typing in microbiology—a technical review. J Hosp Infect. 1996;34:247–265. doi: 10.1016/s0195-6701(96)90106-1. [DOI] [PubMed] [Google Scholar]
- 29.Higuchi R, Dollinger G, Walsh PS. Simultaneous amplification and detection of specific DNA sequences. Biotechnology. 1992;10:413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 30.Heid CA, Stevens J, Livak KJ. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Zehnbauer B, Gnirke A. Fluorescence energy transfer detection as a homogeneous DNA diagnostic method. Proc Natl Acad Sci USA. 1997;94:10756–10761. doi: 10.1073/pnas.94.20.10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 33.Post JC, Ehrlich G. The impact of the polymerase chain reaction in clinical medicine. JAMA. 2000;283:1544–1546. doi: 10.1001/jama.283.12.1544. [DOI] [PubMed] [Google Scholar]
- 34.Louie M, Louie L, Simor A. The role of DNA amplification technology in the diagnosis of infectious disease. CMAJ. 2000;163:301–309. doi: 10.1016/s1381-1169(00)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Black CM, Marrazzo J, Johnson RE. Head-to-head multicenter comparison of DNA probe and nucleic acid amplification tests for Chlamydia trachomatis infection in women performed with an improved reference standard. J Clin Microbiol. 2002;40:3757–3763. doi: 10.1128/JCM.40.10.3757-3763.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Dyck E, Ieven M, Pattyn S. Detection of Chlamydia trachomatis and Neisseria gonorrhoeae by enzyme immunoassay, culture, and three nucleic acid amplification tests. J Clin Microbiol. 2001;39:1751–1756. doi: 10.1128/JCM.39.5.1751-1756.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girdner JL, Cullen AP, Salama TG. Evaluation of the digene hybrid capture II CT-ID test for detection of Chlamydia trachomatis in endocervical specimens. J Clin Microbiol. 1999;37:1579–1581. doi: 10.1128/jcm.37.5.1579-1581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witt DJ, Kemper M, Stead A. Analytical performance and clinical utility of a nucleic acid sequence-based amplification assay for detection of cytomegalovirus infection. J. Clin Microbiol. 2000;38:3994–3999. doi: 10.1128/jcm.38.11.3994-3999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazzulli T, Drew LW, Yen-Lieberman B. Multicenter comparison of the digene hybrid capture CMV DNA assay (version 2.0), the pp65 antigenemia assay, and cell culture for detection of cytomegalovirus viremia. J Clin Microbiol. 1999;37:958–963. doi: 10.1128/jcm.37.4.958-963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Briselden AM, Hillier SL. Evaluation of Affirm VP microbial identification test for Gardnerella vaginalis and. Trichomonas vaginalis. J Clin Microbiol. 1994;32:148–152. doi: 10.1128/jcm.32.1.148-152.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendricks DA, Friesenhahan M, Tanimoto L. Multicenter evaluation of the VERSANT HCV RNA qualitative assay for detection of hepatitis C virus RNA. J Clin Microbiol. 2003;41:651–656. doi: 10.1128/JCM.41.2.651-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trimoulet P, Halfon P, Pohier E. Evaluation of the VERSANT HCV RNA 3.0 assay for quantification of hepatitis C virus RNA in serum. J Clin Microbiol. 2002;40:2031–2036. doi: 10.1128/JCM.40.6.2031-2036.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vargo J, Smith K, Knott C. Clinical specificity and sensitivity of a blood screening assay for detection of HIV-1 and HCV RNA. Transfusion. 2002;42:876–885. doi: 10.1046/j.1537-2995.2002.00130.x. [DOI] [PubMed] [Google Scholar]
- 44.Gleaves CA, Welle J, Campbell M. Multicenter evaluation of the Bayer VERSANT HIV-1 RNA 3.0 assay: analytical and clinical performance. J Clin Virol. 2002;25:205–216. doi: 10.1016/s1386-6532(02)00011-2. [DOI] [PubMed] [Google Scholar]
- 45.Clavel C, Masure M, Bory JP. Human papillomavirus testing in primary screening for the detection of high-grade cervical lesions: a study of 7932 women. Br J Cancer. 2001;84:161623. doi: 10.1054/bjoc.2001.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schachter J, Hook EW 3rd, McCormack WM. Ability of the digene hybrid capture II test to identify Chlamydia trachomatis and Neisseria gonorrhoeae in cervical specimens. J Clin Microbiol. 1999;37:3668–3671. doi: 10.1128/jcm.37.11.3668-3671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altwegg M, Kayser FH. Will cultures survive? The role of molecular tests in diagnostic bacteriology. Infection. 1997;25:265–268. doi: 10.1007/BF01720394. [DOI] [PubMed] [Google Scholar]
- 48.Black CM. Current methods of laboratory diagnosis of chlamydia trachomatis infections. Clin Micro Rev. 1997;10:160–184. doi: 10.1128/cmr.10.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chernesky MA, Jang D, Lee J. Diagnosis of Chlamydia trachomatis infections in men and women by testing first-void urine by ligase chain reaction. J Clin Microbiol. 1994;32:2682–2685. doi: 10.1128/jcm.32.11.2682-2685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chernesky MA., Mahoney JB, Castriciano S. Detection of Chlamydia trachomatis antigens by enzyme immunoassay and immunofluorescence in genital specimens from symptomatic and asymptomatic men and women. J Infect Dis. 1986;154:141–148. doi: 10.1093/infdis/154.1.141. [DOI] [PubMed] [Google Scholar]
- 51.Croft RA, Croft RP. Expenditure and loss of income incurred by tuberculosis patients before reaching effective treatment in Bangladesh. Int J Tuberc Lung Dis. 1998;2:252–254. [PubMed] [Google Scholar]
- 52.Muir P, Kämmerer U, Korn K. Molecular typing of enteroviruses: current status and future requirements. Clin Micro Rev. 1998;11:202–227. doi: 10.1128/cmr.11.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carroll KC, Taggart B, Robison J. Evaluation of the roche AMPLICOR enterovirus PCR assay in the diagnosis of enteroviral central nervous system infections. J Clin Virol. 2000;19:149–156. doi: 10.1016/s1386-6532(00)00115-3. [DOI] [PubMed] [Google Scholar]
- 54.D'Amato RF, Wallman AA, Hochstein LH. Rapid diagnosis of pulmonary tuberculosis by using Roche AMPLIOR Mycobacterium tuberculosis PCR test. J Clin Microbiol. 1995;33:1832–1834. doi: 10.1128/jcm.33.7.1832-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piersimoni C, Callegaro A, Nista D. Comparative evaluation of two commercial amplification assays for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 1997;35:193–196. doi: 10.1128/jcm.35.1.193-196.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woods GL, Bergmann JS, Williams-Bouyer N. Clinical evaluation of the Gen-Probe amplified Mycobacterium tuberculosis direct test for rapid detection of Mycobacterium tuberculosis in select nonrespiratory specimens. J Clin Microbiol. 2001;39:747–749. doi: 10.1128/JCM.39.2.747-749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bergmann JS, Yuoh G, Fish G. Clinical evaluation of the enhanced Gen-Probe amplified Mycobacterium Tuberculosis direct test for rapid diagnosis of tuberculosis in prison inmates. J Clin Microbiol. 1999;37:1419–1425. doi: 10.1128/jcm.37.5.1419-1425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Black CM. Current methods of laboratory diagnosis of Chlamydia trachmatis infections. Clin Microbiol Rev. 1997;10:160–184. doi: 10.1128/cmr.10.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehta SD, Rothman RE, Kelen GD. Unsuspected gonorrhea and chlamydia in patients of an urban adult emergency department. STD. 2000;28:3339. doi: 10.1097/00007435-200101000-00008. [DOI] [PubMed] [Google Scholar]
- 60.Mehta SD, Rothman RE, Kelen GD. Clinical aspects of diagnosis of gonorrhea and chlamydia infection in an acute care setting. CID. 2001;32:655–659. doi: 10.1086/318711. [DOI] [PubMed] [Google Scholar]
- 61.Ogborn CJ, Soulen JL, DeAngelis C. Hospitalization vs. outpatient treatment of young febrile infants. 10-year comparison. Arch Pediatr Adol Med. 1995;149:94–97. doi: 10.1001/archpedi.1995.02170130096023. [DOI] [PubMed] [Google Scholar]
- 62.Dumler JS, Valsamakis A. Molecular diagnostics for existing and emerging infectious. Am J Clin Pathol. 1999;112:S33–S39. [PubMed] [Google Scholar]
- 63.Rotbart HA. Diagnosis of enteroviral meningitis with the polymerase chain reaction. J Pediatr. 1990;117:85–89. doi: 10.1016/s0022-3476(05)82451-5. [DOI] [PubMed] [Google Scholar]
- 64.Smalling TW, Sefers SE, Li H. Molecular approaches to detecting herpes simplex virus and enteroviruses in the central nervous system. J Clin Microbiol. 2002;40:2317–2322. doi: 10.1128/JCM.40.7.2317-2322.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stellrecht KA, Harding I, Woron AM. The impact of an enteroviral RT-PCR assay on the diagnosis of aseptic meningitis and patient management. J Clin Virol. 2002;25(suppl 1):S19–S26. doi: 10.1016/s1386-6532(02)00030-6. [DOI] [PubMed] [Google Scholar]
- 66.Romeroa J, Magelli M, Hinrishs S. Reverse transcription polymerase chain reaction diagnosis of enterovirus infections in febrile children results in reduced health care expenditures. In Program and Abstracts of the 36th Annual Meeting of Infectious Disease Society of America, Denver, CO, November 12–15. 1998 Abstract 668. [Google Scholar]
- 67.Mackay IM, Arden KE, Nitsche A. Real-time PCR in virology. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelley VA, Caliendo AM. Successful testing protocols in virology. Clin Chem. 2001;47:1559–1562. [PubMed] [Google Scholar]
- 69.Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;323:1573–1580. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen K, Niemark H, Rumore P. Broad range DNA probes for detecting and amplifying eubacterial nucleic acids. FEMS Microbiol Lett. 1989;57:19–24. doi: 10.1016/0378-1097(89)90139-0. [DOI] [PubMed] [Google Scholar]
- 71.Greigen K, Loeffelholz M, Purohit A. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin. Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Evertsson U, Monstein HJ, Johannson AG. Detection and identification of fungi in blood using broad-range 28S rDNA PCR amplification and species-specific hybridisation. APMIS. 2000;108:385–392. doi: 10.1034/j.1600-0463.2000.d01-73.x. [DOI] [PubMed] [Google Scholar]
- 73.Hryniewiecki T, Gzyl A, Augustynowicz E. Development of broad-range polymerase chain reaction (PCR) bacterial identification in diagnosis of infective endocarditis. J Heart Valve Dis. 2002;11:870–874. [PubMed] [Google Scholar]
- 74.Rothman RE, Majmudar MD, Kelen GD. Detection of bacteremia in emergency department patients at risk for infective endocarditis using universal 16S rRNA primers in a decontaminated polymerase chain reaction assay. J Infect Dis. 2002;186:1677–1681. doi: 10.1086/345367. [DOI] [PubMed] [Google Scholar]
- 75.McCabe KM, Khan G, Zhang Y-H. Amplification of bacterial DNA using highly conserved sequences: automated analysis and potential molecular triage of sepsis. Pediatrics. 1995;95:165–169. [PubMed] [Google Scholar]
- 76.Lisby G, Gutschik E, Durack DT. Molecular methods for diagnosis of infective endocarditis. Infect Dis Clin North Am. 2002;16:393–412. doi: 10.1016/s0891-5520(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ley BE. Detection of bacteremia in patients with fever and neutropenia using 16S rRNA gene amplification by PCR. Eur J Clin Microbiol Infect Dis. 1998;17:247–253. doi: 10.1007/BF01699981. [DOI] [PubMed] [Google Scholar]
- 78.Kane TD, Alexander JW, Johannigman A. The detection of microbial DNA in the blood: A sensitive method for the detection of bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1997;227:1–9. doi: 10.1097/00000658-199801000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Relman DA, Loutit JS, Schmidt TM. The agent of bacillary angiomatosis: an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:15731580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 80.Relman DA, Schmidt TM, MacDermott RP. Identification of the uncultured bacillus of Whipple's Disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 81.Leinonen M. Interaction of Chlamydia pneumoniae infection with other risk factors of atherosclerosis. Am Heart J. 1999;135:S504–S506. doi: 10.1016/s0002-8703(99)70286-3. [DOI] [PubMed] [Google Scholar]
- 82.Dorsten C, Gunther S, Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 83.Peiris JSM, Lai ST, Poon LLM. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Poon LLM, Wong OK, Luk W. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS) Clin Chem. 2004;49:953–955. doi: 10.1373/49.6.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.CDC Outbreak of severe acute respiratory syndrome—worldwide. MMWR Morb Mortal Wkly Rep. 2003;52:226–228. [PubMed] [Google Scholar]
- 86.Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 2001;14:836–871. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tenover FC, Jones RN, Swenson JM. Methods for improved detection of oxacillin resistance in coagulase-negative staphylococci: results of a multicenter study. J Clin Microbiol. 1999;37:4051–4058. doi: 10.1128/jcm.37.12.4051-4058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Telenti A, Honoré N, Bernasconi C. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.De Beenhouwer H, Lhiang Z, Jannes G. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tubercle Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 90.Marttila HJ, Soini H, Vyshnevskaya E. Line probe assay in the rapid detection of rifampin-resistant Mycobacterium tuberculosis directly from clinical specimens. Scand J Infect Dis. 1999;31:269–273. doi: 10.1080/00365549950163563. [DOI] [PubMed] [Google Scholar]
- 91.Elnifro EM, Ashshi AM, Cooper RJ. Multiplex PCR: optimization and application in diagnostic virology. Clin Microbiol Reviews. 2000;13:559–570. doi: 10.1128/cmr.13.4.559-570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Petrik J. Microarray technology: the future of blood testing? Vox Sanguinis. 2001;80:1–11. doi: 10.1046/j.1423-0410.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 93.DeGruttola V, Dix L, D'Aquila R. The relation between baseline HIV drug resistance and response to antiretroviral therapy: reanalysis of retrospective and prospective studies using a standardized data analysis plan. Antivir Ther. 2000;5:41–48. doi: 10.1177/135965350000500112. [DOI] [PubMed] [Google Scholar]
- 94.Demeter L, Haubrich R. Phenotypic and genotypic resistance assays: methodology, reliability, and interpretations. J Acquir Immune Defic Syndr. 2001;26(suppl 1):S3–S9. doi: 10.1097/00042560-200103011-00002. [DOI] [PubMed] [Google Scholar]
- 95.Hanna GJ, D'Aquila RT. Clinical use of genotypic and phenotypic drug resistance testing to monitor antiretroviral chemotherapy. Clin Infect Dis. 2001;32:774–782. doi: 10.1086/319231. [DOI] [PubMed] [Google Scholar]
- 96.Pavlin JA, Gilchrist MJR, Osweiler GD. Diagnostic analyses of biological agent-causes syndromes: laboratory and technical assistance. Emerg Med Clin North Am. 2002;20:331–350. doi: 10.1016/s0733-8627(01)00004-9. [DOI] [PubMed] [Google Scholar]
- 97.Bell CA, Uhl JR, Hadfield TL. Detection of Bacillus anthracis DNA by LightCycler PCR. J Clin Microbiol. 2002;40:2897–2902. doi: 10.1128/JCM.40.8.2897-2902.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Espy MJ, Uhl JR, Sloan LM. Detection of vaccinia virus, herpes simplex virus, varicella-zoster virus, and Bacillus anthracis DNA by LightCycler polymerase chain reaction after autoclaving: implications for biosafety of bioterrorism agents. Mayo Clin Proc. 2002;77:624–628. doi: 10.4065/77.7.624. [DOI] [PubMed] [Google Scholar]
- 99.Radnedge L, Gamez-Chin S, McCready PM. Identification of nucleotide sequences for the specific and rapid detection of. Yersinia pestis. Appl Environ Microbiol. 2001;67:3759–3762. doi: 10.1128/AEM.67.8.3759-3762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ropp SL, Jin Q, Knight JC. PCR strategy for identification and differnentiation of small pox and other orthopoxviruses. J Clin Microbiol. 1995;33:2069–2076. doi: 10.1128/jcm.33.8.2069-2076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alibek K. Biohazard. Dell Publishing; New York: 1999. [Google Scholar]
- 102.Louie M, Louie L, Simor AE. The role of DNA amplification technology in the diagnosis of infectious diseases. CMAJ. 2000;163:301–309. doi: 10.1016/s1381-1169(00)00220-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Livengood CH 3rd, Wrenn JW. Evaluation of COBAS AMPLICOR (Roche): accuracy in detection of Chlamydia trachomatis and Neisseria gonorrhoeae by coamplification of endocervical specimens. J Clin Microbiol. 2001;39:2928–2932. doi: 10.1128/JCM.39.8.2928-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haberland CA, Benitz WE, Sanders GD. Perinatal screening for group B streptococci: cost-benefit analysis of rapid polymerase chain reaction. Pediatrics. 2002;110:471–480. doi: 10.1542/peds.110.3.471. [DOI] [PubMed] [Google Scholar]
- 105.Fredricks DN and Relman DA. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin Infect Dis. 2000;29:475–486. doi: 10.1086/598618. [DOI] [PubMed] [Google Scholar]
- 106.Longo MC, Berninger MS, Hartley JL. Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene. 1990;93:125–128. doi: 10.1016/0378-1119(90)90145-h. [DOI] [PubMed] [Google Scholar]
- 107.Carroll NM, Adamson P, Okhravi N. Elimination of bacterial DNA from Taq DNA polymerases by restriction endonuclease digestion. J Clin Microbiol. 1999;37:3402–3404. doi: 10.1128/jcm.37.10.3402-3404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Corless CE, Guiver M, Borrow R. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol. 2000;38:1747–1752. doi: 10.1128/jcm.38.5.1747-1752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang S, Lin S, Kelen GD. Quantitative multiprobe PCR assay for the simultaneous detection and species identification of bacterial pathogens. J Clin Microbiol. 2002;40:3449–3454. doi: 10.1128/JCM.40.9.3449-3454.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Burke DT, Burns MA, Mastrangelo C. Microfabrication technologies for integrated nucleic acid analysis. Genome Res. 1997;7:189–197. doi: 10.1101/gr.7.3.189. [DOI] [PubMed] [Google Scholar]
- 111.Mangiapan G, Vokurka M, Schouls L. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J Clin Microbiol. 1996;34:1209–1215. doi: 10.1128/jcm.34.5.1209-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reiss RA, Rutz B. Quality control PCR: a method for detecting inhibitors of Taq DNA polymerase. Biotechniques. 1999;27:920–926. doi: 10.2144/99275bm09. [DOI] [PubMed] [Google Scholar]
- 113.McAdam AJ. Discrepant analysis: how can we test a test? J Clin Microbiol. 2000;38:2027–2029. doi: 10.1128/jcm.38.6.2027-2029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee HH, Chernesky MA, Schachter J. Diagnosis of Chlamydia trachomatis genitourinary infection in women by ligase chain reaction assay of urine. Lancet. 1995;345:213–216. doi: 10.1016/s0140-6736(95)90221-x. [DOI] [PubMed] [Google Scholar]
- 115.Jaschek G, Gaydos CA, Welsh LE. Direct detection of Chlamydia trachomatis in urine specimens from symptomatic and asymptomatic men by using a rapid polymerase chain reaction assay. J Clin Microbiol. 1993;31:1209–1212. doi: 10.1128/jcm.31.5.1209-1212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Green TA, Black CM, Johnson RE. Evaluation of bias in diagnositic-test sensitivity and spceificity estimates computed by discrepant analysis. J Clin Microbiol. 1998;36:375–381. doi: 10.1128/jcm.36.2.375-381.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hadgu A. Discrepant analysis: a biased and an unscientific method for estimating test sensitivity and specificity. J Clin Epidemiol. 1999;52:1231–1237. doi: 10.1016/s0895-4356(99)00101-8. [DOI] [PubMed] [Google Scholar]
- 118.Nikkari S, McLaughlin IJ, Wanli B. Does blood of healthy subjects contain bacterial ribosomal DNA? J Clin Microbiol. 2001;39:1956–1959. doi: 10.1128/JCM.39.5.1956-1959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kennedy N, Gillespie SH, Saruni AO. Polymerase chain reaction for assessing treatment response in patients with pulmonary tuberculosis. J Infect Dis. 1994;170:713–716. doi: 10.1093/infdis/170.3.713. [DOI] [PubMed] [Google Scholar]
- 120.Noordhoek GT, Wolters EC, de Jonge ME. Detection by polymerase chain reaction of Treponema pallidum DNA in cerebrospinal fluid from neurosyphilis patients before and after antibiotic treatment. J Clin Microbiol. 1991;29:1976–1984. doi: 10.1128/jcm.29.9.1976-1984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mellors JW, Rinaldo CR, Gupta P. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 122.Ho DD, Tarsem M, Masud A. Quantification of human immunodeficiency virus type 1 in the blood of infected persons. N Engl J Med. 1989;321:1621–1625. doi: 10.1056/NEJM198912143212401. [DOI] [PubMed] [Google Scholar]
- 123.Baker BL, Di-Bisceglie AM, Kaneko S. Determination of hepatitis B virus DNA in serum using the polymerase chain reaction: clinical significance and correlation with clinical and biochemical markers. Hepatology. 1991;13:632–636. [PubMed] [Google Scholar]
- 124.Berger A, Braner J, Doerr HW. Quantification of viral load: clinical relevance for human immunodeficiency virus, hepatitis B virus, hepatitis C virus infection. Intervirology. 1997;41:24–34. doi: 10.1159/000024912. [DOI] [PubMed] [Google Scholar]
- 125.Yamamoto M, Kimura H, Hironaka T. Detection and quantitation of virus DNA in plasma of patients with Epstein-Barr virus-associated diseases. J Clin Microbiol. 1995;33:1765–1768. doi: 10.1128/jcm.33.7.1765-1768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Walsh KM, Good T, Cameron S. Viral kinetics can predict early response to alpha-interferon in chronic hepatitis C. Liver. 1998;18:191–195. doi: 10.1111/j.1600-0676.1998.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 127.Hackett SJ, Guiver M, Marsh J. Meningococcal bacterial DNA load at presentation correlates with disease severity. Arch Dis Child. 2002;8:44–46. doi: 10.1136/adc.86.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Alifano P, Bruni CB, Carlomagno MS. Control of mRNA processing and decay in prokaryotes. Genetica. 1994;94:157–172. doi: 10.1007/BF01443430. [DOI] [PubMed] [Google Scholar]
- 129.Birch L, Dawson CE, Cornett JH. A comparison of nucleic acid amplification techniques for the assessment of bacterial viability. Lett Appl Microbiol. 2001;33:296–301. doi: 10.1046/j.1472-765x.2001.00999.x. [DOI] [PubMed] [Google Scholar]
- 130.Bej AK, Ng WY, Morgan S. Detection of viable Vibrio cholerae by reverse-transcriptase polymerase chain reaction (RT-PCR) Mol Biotechnol. 1996;5:1–10. doi: 10.1007/BF02762407. [DOI] [PubMed] [Google Scholar]
- 131.Sheridan GEC, Masters CI, Shallcross JA. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313–1318. doi: 10.1128/aem.64.4.1313-1318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jou NT, Yoshimori RB, Mason GR. Single-tube, nested, reverse transcriptase PCR for detection of viable. Mycobacterium tuberculosis. J Clin Microbiol. 1997;25:1161–1165. doi: 10.1128/jcm.35.5.1161-1165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hellyer TJ, DesJardin LE, Hehman GL. Quantitative analysis of mRNA as a marker for viability of. Mycobacterium tuberculosis. J Clin Microbiol. 1999;35:1161–1165. doi: 10.1128/jcm.37.2.290-295.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Patel BKR, Banerjee DK, Butcher PD. Determination of Mycobacterium leprae viability by polymerase chain reaction amplification of 71-kDa heat-shock protein mRNA. J Inf Dis. 1993;168:779–800. doi: 10.1093/infdis/168.3.799. [DOI] [PubMed] [Google Scholar]
- 135.Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sharples GJ, Lloyd RG. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18:6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schouten JP, McElgunn CJ, Waaijer R. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Baner J, Nilsson M, Isaksson A. More keys to padlock probes: mechanisms for high-throughput nucleic acid analysis. Curr Opin Biotechnol. 2001;12:11–15. doi: 10.1016/s0958-1669(00)00174-9. [DOI] [PubMed] [Google Scholar]
- 139.Goldenberger D, Kunzli A, Vogt P. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol. 1997;35:2733–2739. doi: 10.1128/jcm.35.11.2733-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Corless CE, Guiver M, Borrow R. Simultaneous detection of Neisseria meningitides, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J Clin Microbiol. 2001;39:1553–1558. doi: 10.1128/JCM.39.4.1553-1558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramsay G. DNA chips: state-of-the-art. Nat Biotechnol. 1998;16:40–44. doi: 10.1038/nbt0198-40. [DOI] [PubMed] [Google Scholar]
- 142.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 143.Westin L, Miller C, Vollmer D. Antimicrobial resistance and bacterial identification utilizing a microelectronic chip array. J Clin Microbiol. 2001;39:1097–1104. doi: 10.1128/JCM.39.3.1097-1104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Stults JT. Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) Curr Opin Struct Biol. 1995;5:691–698. doi: 10.1016/0959-440x(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 145.Cotter RJ. Time-of-flight mass spectrometry for the structural analysis of biological molecules. Anal Chem. 1992;64:1027A–1039A. doi: 10.1021/ac00045a002. [DOI] [PubMed] [Google Scholar]
- 146.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10 000 daltons. Anal Chem. 1998;60:2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 147.Hurst GB, Weaver K, Doktycz MJ. MALDI-TOF anaylsis of polymerase chain reaction products from methanotrophic bacteria. Anal Chem. 1998;70:2693–2698. doi: 10.1021/ac980044e. [DOI] [PubMed] [Google Scholar]
- 148.Von Wintzingerode F, Bocker S, Schlotelburg C. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. PNAS. 2002;99:7039–7044. doi: 10.1073/pnas.102165899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Koomen JM, Russell WK, Tichy SE. Accurate mass measurement of DNA oligonucleotide ions using high-resolution time-of-flight mass spectrometry. J Mass Spect. 2002;37:357–371. doi: 10.1002/jms.312. [DOI] [PubMed] [Google Scholar]
- 150.Emanuel PA, Bell R, Dang JL. Detection of Francisella tularensis within infected mouse tissues by using a hand-held PCR thermocycler. J Clin Microbiol. 2003;41:689–693. doi: 10.1128/JCM.41.2.689-693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gutierres SL, Welty TE. Point-of-care testing: an introduction. Ann Pharmacother. 2004;38:119–125. doi: 10.1345/aph.1D212. [DOI] [PubMed] [Google Scholar]