Highlights
-
•
An intermediate amount of T cell stimulation induces Foxp3 transcription.
-
•
Treg cell lineage factor Foxp3 cooperates with its partners to promote Treg cell function.
-
•
Cell signaling-regulated Foxo1 is indispensable for Treg cell function.
Keywords: cell signaling, transcription factor, Foxp3, Foxo1
Abstract
Regulatory T (Treg) cells differentiate from thymocytes or peripheral T cells in response to host and environmental cues, culminating in induction of the transcription factor forkhead box P3 (Foxp3) and the Treg cell-specific epigenome. An intermediate amount of antigen stimulation is required to induce Foxp3 expression by engaging T cell receptor (TCR)-activated [e.g., nuclear factor (NF)-κB] and TCR-inhibited (e.g., Foxo) transcription factors. Furthermore, Treg cell differentiation is associated with attenuated Akt signaling, resulting in enhanced nuclear retention of Foxo1, which is indispensable for Treg cell function. These findings reveal that Treg cell lineage commitment is not only controlled by genetic and epigenetic imprinting, but also modulated by transcriptional programs responding to extracellular signals.
Introduction
Capable of mounting potent antigen-specific defense responses against invading pathogens, the adaptive immune system has evolved exquisitely balanced mechanisms to avoid attacking healthy self-tissues. Despite extensive pruning of self-reactive cells through clonal deletion in the primary lymphoid organs, potentially self-destructive lymphocytes escape into the periphery, necessitating additional tolerance mechanisms. Indeed, dominant suppression, by which a specialized suppressor T cell population, referred to as Treg cells, acting in trans to restrain pathogenic immune responses, has emerged as a pivotal mechanism of immune tolerance 1, 2.
The concept of T cell suppression was initially implicated by the finding that neonatal thymectomy leads to the loss of self-tolerance in mice 3, 4. Subsequent studies identified thymus-derived Treg (tTreg) cells as the major Treg population, which appear sufficient for the control of systemic and tissue-specific autoimmunity 1, 2. Furthermore, peripherally generated Treg (pTreg) cells that develop from mature CD4+ T cells may broaden the antigen specificity of Treg cells and promote immune tolerance to environmental antigens 5, 6. During the past decade, much of the Treg cell research explored the genetic and epigenetic programs that promote Treg cell lineage commitment. In this article, we discuss an emerging theme of how signaling pathways integrate host and environmental inputs to the transcriptional control of Treg cell differentiation and function.
tTreg cell differentiation
In the thymus, Foxp3+ Treg cells are generated roughly in sync with or shortly after the positive selection of CD4+ T cells. Extensive studies in the past decade have focused on the molecular events that converge on Foxp3 induction. The expression of Foxp3 gene is controlled by a proximal promoter and the intronic regulatory elements, designated as conserved noncoding sequences (CNS1–3). Experiments using genetically engineered mouse models demonstrate differential roles of the three enhancers: CNS1 is essential for pTreg, but not tTreg cell development (see below); CNS2 regulates the heritable maintenance of Foxp3 expression; and CNS3 acts as a pioneer element to control de novo Foxp3 induction [7]. In addition, recent studies have shown that tTreg cell-specific CpG hypomethylation (tTreg-Me) is induced during tTreg cell specification independent of Foxp3 expression [8]. Although TCR engagement with self-peptide major histocompatibility complex (pMHC) ligands with proper duration appears to elicit tTreg-specific epigenome, the exact mechanism remains to be determined.
TCR and co-stimulatory signals instruct tTreg cell development
Variations in TCR signaling strength and duration have been proposed as key determinants of T cell lineage commitment during thymic differentiation. Studies using transgenic TCRs provided the first direct evidence that TCR signaling directs tTreg cell development 9, 10. Introducing a cognate ligand for the transgenic receptor leads to differentiation of Treg cells bearing the transgenic TCR, whereas expression of a TCR with an intrinsically lower affinity for the same self-peptide fails to select the Treg cell subset [11]. TCR usage analyses revealed that the repertoires of Treg cells and CD4+ conventional T (Tconv) cells are similarly diverse, but only partially overlapped [12]. T cells transduced with Treg cell TCRs undergo homeostatic expansion more rapidly in lymphopenic recipients than cells engineered with receptors cloned from Tconv cells, supporting the hypothesis that Treg cells recognize the self-ligands with higher affinity. Nevertheless, TCR/self-peptide interactions that trigger T cell negative selection likely impose an upper limit on tTreg cell development. Indeed, attenuation of MHC class II expression on medullary thymic epithelial cells (mTECs) results in a shift from T cell clonal deletion to tTreg cell differentiation [13], whereas ablation of the transforming growth factor (TGF)-β cytokine signaling leads to the augmented T cell negative selection and tTreg cell paucity in neonatal mice [14]. Taken together, these observations suggest that tTreg cell selection is instructed by TCRs in an affinity and duration window for self-pMHC ligands between positive selection and negative selection (Figure 1A).
Figure 1.
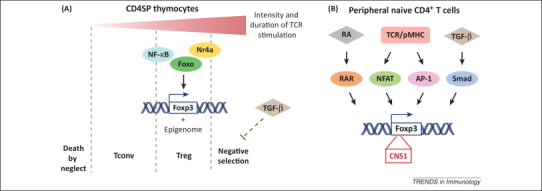
Transcriptional regulation of regulatory T (Treg) cell development. (A) Thymus-derived Treg (tTreg) cell differentiation. In developing thymocytes, T cell receptor (TCR) gene rearrangement generates diverse TCRs that recognize self-peptide major histocompatibility complex (pMHC) ligands at various intensities and durations. Thymocytes bearing TCRs that fail to productively interact with pMHC die by neglect, whereas those with extremely high affinity are eliminated by negative selection. TCR stimulation with relatively high intensity induces forkhead box P3 (Foxp3) expression, which is mediated in part by transcription factors including nuclear factor (NF)-κB, Foxo and nuclear receptor Nr4a. TCR/self-ligand interaction with proper duration induces Treg cell-specific epigenome. In the thymus, transforming growth factor (TGF)-β signaling promotes tTreg cell generation by restraining negative selection rather than direct transcriptional control of Foxp3. (B) Peripherally generated Treg (pTreg) and in vitro differentiated Treg (iTreg) development. Signaling cascades downstream of antigen stimulation, TGF-β, and retinoic acid (RA) signaling regulate Foxp3 gene expression from naive CD4+ T cells in the periphery. Transcription factors binding to the conserved noncoding sequence (CNS)1 element of the Foxp3 locus, nuclear factor of activated T cells Smad (NFAT), activator protein (AP)-1, Smad and, RA receptor (RAR), are shown. Abbreviation: Tconv, conventional T cell.
In addition to TCRs, the co-stimulatory receptor CD28 plays an important role in promoting tTreg cell differentiation. Mice deficient in CD28 or its ligands CD80 and CD86 have significantly impaired Treg cell populations 15, 16, whereas ablation of the co-inhibitory receptor cytotoxic T lymphocyte antigen (CTLA)-4 results in a higher frequency of tTreg cells [17]. TCR and CD28 induce a multitude of intracellular signaling events that culminate in the activation of transcription factors including NF-κB. Genetic perturbation of genes involved in NF-κB activation such as protein kinase (PK)C- θ, Bcl10, CARD-containing MAGUK protein 1 (CARMA1), TAK1, IkappaB kinase (IKKβ) cause defective tTreg cell generation 18, 19, 20, 21, 22, whereas the expression of a constitutively active form of IKKβ triggers ectopic tTreg cell differentiation in transgenic mice bearing conventional TCRs [23], bypassing the requirement of high affinity TCR/self-peptide interaction. These findings revealed a crucial role for NF-κB signaling in tTreg cell lineage commitment, and further prompted studies to determine the function of individual members of the NF-κB family in this process. Indeed, the c-Rel subunit of NF-κB has been reported to bind to the promoter, CNS2 and CNS3 elements of the Foxp3 gene 7, 23, 24, 25. CNS3 is marked by a permissive histone modification (H3K4me1), and c-Rel binding to CNS3 potentially initiates chromatin remodeling in the Foxp3 locus [7]; whereas its recruitment to the Foxp3 promoter may facilitate the formation of a c-Rel enhanceosome to control transcriptional initiation [24].
Aside from the NF-κB pathway, accumulating evidence has pointed to a role of Ca2+ signaling in tTreg cell development 26, 27, yet the function of Ca2+-activated transcription factor nuclear factor of activated T cells (NFAT) is incompletely understood. NFAT is recruited to the Foxp3 promoter in human effector T cells [28], but this binding is barely detectable in mice 29, 30. Using mouse models with constitutive knockout of NFAT1 and NFAT4, as well as CD4 promoter-directed conditional ablation of NFAT2, several studies have revealed that mice lacking one or two of the three family members (NFAT1, NFAT2, and NFAT4) have normal frequencies of tTreg cells 30, 31. However, a recent report using Nfat1 −/− Nfat2 fllfl Lck-Cre mice, in which NFAT2 is deleted at the early stage of T cell differentiation, revealed a defect of tTreg cell differentiation [27]. As NFAT2 has been suggested to play a role in early thymocyte development [32], it remains to be determined whether NFAT proteins directly participate in Foxp3 induction and the potential functional redundancy among the family members. The activator protein (AP)-1 and cAMP response element-binding protein (CREB) transcription factors downstream of the TCR−CD28 signaling pathway have also been implicated in the control of Foxp3 gene expression. AP-1 is recruited to the Foxp3 promoter, and controls the promoter activity in a luciferase reporter assay [28]. CREB binds to the CNS2 element, which inversely correlates with the methylation status of the CpG island [33]. Indeed, CNS2 demethylation is a major component of tTreg-Me with important functions in stabilizing Foxp3 expression 8, 34. Nevertheless, the precise roles of AP-1 and CREB in the control of tTreg cell differentiation are open for future investigation.
In addition to the transcription factors directly activated by TCR–CD28 stimulation, a set of transcription factors is induced by the signal and functions as secondary modulators of Foxp3 expression, which includes the nuclear receptor 4a (Nr4a) family of orphan nuclear receptors (Nr4a1, Nr4a2, and Nr4a3). In mice with transgenic expression of GFP driven by the promoter of Nur77 (encoding Nr4a1), tTreg cells express substantially higher levels of GFP than Tconv cells [35], providing another line of evidence that tTreg cells are exposed to strong TCR signals. Deficiencies of the individual Nr4a factors do not significantly affect tTreg cell differentiation. However, deletion of all three Nr4a members in mice results in a marked loss of tTreg cells and lethal autoimmunity [36]. Nr4a binds to the Foxp3 promoter and induces Foxp3 expression [37]. Forced expression of Nr4a compensates for the suboptimal TCR signaling in the control of tTreg cell differentiation, implying a role for Nr4a in translating the strength of TCR signaling into transcriptional control of Foxp3 [36]. It is important to note that strong TCR stimulation induces high expression of Nr4a that promotes T cell negative selection [38]. Thus, it remains to be determined how Nr4a expression is regulated by TCR signaling, and how differential Nr4a expression distinguishes T cell choices of Foxp3 induction versus clonal deletion.
Common γ-chain cytokines promote tTreg cell differentiation
The fact that TCR repertories from Treg and Tconv cells partially overlap raises the question of what are the additional signals required for tTreg cell development. The findings that the common γ-chain (γc) cytokines play an important role in the control of tTreg cell generation have led to the proposal of a two-step model of tTreg cell differentiation 39, 40. In this model, a high avidity TCR signal instructs the development of Foxp3–CD25+ Treg cell precursors (Step 1). Interleukin (IL)-2, and to a lesser extent IL-7 or IL-15, subsequently induces Foxp3 expression in a TCR-independent manner (Step 2) [40]. Signal transducer and activator of transcription (Stat)5, a key transcription factor activated by γc cytokine receptors, is essential for tTreg cell differentiation. Mice with ablation of Stat5 had a dramatic reduction in tTreg cells, similar to IL-2 IL-7 IL-15 triple deficient mice or mice devoid of the γc receptor 41, 42, 43. Expression of a hyperactive form of Stat5 expands tTreg cells, and broadens the Treg cell TCR repertoire to include TCRs that are normally found in Tconv cells [39]. Mechanistically, Stat5 binds to the Foxp3 promoter and CNS2 element 43, 44, and may directly control Foxp3 transcriptional initiation and the heritable Foxp3 expression in Treg cells.
Phosphoinositide 3-kinase (PI3K)–Akt signaling negatively regulates tTreg cell differentiation
In contrast to the signaling pathways discussed above, activation of the PI3K–Akt pathway downstream of TCR, CD28, γc cytokine, and other cell surface receptors is inhibitory to tTreg cell differentiation. Reduced Akt activation caused by a kinase-inactive version of PI3K p110δ or deficiency in sphingosine 1-phosphate receptor (S1P1) favors tTreg cell development 45, 46, whereas overexpression of a constitutively active form of Akt or S1P1 diminishes tTreg cell frequencies 46, 47. Recent studies have connected these findings with the transcriptional regulation of Foxp3 by aligning Foxo transcription factors in this pathway 48, 49. Foxo1 and Foxo3a translocate to the nucleus under the condition of Akt inhibition, bind to the promoter and CNS2 of the Foxp3 locus, and promote tTreg cell differentiation 48, 50. It appears paradoxical that different signaling pathways downstream of TCR, CD28, and γc cytokine receptors have opposing roles in the control of Foxp3 expression. We have proposed a ‘hit and run’ model that strong antigen stimulation activates NF-κB and inactivates Foxo, whereas cessation of antigen interaction allows Foxo proteins to translocate back to the nucleus and collaborate with NF-κB and other factors to induce Foxp3 transcription. Thereby, only thymocytes with infrequent encounters with high-affinity antigens are selected to differentiate to tTreg cells [51]. Such a kinetic model of tTreg cell differentiation is supported by the finding that transient activation of thymic Tconv cells followed by a resting period induces robust Foxp3 expression [48]. This dynamic model of tTreg cell differentiation is reminiscent of CD4/CD8 lineage choice in that the decision is determined by a plethora of factors, including TCR signaling strength, TCR signaling duration, as well as cytokine signals [52].
pTreg cell differentiation
The requirements for the lineage commitment of pTreg cells differ substantially from those of tTreg cells. Most importantly, CNS1 is critical for Foxp3 induction in peripheral CD4+ T cells but not in thymocytes [7]. With a Foxp3–CNS1-deficient mouse model, recent studies have revealed that pTreg cells play a nonredundant role in the control of inflammation at mucosal interfaces and maternal–fetal conflict, which coincides with the acquisition of CNS1 in placental mammals 53, 54. In vitro differentiated Treg (iTreg) cells appear to lack tTreg-Me, whereas in vivo pTreg cells gradually obtain tTreg-Me [8], which probably contributes to their relative instability compared to tTreg cells.
TCR and co-inhibitory signals promote pTreg cell development
Although tTreg cells develop in a tightly controlled thymic microenvironment, differentiation of extrathymic Treg cells occurs under more diverse conditions, preferentially at environmental interfaces such as in the gut tissue [6]. pTreg cells are induced by high-affinity non-self antigens, likely in adaptation to the environmental niche 55, 56. Indeed, the Treg cell TCR repertoire varies by the anatomical location, and a subset of colonic TCRs recognize antigens derived from the commensal microbiota [57], although a recent study suggests that colonic Treg cells may be predominantly thymically derived [58]. Chronic administration of low-dose agonistic antigens induces pTreg cells that mitigate autoimmune or allergic responses 59, 60, 61, 62. Similar to tTreg cells, TCR-activated or -induced transcription factors NF-κB and Nr4a are required for iTreg and possibly pTreg cell differentiation 23, 24, 36. NFAT, which appears superfluous for tTreg development, is recruited to CNS1, and is also vital for the induction of Foxp3 expression from mature T cells [30]. Binding sites for the TCR-activated transcription factor AP-1 are present in CNS1 [63], but its definitive function in the control of pTreg cell differentiation remains to be determined.
In contrast to tTreg cell development, suboptimal or no co-stimulation favors pTreg and iTreg cell generation. Indeed, CD28 activation suppresses Treg cell differentiation from Tconv cells [64], whereas CTLA-4 is required for the induction of Foxp3 expression [65]. Such a negative effect of co-stimulation could be due to the inhibition of Foxp3 expression by PI3K–Akt signaling that is potently induced by CD28 in mature T cells. In parallel with tTreg cell differentiation, sustained Akt activation, for example, in Cbl-b or phosphatase and tension homolog (PTEN)-deficient T cells, results in profound defects in Foxp3 induction 50, 66, whereas blockade of PI3K–Akt signaling augments iTreg cell differentiation 66, 67. These observations could again be explained by the negative modulation of Akt signaling on Foxo transcription factors, which play a critical role in the control of iTreg and possibly pTreg cell differentiation 48, 49, 50.
Cytokines and environmental cues control pTreg cell differentiation
Additional insights into the control of pTreg cell differentiation have come from studies of cytokine requirements, most notably, TGF-β and IL-2. TGF-β promotes Foxp3 transcription in peripheral CD4+ T cells through binding of Smad3 at CNS1 [29]. The observations that CNS1 deficiency or specific ablation of Smad-binding sites in CNS1 does not impair tTreg cell development argue against direct transcriptional regulation of Foxp3 by TGF-β in tTreg cells 7, 68. Indeed, the tTreg cell differentiation defects in TGF-β-receptor-deficient mice have been attributed to the enhanced negative selection of T cells [14]. IL-2 signaling facilitates TGF-β-mediated induction of iTreg cells [69], probably through Stat5-dependent transactivation of Foxp3 locus. Furthermore, IL-2 may promote iTreg and/or pTreg cell development indirectly by curtailing Th17 cell differentiation and confer a survival advantage to Treg cells [70]. In addition to IL-2, retinoic acid (RA), a vitamin A metabolite, augments TGF-β-induced Treg cell development 64, 71, 72. RA may directly induce Foxp3 transcription via the activation of RA receptor (RAR) followed by its recruitment to CNS1 [63], or indirectly through constraining inflammatory cytokine production by effector cells [73].
Transcriptional control of Treg cell function
The pivotal function of Treg cells in the control of immune tolerance has been best demonstrated by constitutive or inducible depletion of Foxp3+ T cells. These studies revealed that Treg cells are indispensable for restraining lethal inflammatory responses in neonates as well as in adult animals 74, 75. Elucidating the precise mechanisms by which Treg cells function is an area of active research [76]. Recent studies have started to unravel the molecular pathways and the transcriptional circuits that enable Treg cells to suppress pathological immune responses.
Foxp3-dependent functional program
An enormous body of literature has demonstrated that Foxp3, the key lineage-specifying factor of Treg cells, has a crucial role in the control of Treg cell function (Figure 2 ). Foxp3 deficiency results in a lethal lymphoproliferative disorder in scurfy mice, and is associated with immunedysregulation, polyendocrinopathy, enteropathy, and X-linked syndrome (IPEX) in human patients 77, 78, 79, 80. Continuous Foxp3 expression in mature Treg cells is required to sustain their suppressive function [81]. A subset of Treg ‘wannabe’ cells that develop in mice bearing a dysfunctional Foxp3 reporter allele is completely devoid of suppressor activity 82, 83, which is consistent with the idea that Foxp3 is absolutely required for Treg cell function. However, these Foxp3-deficient cells exhibit some phenotypic and molecular characteristics of Foxp3+ Treg cells 82, 84, implying the presence of Foxp3-independent programs such as the Treg cell-specific epigenome that are induced independent of Foxp3 induction during Treg cell differentiation.
Figure 2.
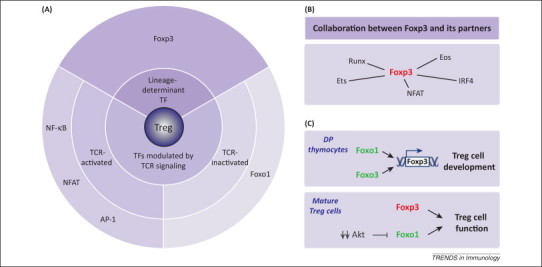
Transcriptional control of regulatory T (Treg) cell function. (A) Forkhead box P3 (Foxp3), the Treg lineage-specifying factor, is essential for the maintenance of Treg cell suppressive function. In addition, transcriptional factors (TFs) modulated by T cell receptor (TCR) signaling, including nuclear factor (NF)-κB, NFAT, activator protein (AP)-1 and Foxo1, may regulate Treg cell function. Foxo1 is unique among these TFs in that TCR engagement negatively regulates its activity. (B) Foxp3 does not function alone. Instead, Foxp3 cooperates with other nuclear factors, such as Runx, Ets, NFAT, interferon regulatory factor (IRF)4 and Eos, to establish the molecular signature and function of Treg cells. (C) In immature thymocytes, the Foxo family proteins Foxo1 and Foxo3 are both highly expressed. They bind to the Foxp3 locus and cooperatively induce Foxp3 gene transcription. The transcript of Foxo1 is upregulated during T cell maturation, whereas that of Foxo3 is downregulated. The nonredundant role of Foxo1 in mature Treg cells has been studied. Foxp3 and Foxo1 regulate largely independent transcription programs; both indispensable for Treg cell function. Notably, Foxo1 activity is subject to modulation of Akt kinase signaling, and Treg cells have dampened Akt signaling in response to TCR stimulation compared with conventional T (Tconv) cells.
Indeed, Foxp3 expression alone might not be sufficient to define the transcriptional landscape of Treg cells. A recent study showed that a set of transcription factors including Eos, interferon (IFN) regulatory factor (IRF)4, special AT-rich sequence-binding protein-1 (Satb1), lymphoid enhancer-binding factor-1 (Lef1), and GATA-1 can act in synergy with Foxp3 to establish the Treg cell-associated transcriptional program [85]. For at least one such factor, Eos, whose expression is enriched in Treg cells, its genomic loci is imprinted with lower CpG methylation in Treg cells than in Tconv cells [8]. Intriguingly, these Foxp3 cofactors physically interact with Foxp3 85, 86, 87, and may stabilize Foxp3 DNA binding [85]. The concept of functional cooperation between Foxp3 and its binding partners was further corroborated by the study of Foxp3 interactome. Foxp3 forms multiprotein complexes with several hundred proteins, a large proportion of which are in fact transcriptional targets of Foxp3 [88]. In addition, mapping of the genome-wide Foxp3-binding sites and Treg cell enhancer elements revealed that Foxp3 exploits a pre-existing enhancer landscape in T cells, and is recruited to the sites enriched for Foxp3 cofactors including the runt-related transcription factor (Runx) and E-twenty six (Ets) family of transcription factors [89]. The Foxp3–Runx–core-binding factor (CBFβ) complex appears to be crucial for recruiting Foxp3 to the CNS2 element in the Foxp3 locus, and maintaining the stable expression of Foxp3 in Treg cells 90, 91. Nevertheless, the precise activity and mechanism by which the elaborate network of transcription factors modulate Foxp3-dependent functional program in Treg cells remain to be fully elucidated.
Among the Foxp3-binding sites are enhancers that are induced upon TCR activation. These findings are in line with the observation that Treg cells recognize self-antigens with high affinity and appear to be constantly stimulated in vivo. It is thus conceivable that transcription factors activated upon TCR stimulation might cooperate with Foxp3 to control Treg cell function (Figure 2A,B). TCR engagement activates downstream transcription factors, including NFAT, NF-κB and AP-1. Indeed, NFAT forms a complex with Foxp3, which has been postulated to recruit Foxp3 to the promoter region of Il2 gene and suppress its expression [92]. Recent studies have also suggested a role for NF-κB in the control of Treg cell function. Treg cell-specific knockout of Ubc13, a Lys63-specific ubiquitin-conjugating enzyme that activates IKK, results in abnormal T cell activation and multiorgan inflammation in mice [93]. Loss of Ubc13 does not affect Foxp3 expression or Treg cell homeostasis, but impairs the in vivo suppressive function of Treg cells. Transgenic expression of a hyperactive form of IKKβ largely corrects the inflammatory disease, supporting a role for NF-κB in the control of Treg cell function [93]. Interestingly, both Rel-A and c-Rel subunits of NF-κB have been shown to physically interact with Foxp3 94, 95. Nevertheless, it remains to be determined whether NF-κB directly collaborates with Foxp3 to control Treg cell function.
Foxo1-dependent functional program
In contrast to the NFAT or NF-κB family of transcription factors that translocate from the cytosol to the nucleus upon TCR stimulation, Foxo proteins are excluded from the nucleus in response to TCR-induced Akt activation (Figures 2A and 3 ). In addition to the well-established function of Foxo1 and Foxo3 in Treg cell differentiation, a recent study has revealed a crucial role for Foxo1 in the control of Treg cell function (Figure 2C) [96]. Among the three Foxo family proteins expressed in T cells, Foxo1 is specifically upregulated during T cell maturation in Tconv cells as well as in tTreg cells [48]. However, compared to Tconv cells, Treg cells maintain high amounts of nuclear Foxo1 in response to TCR stimulation, which is associated with substantially reduced Akt activation [96]. Treg cell-specific ablation of Foxo1 results in a fatal inflammatory disease that is comparable in severity to Foxp3-deficient mice, but without the loss of Treg cells [96]. The lethal inflammatory phenotype can be rescued by Treg cell expression of an Akt-insensitive Foxo1 mutant that is constitutively localized in the nucleus, supporting a critical role for Foxo1-dependent transcription in the control of Treg cell function.
Figure 3.
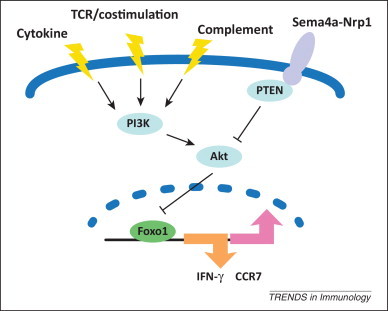
Forkhead box (Fox)o-dependent program in T regulatory (Treg) cells. Akt kinase residues in a major node of T cell signaling that senses diverse extracellular inputs, such as antigen, co-stimulation, cytokine, and complement. These stimuli signal through phosphoinositide 3-kinase (PI3K) and activate Akt, which is counteracted by PTEN (phosphatase and tension homolog). Ligation of Treg cell-expressed receptor neuropilin-1 (Nrp1) with semaphorin-4a (Sema4a) restrains Akt activation via recruitment of PTEN. Activated Akt can inhibit Foxo transcriptional activity through phosphorylation at three conserved residues, resulting in cytoplasmic retention of Foxo. In the context of Treg cells, nuclear Foxo1 represses transcription of the proinflammatory cytokine interferon (IFN)-γ and promotes expression of chemokine CC receptor (CCR)7, which may collectively promote Treg cell function in vivo.
The precise Foxo1-dependent transcriptional program in Treg cells has started to be unraveled. Genome-wide Foxo1 binding coupled with transcriptome analysis of Foxo1-deficient Treg cells have revealed >300 putative Foxo1 direct target genes that appear mostly distinct from the putative direct target genes of Foxp3, suggesting that Foxo1 and Foxp3 control largely nonoverlapping genetic programs [96]. Indeed, although Foxp3 is essential for Treg cell suppression of Tconv cell proliferation in vitro, possibly via its induction of Cd25 and Ctla4 expression, Foxo1-deficient Treg cells have no defects in such assays [96]. In the absence of Foxo1, Treg cells abnormally produce high amounts of the proinflammatory cytokine IFN-γ that contributes to their loss of suppressor function in vivo [96]. Although Foxo1-deficient Tconv cells express comparable amounts of IFN-γ to wild type Tconv cells under the type I polarization conditions [96], ectopic activation of an Akt-insensitive Foxo mutant inhibits IFN-γ expression in these cells [97]. These observations, together with the finding that Foxo1 directly binds to the Ifng locus in Treg cells, support the conclusion that high amounts of nuclear Foxo1 are required to suppress Ifng transcription in T cells. Nevertheless, even under the optimal type I polarization conditions, Foxo1-deficient Treg cells produce lower amounts of IFN-γ than Tconv cells [96], implying additional Foxo1-independent mechanisms of IFN-γ suppression in Treg cells. Compared to Tconv cells, Treg cells are refractory to STAT1-induced IL-12Rβ2 expression, and are less sensitive to IL-12-triggered IFN-γ expression [98]. In Treg cells, the Il12rb2 locus is marked preferentially with the repressive histone marker H3K27me3 [98], but the exact mechanisms of such epigenetic imprinting remain to be determined.
Foxo1 has a well-established function in the control of naïve Tconv cell homeostasis and migration in part via its regulation of chemokine CC receptor (CCR)7 expression 99, 100. Interestingly, Ccr7 has been identified as a Foxo1-bound target gene in Treg cells as well [96]. Although CCR7 was initially proposed to orchestrate naïve T cell priming by mature dendritic cells (DCs) and the initiation of adaptive immune responses [101], recent studies have shown that CCR7 plays a more critical role in Treg cell-mediated immune suppression through its control of Treg cell homing to yet-to-be-defined functional niches 102, 103. In agreement with these observations, CCR7-deficient mice spontaneously develop a multiorgan autoimmune disease 104, 105. These findings suggest that in addition to suppress Treg cell acquisition of effector T cell characteristics, Foxo1 might promote Treg cell function through its regulation of CCR7 expression and Treg cell trafficking (Figure 3).
The pivotal role of Foxo1 in promoting Treg cell function is supported by reduced Akt activation, and the consequent enhanced nuclear Foxo1 activity in Treg cells. How the attenuated Akt signaling in Treg cells is established during Treg cell lineage commitment remains to be fully elucidated. A recent study showed that activation of the Treg cell-enriched receptor neuropilin-1 by semaphorin-4a potentiates Treg cell function by recruiting PTEN at the immunological synapse, which limits Akt phosphorylation and retains nuclear localization of Foxo proteins (Figure 3) [106]. Intriguingly, using a Treg cell-specific neutropilin-1-deficient mouse strain, neuropilin-1 has been revealed as essential to potentiate Treg cell-dependent suppression in cancer and colitis models [106]. These findings imply an exciting possibility that the Foxo-dependent program could be targeted for the treatment of inflammatory diseases.
Importantly, the PI3K–Akt pathway in T cells integrates a plethora of environmental inputs including TCR, co-stimulatory receptors, proinflammatory cytokines, as well as complement molecules (Figure 3). Recent studies have revealed that signaling through complement receptors represses Treg differentiation as well as Treg cell function 107, 108, which is associated with the enhanced Akt activation and Foxo1 phosphorylation [108]. Such dynamic regulation of Akt/Foxo signaling raises the possibility that excessive Akt stimulation, for instance, under extreme inflammatory conditions, might impair the Foxo1-dependent transcriptional program in Treg cells. Intriguingly, Treg cells with anomalous expression of IFN-γ, concomitant with the loss of suppressive activity, have been observed in mice infected with Toxoplasma gondii or coronavirus, as well as in multiple sclerosis and diabetes patients 109, 110, 111, 112. Future studies will unravel whether the loss of Foxo1-dependent Treg cell suppression accounts for Treg cell plasticity under these pathological conditions. Crosstalk between Akt/Foxo and the mechanistic target of rapamycin (mTOR) signaling pathways has been well documented. mTORC2 contributes to Akt activation, whereas activated Akt phosphorylates mTORC1. Treg-specific deletion of raptor, the essential component of mTORC1, results in a loss of Treg cell suppressive activity [113]. Elevated Akt activation and Foxo phosphorylation is observed in raptor-deficient Treg cells, and it is corrected by additional depletion of rictor, the defining element of mTORC2 [113]. However, compound ablation of mTORC1 and mTORC2 marginally ameliorates the inflammatory disorder, suggesting that mTORC1 promotes Treg cell function mostly through Foxo1-independent mechanisms.
Concluding remarks
With the identification of Foxp3 as a lineage marker, the Treg cell field has flourished during the past decade. Recent studies have revealed that, in addition to Foxp3 expression, Treg cell lineage specification is accompanied with epigenetic alterations and possibly rewiring of cellular signaling pathways. Furthermore, Foxp3 cooperates with a large number of binding partners to control Treg cell stability and function. A recent report showed that a subset of Treg cells has unstable expression of the Foxp3-binding protein, Eos. Such Eos-labile Treg cells are capable of acquiring T helper activity upon immunization without Foxp3 downregulation [114]. Therefore, understanding the intricate network of transcription factors in the control of Treg cell differentiation and function will shed new insights on Treg cell biology, and pave the way for targeting Treg cells for immunotherapy.
References
- 1.Sakaguchi S. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz S.Z. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishizuka Y. Thymus and reproduction: Sex-linked dysgnesia of the gonad after neonatal thymectomy in mice. Science. 1969;166:753–755. doi: 10.1126/science.166.3906.753. [DOI] [PubMed] [Google Scholar]
- 4.Sakaguchi S. Study on cellular events in post-thymectomy autoimmune oophoritis in mice. II. Requirement of Lyt-1 cells in normal female mice for the prevention of oophoritis. J. Exp. Med. 1982;156:1577–1586. doi: 10.1084/jem.156.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belkaid Y., Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annu. Rev. Immunol. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 6.Curotto de Lafaille M.A., Lafaille J.J. Natural and adaptive Foxp3+ regulatory T cells: more of the same or a division of labor? Immunity. 2009;30:626–635. doi: 10.1016/j.immuni.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Y. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohkura N. T cell receptor stimulation-induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity. 2012;37:785–799. doi: 10.1016/j.immuni.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 9.Olivares-Villagómez D. Regulatory CD4+ T cells expressing endogenous T cell receptor chains protect myelin basic protein–specific transgenic mice from spontaneous autoimmune encephalomyelitis. J. Exp. Med. 1998;188:1883–1894. doi: 10.1084/jem.188.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hori S. Specificity requirements for selection and effector functions of CD25+ 4+ regulatory T cells in anti-myelin basic protein T cell receptor transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8213–8218. doi: 10.1073/pnas.122224799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jordan M.S. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat. Immunol. 2001;2:301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh C-S. Recognition of the peripheral self by naturally arising CD25+CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Hinterberger M. Autonomous role of medullary thymic epithelial cells in central CD4+ T cell tolerance. Nat. Immunol. 2010;11:512–519. doi: 10.1038/ni.1874. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang W. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity. 2010;32:642–653. doi: 10.1016/j.immuni.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salomon B. B7/CD28 costimulation is essential for the homeostasis of the CD4(+)CD25(+) immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12:431–440. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 16.Tai X. CD28 costimulation of developing thymocytes induces Foxp3 expression and regulatory T cell differentiation independently of interleukin 2. Nat. Immunol. 2005;6:152–162. doi: 10.1038/ni1160. [DOI] [PubMed] [Google Scholar]
- 17.Verhagen J. Enhanced selection of FoxP3+ T-regulatory cells protects CTLA-4-deficient mice from CNS autoimmune disease. Proc. Natl. Acad. Sci. U.S.A. 2009;106:3306–3311. doi: 10.1073/pnas.0803186106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt-Supprian M. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S. Differential requirement of PKC-θ in the development and function of natural regulatory T cells. Mol. Immunol. 2008;46:213–224. doi: 10.1016/j.molimm.2008.08.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medoff B.D. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur. J. Immunol. 2009;39:78–84. doi: 10.1002/eji.200838734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan Y.Y. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat. Immunol. 2006;7:851–858. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
- 22.Barnes M.J. Commitment to the regulatory T cell lineage requires CARMA1 in the thymus but not in the periphery. PLoS Biol. 2009;7:e51. doi: 10.1371/journal.pbio.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long M. Nuclear factor-κB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 24.Ruan Q. Development of Foxp3+ regulatory T cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isomura I. c-Rel is required for the development of thymic Foxp3+ CD4 regulatory T cells. J. Exp. Med. 2009;206:3001–3014. doi: 10.1084/jem.20091411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh-hora M. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat. Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh-hora M. Agonist-selected T cell development requires strong T cell receptor signaling and store-operated calcium entry. Immunity. 2013;38:881–895. doi: 10.1016/j.immuni.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mantel P-Y. Molecular mechanisms underlying FOXP3 induction in human T cells. J. Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 29.Tone Y. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 30.Vaeth M. Dependence on nuclear factor of activated T-cells (NFAT) levels discriminates conventional T cells from Foxp3+ regulatory T cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16258–16263. doi: 10.1073/pnas.1203870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bopp T. NFATc2 and NFATc3 transcription factors play a crucial role in suppression of CD4+ T lymphocytes by CD4+ CD25+ regulatory T cells. J. Exp. Med. 2005;201:181–187. doi: 10.1084/jem.20041538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canté-Barrett K. Selective role of NFATc3 in positive selection of thymocytes. J. Immunol. 2007;179:103–110. doi: 10.4049/jimmunol.179.1.103. [DOI] [PubMed] [Google Scholar]
- 33.Kim H.P., Leonard W.J. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J. Exp. Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polansky J.K. DNA methylation controls Foxp3 gene expression. Eur. J. Immunol. 2008;38:1654–1663. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 35.Moran A.E. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekiya T. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 37.Sekiya T. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fassett M.S. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc. Natl. Acad. Sci. U.S.A. 2012;109:3891–3896. doi: 10.1073/pnas.1200090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burchill M.A. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lio C-W.J., Hsieh C-S. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontenot J.D. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 42.Vang K.B. IL-2,-7, and-15, but not thymic stromal lymphopoeitin, redundantly govern CD4+ Foxp3+ regulatory T cell development. J. Immunol. 2008;181:3285–3290. doi: 10.4049/jimmunol.181.5.3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Z. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burchill M.A. IL-2 receptor β-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 45.Patton D.T. Cutting edge: The phosphoinositide 3-kinase p110 delta is critical for the function of CD4(+)CD25(+)Foxp3(+) regulatory T cells. J. Immunol. 2006;177:6598–6602. doi: 10.4049/jimmunol.177.10.6598. [DOI] [PubMed] [Google Scholar]
- 46.Liu G. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat. Immunol. 2009;10:769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haxhinasto S. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J. Exp. Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouyang W. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 49.Kerdiles Y.M. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harada Y. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J. Exp. Med. 2010;207:1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouyang W., Li M.O. Foxo: in command of T lymphocyte homeostasis and tolerance. Trends Immunol. 2011;32:26–33. doi: 10.1016/j.it.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singer A. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Josefowicz S.Z. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482:395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samstein R.M. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kretschmer K. Inducing and expanding regulatory T cell populations by foreign antigen. Nat. Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 56.Gottschalk R.A. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J. Exp. Med. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lathrop S.K. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478:250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cebula A. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497:258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mucida D. Oral tolerance in the absence of naturally occurring Tregs. J. Clin. Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Curotto de Lafaille M.A. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 61.Verginis P. Induction of antigen-specific regulatory T cells in wild-type mice: visualization and targets of suppression. Proc. Natl. Acad. Sci. U.S.A. 2008;105:3479–3484. doi: 10.1073/pnas.0800149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Daniel C. Prevention of type 1 diabetes in mice by tolerogenic vaccination with a strong agonist insulin mimetope. J. Exp. Med. 2011;208:1501–1510. doi: 10.1084/jem.20110574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu L. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33:313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benson M.J. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J. Exp. Med. 2007;204:1765–1774. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng S.G. TGF-β requires CTLA-4 early after T cell activation to induce FoxP3 and generate adaptive CD4+ CD25+ regulatory cells. J. Immunol. 2006;176:3321–3329. doi: 10.4049/jimmunol.176.6.3321. [DOI] [PubMed] [Google Scholar]
- 66.Sauer S. T cell receptor signaling controls Foxp3 expression via PI3K, Akt and mTOR. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Delgoffe G.M. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schlenner S.M. Smad3 binding to the Foxp3 enhancer is dispensable for the development of regulatory T cells with the exception of the gut. J. Exp. Med. 2012;209:1529–1535. doi: 10.1084/jem.20112646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davidson T.S. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J. Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 70.Laurence A. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 71.Sun C.M. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mucida D. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 73.Hill J.A. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim J.M. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2006;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 75.Lahl K. Selective depletion of Foxp3+ regulatory T cells induces a scurfy-like disease. J. Exp. Med. 2007;204:57–63. doi: 10.1084/jem.20061852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shevach E.M. Mechanisms of Foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Khattri R. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003;4:337–342. doi: 10.1038/ni909. [DOI] [PubMed] [Google Scholar]
- 78.Chatila T.A. JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J. Clin. Invest. 2000;106:R75–R81. doi: 10.1172/JCI11679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bennett C.L. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 80.Wildin R.S. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 81.Williams L.M., Rudensky A.Y. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 82.Gavin M.A. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 83.Lin W. Regulatory T cell development in the absence of functional Foxp3. Nat. Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 84.Hill J.A. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 85.Fu W. A multiply redundant genetic switch ‘locks in’ the transcriptional signature of regulatory T cells. Nat. Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pan F. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng Y. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudra D. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Samstein R.M. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rudra D. Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kitoh A. Indispensable role of the Runx1-Cbfβ transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 92.Wu Y. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 93.Chang J.H. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat. Immunol. 2012;13:481–490. doi: 10.1038/ni.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Loizou L. Foxp3 interacts with c-Rel to mediate NF-kappaB repression. PLoS ONE. 2011;6:e18670. doi: 10.1371/journal.pone.0018670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Camperio C. Forkhead transcription factor FOXP3 upregulates CD25 expression through cooperation with RelA/NF-kappaB. PLoS ONE. 2012;7:e48303. doi: 10.1371/journal.pone.0048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ouyang W. Novel Foxo1-dependent transcriptional programs control Treg cell function. Nature. 2012;491:554–559. doi: 10.1038/nature11581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rao Rajesh R. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity. 2012;36:374–387. doi: 10.1016/j.immuni.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Koch M.A. T-bet(+) Treg cells undergo abortive Th1 cell differentiation due to impaired expression of IL-12 receptor beta2. Immunity. 2012;37:501–510. doi: 10.1016/j.immuni.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kerdiles Y.M. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat. Immunol. 2009;10:176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ouyang W. An essential role of the Forkhead-box transcription factor Foxo1 in control of T cell homeostasis and tolerance. Immunity. 2009;30:358–371. doi: 10.1016/j.immuni.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Förster R. CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 102.Schneider M.A. CCR7 is required for the in vivo function of CD4+ CD25+ regulatory T cells. J. Exp. Med. 2007;204:735–745. doi: 10.1084/jem.20061405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kocks J.R. Regulatory T cells interfere with the development of bronchus-associated lymphoid tissue. J. Exp. Med. 2007;204:723–734. doi: 10.1084/jem.20061424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kurobe H. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 2006;24:165–177. doi: 10.1016/j.immuni.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 105.Davalos-Misslitz A. Generalized multi-organ autoimmunity in CCR7-deficient mice. Eur. J. Immunol. 2007;37:613–622. doi: 10.1002/eji.200636656. [DOI] [PubMed] [Google Scholar]
- 106.Delgoffe G.M. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. 2013 doi: 10.1038/nature12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Strainic M.G. Absence of signaling into CD4+ cells via C3aR and C5aR enables autoinductive TGF-β1 signaling and induction of Foxp3+ regulatory T cells. Nat. Immunol. 2012;14:162–171. doi: 10.1038/ni.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kwan W.H. Signaling through C5a receptor and C3a receptor diminishes function of murine natural regulatory T cells. J. Exp. Med. 2013;210:257–268. doi: 10.1084/jem.20121525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Oldenhove G. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity. 2009;31:772–786. doi: 10.1016/j.immuni.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhao J. IFNgamma- and IL-10-expressing virus epitope-specific Foxp3+ T reg cells in the central nervous system during encephalomyelitis. J. Exp. Med. 2011;208:1571–1577. doi: 10.1084/jem.20110236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dominguez-Villar M. Identification of T helper type 1–like, Foxp3+ regulatory T cells in human autoimmune disease. Nat. Med. 2011;17:673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McClymont S.A. Plasticity of human regulatory T cells in healthy subjects and patients with type 1 diabetes. J. Immunol. 2011;186:3918–3926. doi: 10.4049/jimmunol.1003099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zeng H. mTORC1 couples immune signals and metabolic programming to establish Treg-cell function. Nature. 2013 doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sharma Madhav D. An inherently bifunctional subset of Foxp3+ T helper cells is controlled by the transcription factor Eos. Immunity. 2013;38:998–1012. doi: 10.1016/j.immuni.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]


