Summary
Sporadic human infection with avian influenza viruses has raised concern that reassortment between human and avian subtypes could generate viruses of pandemic potential. Vaccination is the principal means to combat the impact of influenza. During an influenza pandemic the immune status of the population would differ from that which exists during interpandemic periods. An emerging pandemic virus will create a surge in worldwide vaccine demand and new approaches in immunisation strategies may be needed to ensure optimum protection of unprimed individuals when vaccine antigen may be limited. The manufacture of vaccines from pathogenic avian influenza viruses by traditional methods is not feasible for safety reasons as well as technical issues. Strategies adopted to overcome these issues include the use of reverse genetic systems to generate reassortant strains, the use of baculovirusexpressed haemagglutinin or related non-pathogenic avian influenza strains, and the use of adjuvants to enhance immunogenicity. In clinical trials, conventional surfaceantigen influenza virus vaccines produced from avian viruses have proved poorly immunogenic in immunologically naive populations. Adjuvanted or whole-virus preparations may improve immunogenicity and allow sparing of antigen.
Few infectious diseases cause such a huge annual toll of morbidity, mortality, and economic loss as influenza. In addition, influenza can unpredictably emerge to cause pandemics. The 1918 Spanish pandemic spread around the world within 9 months causing up to 40 million deaths.1 The transmission of avian influenza H5N1 to at least 32 people during the 2004 Asian H5N1 epizoonotic period2, 3 prompted concerns that the next pandemic is imminent. As highlighted by the recent severe acute respiratory syndrome (SARS)- coronavirus outbreak, international air travel increases global vulnerability to infectious respiratory pathogens. Our ability to combat influenza and its complications depends primarily on vaccination. Annual influenza vaccine production is a well-planned process that takes up to 6 months. Current facilities may not be suitable for rapid bulk manufacture of avian influenza virus vaccines in response to a world threat.
Virology
Influenza viruses are enveloped negative-sense RNA viruses with a segmented genome belonging to the orthomyxoviridae family. They are classified on the basis of their core proteins into three distinct types: A, B, and C.4 Influenza A viruses infect a range of mammalian and avian species, whereas type B and C are essentially restricted to human beings. Influenza A viruses are responsible for annual epidemics and occasional pandemics, whereas influenza B viruses cause outbreaks every 2–4 years, but are not associated with pandemics. The main antigenic determinants of influenza A and B viruses are two surface glycoproteins: the neuraminidase and the haemagglutinin, both capable of eliciting immune responses in human beings. The haemagglutinin is involved with receptor binding and membrane fusion. The neuraminidase facilitates cleavage of virus progeny from infected cells, prevents viral aggregation, and aids movement through the mucosal respiratory-tract epithelium. Virus strains are classified according to host species of origin, geographic site and year of isolation, serial number, and, for influenza A, by serological properties of subtypes of haemagglutinin and neuraminidase.
Antigenic shift and drift
Influenza A H1 and H3 subtypes circulating in human beings evolve and undergo antigenic variability continuously. A lack of effective proofreading by the viral RNA polymerase leads to a high rate of transcription errors that can result in aminoacid substitutions in surface glycoproteins. Virus variants with substitutions in the antibody-binding sites can evade humoral immunity and reinfect individuals. This is termed “antigenic drift”. The segmented viral genome allows for a second type of antigenic variation. If two influenza viruses simultaneously infect a host cell, genetic reassortment may generate a novel virus with new surface or internal proteins. Pandemic influenza viruses arise by this process of “antigenic shift”, when a virus with a new haemagglutinin subtype emerges and spreads efficiently in a naive human population. Comparisons of pandemic and interpandemic influenza are shown in table 1 .
Table 1.
Features of pandemic and interpandemic influenza
| Type | Caused by | Immunity in population | Result |
|---|---|---|---|
| Pandemic influenza A | Antigenic shift: emergence of novel or re-emerging subtype of influenza A | Little or no background immunity (maybe partial immunity in older people if re-emerging virus) | High attack rates, excess mortality and morbidity in all age groups |
| Interpandemic influenza | Antigenic drift: evolution of existing influenza (A or B) strains | Little immunity in infants. Partial immunity in adults by cross-reacting antibody to previously seen and related strains | Variable outbreaks or epidemics with variable morbidity and mortality, usually in elderly and young (H3 greatest severity) |
Natural reservoir of influenza
Aquatic birds are the natural reservoir of influenza A viruses. All of the 15 haemagglutinin and nine neuraminidase subtypes currently identified are maintained in wild water-bird populations.5, 6 Avian influenza A viruses generally do not cause disease in these natural hosts. The principal site of influenza virus replication in aquatic birds is the gastrointestinal tract resulting in high faecal viral titres and viral transmission in migratory feeding areas.
Despite the range of virus subtypes, only a few haemagglutinin (H1, H2, H3) and neuraminidase subtypes (N1, N2) have established in human beings and have caused widespread respiratory disease. The haemagglutinin of human influenza viruses preferentially binds to sialic acid receptors containing α2,6-galactose linkages, whereas avian influenza viruses preferentially bind to those containing α2,3-galactose linkages. These binding preferences correlate with the predominance of sialic acid α2,6-galactose linkages on human epithelial cells, and α2,3-galactose linkages on avian intestinal epithelial cells.7, 8, 9 Although the molecular mechanisms responsible for receptor-binding specificity are poorly defined, it is believed that haemagglutinin of avian origin must acquire human receptor-binding specificity to generate influenza strains capable of sustained human-to-human transmission. Site-directed mutagenesis studies have shown that only one or two aminoacid mutations are required for this change.10 Limited passage in human beings of a virus possessing an avian haemagglutinin, such as occurring in Asia currently,2, 3 may be sufficient to generate such a change.
During the 20th century, an H1N1 virus in 1918, an H2N2 virus in 1957, and an H3N2 virus in 1968 caused influenza pandemics. Human-avian reassortant viruses seem to have caused the pandemics of 1957 and 1968. The 1957 H2N2 virus differed by three genes (haemagglutinin, neuraminidase, and-the RNA polymerase PB1) from the H1N1 virus that infected people between 1918 and 1957. The 1968 H3N2 virus differed by two genes (haemagglutinin and PB1) from the H2N2 virus that infected people between 1957 and 1968. In both cases, the H2 and H3 haemagglutinin genes were contributed by avian viruses.11 Sequence analyses of early H2 and H3 isolates indicate receptor-binding specificity was altered by a single aminoacid substitution soon after human introduction.10 Since pig trachea contains receptors for both avian and human influenza viruses, and can support replication of viruses of both human and avian origin, it has been suggested that genetic reassortment between avian, swine, and human influenza viruses may occur in pigs, and that they represent a “mixing bowl” for the evolution of human pandemic strains.12 Haemagglutinin and neuraminidase of H5N1 viruses isolated from human beings, poultry, and wild ducks have distinguishable properties.13 Chickens seem to support a separate natural reservoir of influenza viruses, indicating a possible role as intermediate hosts in zoonotic transmission. Some avian H9 viruses established in poultry are capable of two-way transmission between domestic ducks, where they are able to generate multiple reassortants with other cocirculating viruses.14 These reassortant viruses have haemagglutinin receptor-binding sequences potentially capable of human infection, suggesting that new viruses may emerge directly from the avian pool. The close proximity of people to high concentrations of waterfowl, poultry, and swine in southeast Asia, and avian influenza activity, has identified this region as a hypothetical influenza epicentre (figure 1 ).15
Figure 1.

Returning from a shopping trip in Hanoi, Vietnam (photo JM Katz).
Avian influenza in people
Avian influenza viruses generally do not replicate efficiently in human beings, even after experimental infection.16 Before 1997, direct transmission of avian influenza viruses to the human respiratory system was not considered possible. However, it is now recognised that at least some subtypes of avian influenza viruses can replicate within the human respiratory tract. Although it is unclear whether the recent reported increase in transmission of avian influenza to people (table 2 ) is the result of heightened surveillance, the geographical expansion of H5N1 poultry outbreaks across Asia is an unprecedented and new event. More than 30 confirmed cases of transmission of avian H5N1 virus to human beings has increased the possibility that an avianhuman reassortant virus may emerge to effectively transmit among people.2, 3
Table 2.
Confirmed cases of avian-to-human transmission of influenza A subtypes
| Year and place | Major strain designations | Infections and deaths | Symptoms | Comments and references |
|---|---|---|---|---|
| 1995 UK | A/Eng/268/95 (H7N7) | 1 | Conjunctivitis | Close contact of patient with infected duck17 |
| 1997 Hong Kong | A/HK/156/97 (H5N1) | 18 (6 deaths) | Respiratory | High mortality rate (6/18, 33%) associated with high rates of pneumonia (61%), intensive care (55%), and prolonged hospitalisation.18, 19, 20 Of 16 H5N1 human isolates, phylogenetic and antigenic analyses identified two closely related groups represented by A/Hong Kong/156/97 and A/Hong Kong/483/9718, 21 Fortunately, serological surveillance revealed little evidence of human-to-human transmission20, and no additional cases occurred following depopulation of poultry in Hong Kong |
| A/HK/148/97 (H5N1) | ||||
| 1999 Hong Kong | A/HK/1073/99 (H9N2) | 2 | Respiratory | G1-like H9N2 virus isolated from 2 hospitalised children.22 No human-to-human transmission identified23 |
| 2003 Hong Kong | A/HK/213/03 (H5N1) | 2 (1 death) | Respiratory | A father and son, returning from a visit to relatives in Fujian province were hospitalised with pneumonia.24 The father died, while the son recovered. A young female family member had died of pneumonia in Fujian but the aetiology of her death was not determined. The H5N1 viruses isolated from the 2 patients were antigenically distinct from H5N1 viruses isolated in 199725 |
| 2003 Netherlands | A/Neth/33/03 (H7N7) | 83 | Conjunctivitis | There were 83 cases of viral conjunctivitis, of which 5 had influenza-like illness, and 2 cases of isolated respiratory illness.26 |
| A/Neth/219/03 (H7N7) | 1 (1 death) | Respiratory | Human-to-human transmission to 3 household contacts was confirmed. All human viruses had internal gene segments from avian influenza A viruses.27 A veterinarian developed fatal respiratory disease and virus sequencing revealed a number of mutations compared with the strains responsible for eye infections. Destruction of infected flocks, quarantining of farms, and administration of antiviral prophylaxis to workers stopped H7N7 transmission to people | |
| 2003 Hong Kong | A/HK/2018/03 (H9N2) | 1 | Respiratory | Y280 (G9-like) H9N2 isolated from respiratory secretions of hospitalised 5 year old boy.28 No further cases identified |
| 2004 Vietnam | A/VN/1203/04 (H5N1) | 22 (15 deaths) | Respiratory | Extensive simultaneous highly pathogenic H5N1 outbreaks emerged across Asian countries in 2004.2, 3, 29, 30 Human infections are associated with direct exposure to dead or ill birds, although one cluster of possible human-to-human transmission occurred in Vietnam. Clinical infection is rapid with features of respiratory distress, pneumonia and multi-systemic organ failure and seem to be predominantly among younger age groups (<25 years). H5N1 viruses have molecular changes associated with adamantane resistance29 |
| A/VN/1194/04 | ||||
| 2004 Thailand | A/Thai/16/04 (H5N1) | 12 (8 deaths) | Respiratory | They are phylogenetically and antigenically distinct from 1997 and 2003 human isolates29 |
| 2004 Canada | Avian H7N3 | 2 | Conjunctivitis | Two laboratory-confirmed cases among poultry workers associated with culling activities in the control of an influenza A(H7N3) outbreak in poultry in British Columbia, Canada30 |
| Respiratory (1) | ||||
| 2004 Egypt | Avian H10N7 | 2 | Respiratory | Two infants, presenting with fever and cough, had virus isolated from specimens. The father of one of the children was a poultry merchant who had recently visited a market in which 5 isolations of avian influenza H10N7 has been reported31 |
The first association of avian influenza viruses with respiratory illness in human beings was during 1997 when six deaths from 18 human cases of highly pathogenic influenza H5N1 occurred during an outbreak among live-bird markets (table 2).18, 19, 20 All viral genes were of avian origin, indicating that H5N1 had crossed the species barrier without adaptation or reassortment with human viruses. Despite the elimination of ducks, geese, and quail (sources of H5N1 and its donor genes) and cleaning days in the markets, H5N1 viruses have subsequently reemerged in Hong Kong poultry markets,32 although Hong Kong remains free from infection in the 2004 H5N1 outbreak across Asia.29, 33 In late 2002, highly pathogenic H5N1 viruses were isolated from dead waterfowl in Hong Kong (personal communication, M Peiris, University of Hong Kong), which was notable since H5N1 viruses do not typically cause disease in waterfowl. In February 2003, H5N1 reemerged in Hong Kong in two family members returning from a trip to China.24 In 2004, pathogenic H5N1 viruses are causing extensive poultry outbreaks in Asia with 23 deaths (68%) of 34 human cases reported in Vietnam and Thailand.2, 3, 30
In Hong Kong in 1999, and again in 2003, influenza H9N2 viruses were isolated from children with mild respiratory illnesses.22, 28 As with H5N1, no human-to-human transmission was evident.23 Additional human H9 infections in China,34 and detection of antibody to H9 in human serum samples in China and Hong Kong35 suggest that further human H9 infection has occurred. Eurasian H9 viruses circulating since the late 1990s have been classified into three phylogenetic sub-lineages: G1, Y280 (G9-like), and Y439.36, 37 Those established in live-bird markets (predominantly chickens and quail) are Y280 (G9-like) and G1-like viruses, respectively. G1-like H9 viruses caused the 1999 human infections and contained internal genes homologous to those of the 1997 human H5N1 virus, suggesting that reassortment between the strains had already taken place,36, 37 whereas the 2003 human isolate was of Y280 (G9-like) lineage (personal communication W Lim, Government Virus Unit, Queen Mary Hospital, Department of Health, Hong Kong, China). Avian H9N2 viruses are now widespread in poultry,38 and have also transmitted to swine in Hong Kong and China.38, 39, 40 Some avian H9 viruses have acquired receptor-binding characteristics typical of human strains, increasing the potential for reassortment within both human and pig respiratory tracts.41
In early 2003, outbreaks of highly pathogenic avian influenza H7 among poultry occurred in the Netherlands and extended to Belgium and Germany.26, 27 More than 80 workers involved in control of the outbreak developed viral H7N7 conjunctivitis and a few developed respiratory illness. Evidence of limited human-to-human spread, and a fatal respiratory infection, highlighted a significant threat to human health. More recently, cases of avian influenza (H7N3) infections have been reported in two cullers during control of an H7N3 poultry outbreak in Canada.30 Avian influenza (H10N7) seems to have crossed the species barrier from poultry to people for the first time. In Egypt in April 2004 two infants presenting with mild febrile respiratory symptoms had H10N7 influenza viruses isolated from respiratory samples.31
Other virus subtypes of concern
As well as H5, H7, and H9 viruses, the H6 subtype has acquired the ability to infect chickens and is rapidly becoming endemic in poultry populations. Phylogenetic analyses of H6N1 viruses isolated from wildfowl, showing high nucleotide homology of the internal genes to human H5N1 and H9N2 viruses, suggests these subtypes can transfer genetic material between each other and are potential sources of new strains.42
H2N2 viruses were responsible for the 1957 influenza pandemic and circulated as the only human subtype until-
1968 when it was replaced by H3N2 subtype. The H2 haemagglutinin gene, along with the neuraminidase and PB1 genes, were derived from avian sources.11 Since people born after 1968 lack immunity to the H2 subtype, H2 viruses pose a pandemic threat to this susceptible population. H2 viruses continue to circulate in wild ducks,43 although the conditions required for the re-emergence of human H2 influenza viruses are unclear.
Pathogenesis of avian influenza viruses
Although the virulence of avian influenza viruses has been well studied in avian species, their virulence in mammals is not well understood. Infection with avian influenza A viruses in birds causes a wide spectrum of disease ranging from subclinical to overwhelming systemic illness (figure 2 ). Both H5 and H7 subtypes have the ability to evolve into highly pathogenic forms. Although virulence is a polygenic trait, a major contributing factor in birds is the haemagglutinin. Cleavage of the haemagglutinin into two subunits is essential for viral infectivity.44 Haemagglutinin from strains of low pathogenicity is cleaved by proteases limited to the respiratory tract of mammalian species and the intestinal tract of avian species. By contrast, haemagglutinin from highly pathogenic viruses can be cleaved by proteases present in a range of tissues, resulting in multi-system infection. Structural features at the cleavage site determine the cleavability of the haemagglutinin. The acquisition of multiple basic aminoacids in highly pathogenic H5 or H7 haemagglutinin enables cleavage of the protein by these ubiquitous proteases and confers virulence. Carbohydrate side chains in the vicinity of the cleavage site may also affect the access of the proteases and hence virulence.45
Figure 2.
The effect of highly pathogenic H5N1 virus on ducklings in Vietnam (photo T Tumpey).
Other factors in addition to the haemagglutinin cleavage site are likely to contribute to virus pathogenicity in mammals. Virulence of mouse-adapted influenza A viruses have been associated with the interferon antagonist properties of the NS1 protein46 or the ability of the neuraminidase glycoprotein to sequester circulating plasminogen and promote haemagglutinin cleavage.47 BALB/c mice47, 48, 49, 50 and ferrets51 are useful mammalian hosts for the evaluation of human H5N1 pathogenesis. In these mammalian hosts, the multibasic aminoacid motif in the haemagglutinin is necessary but not sufficient for virulence.47, 52, 53 A single aminoacid substitution in PB2 is associated with high pathogenicity of human H5N1 viruses in mice52 although substitutions in other gene products are likely to play a part.49
In human H5N1 infection, disease progression to respiratory failure is unusually severe, with features of haemophagocytosis, leucopenia, and multiple organ failure.3, 19, 30 In-vitro infection of human macrophages with 1997 H5N1 human viruses induces high levels of cytokines compared with some human virus strains.54 In pig respiratory epithelial cells, the 1997 H5N1 human viruses were also shown to be relatively resistant to the inhibitory effects of host antiviral cytokines such as interferons.55 Thus, the severity of H5N1 infection in people is likely to be related to the induction of excessive proinflammatory responses that exacerbate tissue injury. It has been suggested that the nonstructural gene has a role.54, 55
Gene sequence analyses of the 1918 pandemic virus, which displayed enhanced virulence in human beings has not yet uncovered molecular determinants previously associated with influenza virus virulence.56, 57 The molecular determinants and gene constellations that confer virulence of avian, swine, and human viruses, and the circumstances under which virulent phenotypes emerge remains unclear. Understanding the basis of virulence is important for vaccine design, particularly of live vaccines, so that viruses can be attenuated.
Influenza vaccines
Current inactivated influenza vaccines are produced from virus grown in embryonated hens' eggs, and are of three types: whole-virus, “split-product”, or subunit “surface-antigen” formulations. Whole-virus vaccines are associated with increased adverse reactions, especially in children, and are little used. Most influenza vaccines are split-product vaccines, produced from detergent-treated, highly purified influenza virus, or surface-antigen vaccines containing purified haemagglutinin and neuraminidase.58 Vaccines are usually trivalent, containing 15μg each of two influenza A subtypes (H1N1 and H3N2) and one influenza B strain. Vaccines elicit a relatively strain-specific humoral response, have reduced efficacy against antigenically drifted viruses, and are ineffective against unrelated strains. The WHO reviews vaccine composition biannually and updates antigenic content depending on prevalent circulating subtypes to provide antigenically well-matched vaccines. Protective efficacy of 70–95% in healthy young adults is obtained when there is a good antigenic match between the vaccine and circulating strains.59 Vaccination of the elderly is associated with 19–63% reductions in hospitalisation for pneumonia and influenza, 17–39% reductions for all respiratory conditions, and 27–75% reductions in all-cause mortality.60
New influenza vaccines must elicit protective immunity. The haemagglutinin-inhibition test is most commonly used for the detection of antibody to influenza, although single radial haemolysis (SRH) may also be used, since both are correlated with immune protection.61, 62 In the European Union, interpandemic influenza vaccines should fulfil certain criteria, prepared by the Committee for Proprietary Medicinal Products (CPMP),63 which are usually assessed by haemagglutinin-inhibition tests in limited annual clinical studies (panel 1 ).
Panel 1. EU criteria for the licensing of annual interpandemic influenza vaccines. Vaccines should be assessed in groups of >50 patients of 18–59 years and ≥60 years of age. At least one of three criteria must be fulfilled.
Mean geometric increase in antibody >2·5 (>2 in the ≥60 years group) Number of seroconversions or significant rises in anti-haemagglutinin antibody (ie, four-fold increase in post-vaccination HI titre or 50% increase in SRH zone) should be >40% (>30% in the ≥60 years group) Proportion of patients achieving a seroprotective HI titre of ≥1/40, or SRH titre of >25 mm2 post vaccination should be >70% (>60% in the >60 years group)
Vaccines for pandemic use In the event of pandemic influenza, vaccine demand would soar. Savings made using monovalent rather than trivalent vaccine (15μ haemagglutinin per dose instead of 45μ) would possibly be offset by a two-dose schedule, increased demand, and difficulties with production of egg-grown viruses. New developments include the use of mammalian cell lines to culture influenza virus for vaccines to provide increased flexibility of production at times of heightened demand.64 Immunopotentiating effects of adjuvants and whole-virus vaccine may increase antigenicity, allowing dose content reduction enabling maximum efficient use of limited supplies.
The population immune status in a pandemic situation differs from that seen during the interpandemic period. At the onset of the previous pandemics, younger adults were immunologically naive to the new strains, whereas older populations may have been primed by previous infections of related strains that circulated in earlier times. Global immune susceptibility to avian influenza subtypes would be expected. The quantity of antigen required to elicit satisfactory immune responses in naive individuals is unclear since few studies have been done after the emergence of a novel virus. Current events suggest the urgent need to develop a clearly defined strategy for clinical assessment of safety and immunogenicity of pandemic vaccines.
Experience with pandemic human influenza virus vaccines
In 1976 and 1977, the emergence of influenza A/New Jersey/76 and A/USSR/92/77 (H1N1) triggered pandemic alerts, and afforded the opportunity for vaccine trials in immunologically naive and primed populations. A series of whole-virus vaccine studies65, 66, 67, 68, 69 reported differences between naive populations (those aged 24 years and not exposed to previous H1N1 strains) and primed populations (older than 24). In naive patients, if one dose of vaccine was administered, large doses (in excess of 60μ haemagglutinin) were required to fulfil CPMP criteria. However, if two doses of vaccine were given, lower antigen doses (5 μg) were needed. Whole-virus vaccine was significantly more immunogenic than subunit or split-product vaccines. In primed patients, as is the case during interpandemic periods, no difference in immunogenicity between whole-virus vaccine and subunit or split-product vaccines was reported. However, a consistent finding was that whole-virus vaccine was associated with increased reactogenicity, particularly in children, who developed febrile complications even with low doses.67
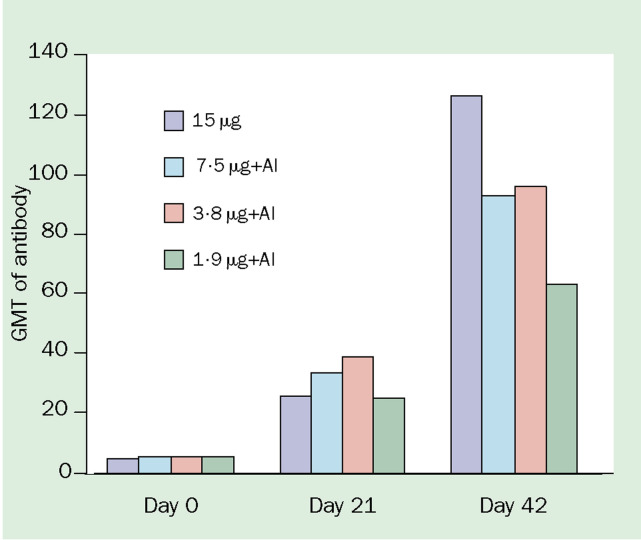
Although licensed for use in human influenza vaccines, aluminium salts are rarely used since studies have indicated little clinical benefit70 However, encouraging findings were more recently reported in a study of whole-virus A/Singapore/1/57 (H2N2) vaccine in immunologically naive people.71 Monovalent alumadjuvanted vaccine containing either 7·5, 3·8, or 1·9 μg H2 haemagglutinin per dose was compared with unadjuvanted whole-virus 15μg vaccine. Although a single dose of any vaccine was unable to elicit responses associated with protection, a second dose of vaccine boosted responses to rates that fulfilled CPMP licensing criteria across all doses, suggesting that up to an eight-fold reduction in antigen content could be achieved with the addition of alum (figure 3 ).
Figure 3.
Effect of alum-adjuvant on immunogenicity of monovalent influenza A/Singapore/1/57 (H2N2) in immunologically naive people aged 18–30 years (data from reference 71). Vaccine administered on day 0 and day 21. GMT=geometric mean titre. Al=aluminum mineral adjuvant.
Vaccines against avian influenza virus
Since influenza A viruses possess a segmented genome, simultaneous infection of eggs with two different viruses may result in reassortment of segments to produce a desired vaccine seed strain. The influenza A virus components of annual influenza vaccines are typically derived from egggrown reassortment viruses that have the relevant haemagglutinin and neuraminidase genes of the antigenically relevant strain, and the six remaining gene segments from A/Puerto Rico/8/34 (H1N1). These PR/8/34 segments confer high growth properties in eggs favoured for inactivated vaccine production.58 This process requires large numbers of eggs, and in many companies lacks the flexibility to respond rapidly to a pandemic event. Highly pathogenic H5 and H7 viruses cannot be grown in large quantities because they are lethal to chicken embryos.72 Such pathogenic strains also impose regulatory and safety issues. As the multibasic sequence cleavage site is believed to contribute to the pathogenesis of human H5N1 infection, vaccine preparation from wild-type H5N1 virus would require heightened biocontainment to protect workers and eliminate the possibility of environmental contamination and infection of susceptible animals. Thus, several approaches have been attempted for avian influenza vaccine development. These include: (1) the production of inactivated vaccine from wildtype virus; (2) the selection of an antigenically related nonpathogenic vaccine strain; (3) the use of baculoviruses to express recombinant haemagglutinin; (4) DNA-based vaccines; (5) the use of plasmid-based reverse genetics systems to construct vaccine seed strains possessing attenuated haemagglutinin; and (6) plasmid-based reverse genetic systems to construct attenuated donor strain recombinants. Reverse genetics is likely to produce the most rapid response in an emerging pandemic. Much of the preclinical and clinical development of highly pathogenic avian influenza vaccines has used H5 virus as a model.
Vaccines for highly pathogenic subtypes (H5 and H7)
After the 1997 H5N1 outbreak, inactivated vaccines from wild-type A/Hong Kong/156/97 (H5N1) virus were prepared in the UK and the Netherlands.73, 74 Whole-virus vaccine was effective in protecting mice against lethal challenge with H5N1 virus.73 Conventional surface-antigen vaccine was poorly immunogenic in chickens and did not protect against lethal dose challenge, although an ISCOM formulation (antigen as immune-complex stimulators) boosted immune responses and protected against lethal H5N1 challenge.74
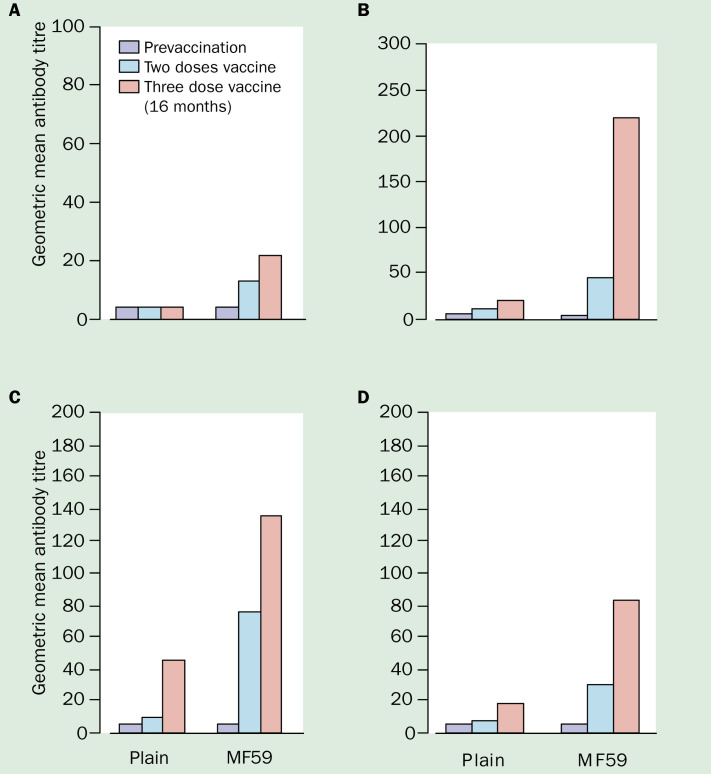
Because the use of highly pathogenic strains has safety restrictions, the selection of a surrogate non-pathogenic virus capable of evoking crossreactive immunity to the 1997 Hong Kong H5N1 viruses was investigated. Both A/duck/Hokkaido/67/96 (H5N4) and A/duck/Singapore/97 (H5N3) have haemagglutinin proteins antigenically similar to A/Hong Kong/ 156/97 (H5N1) and were used in experimental vaccines.72, 73 Although antibody titres to H5N1 induced by A/duck/Singapore (H5N3) vaccine were four-fold lower than the homologous strain,73 inactivated whole-virus vaccine72, 73 and alum-adjuvanted subunit vaccine48 were capable of protecting mice against lethal H5N1 challenge. Although attempts to reassort A/duck/Singapore (H5N3) with A/PR/8/34 to produce a high-growth virus suitable for vaccine production failed,73 conventional and MF59-adjuvanted A/duck/ Singapore/97 (H5N3) surface-antigen vaccines were clinically assessed in a randomised phase I trial.75 Two doses of 7·5, 15, or 30μg H5 haemagglutinin were given 3 weeks apart. Antibody responses were measured by haemagglutinininhibition, virus microneutralisation, and SRH. Although both vaccines were well tolerated, non-adjuvanted vaccine was poorly immunogenic, with only one of 11 (9%) recipients seroconverting by haemagglutinin-inhibition and SRH H5N1, and four (36%) by microneutralisation and SRH H5N3 after two 30μg doses. The addition of MF59 gave significantly higher antibody responses (figure 4 ), and two doses achieved seroconversion rates of 13/31 (42%), 29(94%), 31(100%), and 26(84%) by haemagglutinin-inhibition, microneutralisation, SRH H5N3, and SRH H5N1, respectively. Antibody titres by SRH to H5N1 were about half those to H5N3, showing the need for close antigenic matching between vaccine and pandemic strains to ensure maximum vaccine efficacy. Antibody responses after H5N3 revaccination 16 months later were boosted significantly above those achieved after two doses.76
Figure 4.
Geometric mean titres of antibody for MF59-adjuvanted and conventional surface-antigen H5N3 vaccine before and after two and thre doses of vaccine (data from 75, 76 A=Haemagglutination-inhibition (H5N3); B=Microneutralisation (H5N3); C=Single radial haemolysis (H5N3); D=single radial haemolysis (H5N1)
It is desirable for vaccines to boost responses after initial priming, since second waves occur 3–9 months after the first pandemic wave of infection.77 One problem in assessing vaccine responses was the insensitivity of the haemagglutinininhibition test, routinely used in assessment of influenza vaccines, for the detection of antibody to H5 when compared with neutralisation tests.78, 79 Haemagglutinin-inhibition relies on the ability of antibody to disrupt the haemagglutinin -sialic-acid-receptor binding interaction between virus and erythrocytes. If the erythrocytes used in the test are not optimised for expression of α2,3- galactose linkages that are necessary for avian influenza virus binding, the test is insensitive.80 However, there are no recognised clinical correlates of immune protection for neutralisation antibody. It was only possible to assess vaccine responses with respect to CPMP licensing criteria after development of a specific SRH assay.81 A modified haemagglutinin-inhibition test, using enzymatically altered turkey erythrocytes or horse erythrocytes, was developed to increase the sensitivity for detecting antibody to avian influenza virus antigens.82
An alternative to egg-derived vaccines involves the use of haemagglutinin protein expressed in insect cells by recombinant baculovirus. Potential difficulties include the use of uncleaved rather than cleaved haemagglutinin and differences in glycosylation in insect cells that may effect immunogenicity. Nonetheless, antibody responses to 15–45 ++g doses of recombinant H1 and H3 antigens are similar to those induced by licensed vaccines.83, 84 Baculovirus-derived H5 and H7 haemagglutinin vaccines protect against lethal virus challenge in chickens, even when the haemagglutinin sequence homology differs by up to 16%.85 However, when clinically assessed in people, a recombinant baculovirus-expressed H5 vaccine was suboptimal. Even after two doses of 90 ++g, only 52% subjects seroconverted by microneutralisation, suggesting improvements in immunogenicity are needed.86
Reverse genetics systems can be used to generate attenuated avian influenza viruses, and are likely to prove pivotal in pandemic vaccine development. The appropriate haemagglutinin and neuraminidase genes from a virus can be cloned and, if necessary, the haemagglutinin may be attenuated by removal of the multi-basic cleavage site sequence, and inserted into plasmids. The plasmids are transfected into a cell line together with plasmids encoding the internal genes from the A/PR8/34 virus to generate an appropriate non-pathogenic vaccine seed strain. Vaccine candidates expressing the target haemagglutinin from highly pathogenic viruses could potentially be produced within weeks of an emerging event. Recombinant H5 viruses showing desirable properties for H5 vaccine formulation—including loss of egg lethality, virulence, and infectivity in animal models—have been produced,87, 88 although one attenuated recombinant virus showed limited neurovirulence in mice.87 Reverse genetics is thus capable of generating attenuated viruses from pathogenic strains suitable for vaccine production with only limited enhancement of biosecurity measures and using pre-existing equipment and facilities. It should also be possible to prepare panels of reassortant viruses and vaccine seed candidates containing target genes of potential pandemic viruses in advance of any specific threat. However, there are regulatory, safety, and legal problems to overcome before the technology can be used for vaccine development. Mammalian cell lines (eg, Vero cells) used for transfection must be of certified quality for human vaccine production. Viruses generated by reverse genetics may be considered to be “genetically modified organisms”, imposing local and national safety regulations regarding research and development. In addition, intellectual property rights on reverse genetics technology are held, and licences may need to be granted for commercial use of vaccines.
Inactivated influenza vaccines are poor inducers of cytotoxic T-cell (CTL) responses, which aid in recovery from influenza infection. It has been suggested that crossreactive immunity to several influenza virus subtypes could be induced by CTL responses to conserved epitopes in internal proteins.89 DNA vaccines integrate gene sequences for the antigenic protein of interest into bacterial plasmids that are inoculated into the host. The expression of plasmid DNA produces the antigen in its natural configuration, which is more likely to stimulate neutralising antibody and undergo HLA class I expression inducing CTL responses.90 H5 and H7 haemagglutinin-expressing DNA vaccine protects mice and chickens against lethal dose virus challenge.91, 92 DNA vaccine expressing conserved internal proteins including nucleoprotein give partial protection to mice against H5N1 infection.93 Intradermal DNA influenza vaccines are beginning clinical evaluation. ISCOM-formulated H1N1 vaccine can induce CTL responses and greater longer-lasting antibody responses than conventional vaccine.94 It offers broad protection against virus challenge with H2, H3, H9, and virulent H5 viruses in mice. ISCOM vaccines are tolerated in human beings and induce broad CTL and rapid humoral responses.95 Mucosal delivery of inactivated influenza H3N2 vaccine adjuvanted with modified heat-labile enterotoxin from Escherichia coli induces B-cell-dependent heterosubtypic immunity against lethal H5N1 virus challenge in mice.96 In the absence of an antigenically matched vaccine, alternative vaccine strategies that induce crossreactive immunity by ISCOM, DNA, or mucosal vaccines may provide useful firstline defence against an emerging pandemic strain.
Vaccines for H9 influenza
Since H9 viruses do not have a multibasic haemagglutinin cleavage site, they show low pathogenicity for avian species and may be grown to high titres in eggs. Both G1-like (A/Hong Kong/1073/99) and Y280 (G9-like; A/Hong Kong/2018/03) H9N2 viruses are capable of human infection.22, 28 For an effective H9 vaccine strategy, an understanding of the relative immunogenicity and crossprotection induced by these lineages is required. Although infection of mice with G1 or G9 group H9 viruses did not evoke detectable crossreacting neutralising antibody, they were protected from subsequent rechallenge with the homologous or heterologous virus lineage.97 Whole-virus G1 H9 vaccine produced crossreactive antibody responses to both G1 and G9 viruses, and protected mice against rechallenge with either virus. By contrast, whole-virus G9 H9 vaccine induced homologous antibody titres only and was able to protect against G9 challenge, but showed reduced protective efficacy against the heterologous G1 lineage. Here, a single dose of H9 vaccine induced adequate immune responses in mice, by contrast with findings with H5N1 vaccine that required a two-dose schedule to elicit adequate immune responses.74, 97 Since some Y280 (G9-like) H9 viruses do not grow well in eggs, an A/PR/8/34 reassortant has been produced.98 One dose of inactivated G9/PR8 vaccine protected mice against G9 challenge. Two doses of vaccine increased antibody responses capable of protecting mice against both G1 and G9 H9 challenge.
Whole-virus and subunit A/Hong Kong/1073/99 (H9N2) vaccines were clinically evaluated in the UK.99 60 adults were randomly assigned two doses, administered 3 weeks apart, of 7–5, 15, or 30 ++g H9 haemagglutinin content. Although well tolerated, whole-virus vaccine was more reactive, in keeping with H1N1 vaccines. There was little detectable crossreactive immune response to an antigenically distinct G9 H9 virus (unpublished findings). More than 40% of the prevaccination serum samples showed reactivity to H9N2 by neutralisation and haemagglutinin-inhibition, suggesting preexisting crossreacting antibody from exposure to earlier haemagglutinins. It is unlikely that H9 influenza has circulated widely in the UK. Further serological testing correlated H9 reactivity with antibody responses to H2, but not H1 or H3 haemagglutinin. People with baseline reactivity to H9 were born before 1969, and thus had been potentially exposed to H2 during its period of circulation in human beings. Subjects were immunologically divided into naive and primed recipients. In truly naive subjects, one dose of either vaccine was poorly immunogenic. Although whole-virus vaccine was more immunogenic than subunit vaccine, two doses still left a significant number of vaccinees with serological responses below the protective threshold (table 3 ) Among primed individuals, one dose of either vaccine boosted anti-H9 responses, fulfilling CPMP criteria. Since the second dose was of questionable value, to preserve limited vaccine supplies during the first wave of an emerging pandemic, different schedules in different populations could be considered. A German study among 18–60 year-olds reported one dose of 15 μg whole-virus vaccine A/Hong Kong/1073/99 (H9N2) capable of fulfilling at least one CPMP criterion and that a second dose of vaccine significantly improved responses.71 However, age-related responses or the effect of pre-existing reactivity to H9N2 were not analysed. In keeping with the H2N2 experience, alum-adjuvant allowed a reduction in H9 content to 1·3 μg per dose while maintaining immunogenicity.
Table 3.
Haemagglutinin-inhibition results for A/Hong Kong/1073/99 (H9N2) in relation to the CPMP criteria (data from reference 99)
| CPMP criteria | Day | <32 years of age (naive) | >32 years of age (primed) | ||
|---|---|---|---|---|---|
| Whole virus (n=14) | Subunit (n=14) | Whole virus (n=12) | Subunit (n=16) | ||
| Geometric mean titre increase | 21 | 2·3 (1·4–3·6) | 1·7 (0·8–3·3) | 2·2 (1·5–3·2) | 5·4* (3·0–9·6) |
| 42 | 6·9* (4·6–10·5) | 2·8* (1·5–5·4) | 3·0* (2·1–4·3) | 4·7* (2·7–8·4) | |
| Seroconversions | 21 | 36% | 15% | 50%* | 56%* |
| 42 | 64%* | 36% | 75%* | 56%* | |
| Seroprotection rate (≥1/40) | 0 | 0% | 0% | 17% | 25% |
| 21 | 21% | 14% | 50% | 75%* | |
| 42 | 43% | 14% | 66% | 75%* | |
Data are percentage of participants. 95% confidence intervals shown for geometric mean titre increase.
Fulfilled criteria
Live attenuated influenza vaccines
Intranasally delivered live, attenuated cold-adapted influenza vaccines elicit systemic and local mucosal immune responses and display protective efficacy.100 Attenuated cold-adapted strains are generated by reassortment between a wild-type virus expressing target haemagglutinin and neuraminidase, and a cold-adapted donor such as influenza A/Ann Arbor/6/60 (H2N2). Donor strains are cold adapted, temperature-sensitive, and attenuated. These properties are associated with polygenic mutations. These live attenuated viruses display high levels of phenotypic and genotypic stability and are not transmissible to close seronegative contacts.101 Both attenuated H5N1 and H9N2/Ann Arbor cold-adapted recombinant viruses have been generated and are seen to be non-pathogenic in mammalian and chicken models.98, 102 Concerns over the generation of a reassortant between a live virus vaccine containing an avian influenza virus and a co-infecting human strain, and the possibility of spontaneous genetic change may limit the use of such vaccines in the interpandemic period. While current intramuscular influenza vaccines are effective at inducing relatively strainspecific serum haemagglutination-inhibition IgG, they are poor at stimulating secretory IgA in nasal wash fluid.103, 104 As secretory IgA exhibits potential heterotypic crossreactivity to influenza virus strains at the point of entry,104, 105 live attenuated virus vaccines may offer wider protection against vaccinedrifted variants that could be advantageous once a pandemic is underway.
Role of antiviral therapy
Specific influenza antiviral agents are available for early treatment and prophylaxis of influenza.106 The adamantanes, amantadine and rimantadine, have been available for more than 30 years and inhibit strains of influenza A including nonhuman subtypes. However, rapid emergence and transmission of drug-resistant virus after treatment may render prophylaxis ineffective. The genetic basis for resistance seems to be single aminoacid substitutions in the viral M2 ion channel. The H5N1 strains isolated from poultry and human cases in 2004 had genotypic changes in the M2 gene associated with resistance,29 suggesting these agents would be of little clinical value should these strains become capable of humanto-human transmission.
Neuraminidase inhibitors, such as zanamivir and oseltamivir, are effective in prevention studies106 and are highly active against a broad range of influenza A viruses of both human and avian origin, including amantadine-resistant strains. Although strains with reduced susceptibility to neuraminidase inhibitors have been isolated after sequential passage of virus in presence of drugs, clinically significant resistant strains have not, as yet, been identified. Antiviral drugs may be of benefit in protecting individuals in essential services whilst waiting for an effective vaccine to be prepared; however, supply and cost issues would limit their effect on the course of a pandemic. A sufficiently large supply of antivirals to curb pandemic influenza would require international or national stockpiling before the onset of such an event; this would require considerable expense, and vaccination is likely to remain the principal means of combating pandemic influenza.
Concluding remarks
Although pandemic planning and understanding is greater since the first H5 outbreak in 1997, our ability to respond rapidly remains less than optimal. The 2004 Asian H5N1 epizoonotic outbreak indicates the urgent need for vaccines against avian influenza viruses. However, regulatory and safety considerations confront their development (panel 2 ). It is necessary to improve our understanding of the virulence determinants in mammalian systems to be able to attenuate viruses to select appropriate and safe vaccine strains that can generate broad crossreactivity. National and international authorities must urgently confront regulatory issues to allow production and clinical assessment of newly generated virus vaccine candidates. There are important observations from our clinical experience with vaccines for pandemic influenza (panel 3 ). Despite increased reactogenicity, the greater immunogenicity of whole-virus vaccines could be beneficial in a pandemic. Vaccines containing avian H5 and H9 haemagglutinin seem to be less immunogenic in human beings than vaccines based on H1 and H2 haemagglutinin. Whether this is a general event associated with avian subtypes is at present unclear. Enhancement with MF59 or alum salts may provide best antigen use and enhance immunogenicity. Overall, so far, clinical trials of avian influenza vaccine candidates have given disappointing results. It remains to be seen how plasmid-derived reverse-genetics influenza vaccines will perform once regulatory hurdles have been overcome.
Panel 2. Avian influenza virus vaccine development.
Surveillance
Need for robust surveillance programmes in human and animal populations and sharing of information between animal and human surveillance systems Surveillance information to be open and shared in a timely fashion to assess potential threats Potential pandemic strains come from animal reservoir.
Selection of a vaccine strain
Improved understanding of the antigenic and molecular associations between potential pandemic strains of same subtype Improved understanding of immunogenicity against drifted avian influenza strains is required as the ability to generate broad crossprotective immunity is desirable in vaccine candidate.
Manufacturing of vaccine
Intellectual property rights of attenuated viruses produced by reverse genetics must be addressed in advance because licences for commercial use may be required Vaccine virus candidate needs to be able to grow well in eggs (or approved cell culture) to improve ability to respond rapidly to emerging threat Improved understanding of virulence determinants in mammalian models to be able to attenuate viruses used for vaccine manufacture Safety of virus handling for workers involved in preparation of vaccine
Regulatory issues
To assess and approve mammalian cell lines of human vaccine quality Ensure that reagents from animal sources are transmissible spongiform encephalopathies compliant “Reverse genetics” generated viruses are labelled as genetically modified organisms—implications for national and local regulatory authorities Clinical assessment of vaccines derived by reverse genetics Ability to organise antigenicity studies rapidly in response to emerging threat may require prepared approved protocols that can be readily adapted.
Immunogenicity
Improvement in assessment of antibody responses to avian influenza vaccines to establish licensing criteria Standardisation of assays for detection of neutralising antibody to avian influenza Establish correlates of immune protection of neutralising antibody
Panel 3. Clinical findings with pandemic vaccine candidates against human and avian influenza subtypes.
Whole-virus vaccine more immunogenic than subunit or split-product vaccine in immunological naive populations (H1N1, H9N2) Two doses of vaccine required in immunologically naive populations, the first to prime and the second to boost responses (H1N1, H2N2, H5N3, H9N2)
In primed populations, a single dose of vaccine can potentially induce responses associated with protection (H1N1, H5N3, H9N2) Addition of adjuvants such as MF59 and aluminium salts have the potential to significantly enhance immunogenicity and spare antigen use (H5N3, H9N2 and H2N2)
Avian haemagglutinin (H5 and to a lesser extent H9) seems to be less immunogenic in people than H1 and H2 Assessment of antibody responses to avian influenza may require additional serological methods other than the standard haemagglutinininhibition test (H5N3)
Potential crossreactivity with pre-existing antibodies complicates interpretation of immune responses in people (H9N2) Need to develop understanding of improving vaccine candidates to enhance heterosubtypic crossreactivity and protection Need to assess vaccine candidates in advance of pandemic to identify difficulties and establish dosing schedules in different populations
Search strategy and selection criteria
Searches of Medline, PubMed, Current Contents, and references from relevant articles, as well as the extensive files of the authors identified data for this review. Search terms were “avian influenza”, vaccine”, “pandemic influenza”, “H5 influenza”, “H9 influenza”, “H7 influenza”, “influenza vaccines”, “pathogenesis” and “virulence”. English language articles were reviewed.
Acknowledgments
Acknowledgments
Iain Stephenson is supported by an appointment to the Emerging Infectious Diseases Fellowship Programme administered by the Association of Public Health Laboratories and funded by the US Centers for Disease Control and Prevention.
Conflicts of interest
We have no conflicts of interest.
References
- 1.Potter CW. Chronicle of influenza pandemics. In: Nicholson KG, Webster RG, Hay AJ, editors. The textbook of influenza. Blackwell; Oxford: 1998. pp. 3–18. [Google Scholar]
- 2.WHO disease alert. 2004. Confirmed human cases of avian influenza H5N1. http://www.who.int/csr/disease/avian_influenza/en/ (accessed June 30, 2004).
- 3.Hien TT, Liem NT, Dung NT. Avian influenza (H5N1) in 10 patients in Vietnam. N Engl J Med. 2004;350:1179–1188. doi: 10.1056/NEJMoa040419. [DOI] [PubMed] [Google Scholar]
- 4.Ruigrok RWH. Structure of influenza A, B and C. In: Nicholson KG, Webster RG, Hay AJ, editors. The textbook of influenza. Blackwell Science; Oxford: 1998. pp. 29–42. [Google Scholar]
- 5.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohm C, Zhou NA, Suss JC, Mackenzie J, Webster RG. Characterisation of a novel influenza haemagglutinin H15: criteria for determination of influenza A subtypes. Virology. 1996;218:253–257. doi: 10.1006/viro.1996.0145. [DOI] [PubMed] [Google Scholar]
- 7.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 8.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 9.Ito T, Suzuki Y, Mitnaul L, Vines A, Kida H, Kawaoka Y. Receptor specificity of influenza A viruses correlate with agglutination of erythrocytes from different animal species. Virology. 1997;227:492–499. doi: 10.1006/viro.1996.8323. [DOI] [PubMed] [Google Scholar]
- 10.Matrosovich M, Tuzikov A, Bovin N. Early alterations of the receptor binding properties of the H1, H2 and H3 avian influenza virus haeamgglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawaoka Y, Krauss S, Webster RG. Avian to human transmission of the PB1 gene of influenza A virus in the 1957 and 1968 pandemic. J Virol. 1989;63:4603–4608. doi: 10.1128/jvi.63.11.4603-4608.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholtisrek C, Hinshow VS, Olsen CW. Influenza in pigs and their role as the intermediate host. In: Nicholson KG, Webster RG, Hay AJ, editors. The textbook of influenza. Blackwell; Oxford: 1998. pp. 137–144. [Google Scholar]
- 13.Matrosovich M, Zhou N, Kawaoka Y, Webster RG. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens and wild aquatic birds have distinguishable properties. J Virol. 1999;73:1146–1155. doi: 10.1128/jvi.73.2.1146-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li KS, Xu KM, Peiris JSM. Characterisation of H9 subtype influenza viruses from the ducks of southern china: a candidate for the next influenza pandemic in humans? J Virol. 2003;77:6988–6994. doi: 10.1128/JVI.77.12.6988-6994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shortridge KF, Stuart-Harris CH. An influenza epicenter? Lancet. 1982;2:812–813. doi: 10.1016/s0140-6736(82)92693-9. [DOI] [PubMed] [Google Scholar]
- 16.Beare AS, Webster RG. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 17.Kurtz J, Manvell RJ, Banks J. Avian influenza isolated from a woman with conjunctivitis. Lancet. 1996;348:901–902. doi: 10.1016/S0140-6736(05)64783-6. [DOI] [PubMed] [Google Scholar]
- 18.Claas ECJ, Osterhaus ADME, van Beek R. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet. 1998;351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 19.Yuen KY, Chan PKS, Peiris MK. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351:467–471. doi: 10.1016/s0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 20.Katz JM, Lim W, Buxton-Bridges C. Antibody responses in individuals infected with avian influenza A (H5N1) viruses and detection of anti-H5 antibody among household and social contacts. J Infect Dis. 1999;180:1763–1770. doi: 10.1086/315137. [DOI] [PubMed] [Google Scholar]
- 21.Bender C, Hall H, Huang J. Characterization of the surface proteins of influenza A H5N1 viruses isolated from humans in 1997–1998. Virology. 1999;254:115–123. doi: 10.1006/viro.1998.9529. [DOI] [PubMed] [Google Scholar]
- 22.Peiris JSM, Yuen KY, Leung CW. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 23.Uyeki TM, Chong YH, Katz JM. Lack of evidence for human-to-human transmission of avian influenza A (H9N2) viruses in Hong Kong, China 1999. Emerg Infect Dis. 2002;8:154–159. doi: 10.3201/eid0802.010148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peiris JSM, Yu WC, Leung CW. Re-emergence of fatal human influenza A subtype H5N1 disease. Lancet. 2004;363:617–619. doi: 10.1016/S0140-6736(04)15595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards L, Nguyen D, Lu X, et al. Antigenic characterization of recent avian influenza A H5N1 viruses isolated from humans. International Congress Series 2004; in press.
- 26.Fouchier RA, Schneeberger, Rozendall FW. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory respiratory distress syndrome. Proc Nat Acad Sci USA. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopmans M, Wilbrink B, Conyn M. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 28.WHO disease alert: 10 December 2003. Influenza A (H9N2) in Hong Kong Special Administrative Region of China. http://www.who.int/csr/disease/avian_influenza/updates/en (accessed July 5, 2004)
- 29.WHO disease alert, 22 January 2004 Avian influenza H5N1 infection in humans: urgent need to eliminate the animal reservoir-update 5. http://www.who.int/csr/disease/avian_influenza/updates/en (accessed July 5, 2004)
- 30.Chotpirayasunondh T, Lochindarat S, Srisan P. Cases of influenza A (H5N1)-Thailand 2004. MMWR Morb Mortal Wkly Rep. 2004;53:101–103. [Google Scholar]
- 31.Avian influenza virus A (H10N7) circulating among humans in Egypt. http://www.promedmail.org Archive number: 20040524.1392.
- 32.Sims LD, Guan Y, Ellis TM. An update on avian influenza in Hong Kong in 2002. Avian Dis. 2003;47:1083–1086. doi: 10.1637/0005-2086-47.s3.1083. [DOI] [PubMed] [Google Scholar]
- 33.Guan Y, Peiris M, Kong KF. H5N1 influenza viruses isolated from geese in Southeastern China: evidence for genetic reassortment and interspecies transmission to ducks. Virology. 2002;292:16–23. doi: 10.1006/viro.2001.1207. [DOI] [PubMed] [Google Scholar]
- 34.Guo YJ, Li J, Cheng X, Wang M, Zhou Y. Discovery of man infected by avian influenza virus. Chin J Exp Clin Virol. 1999;13:105–108. [PubMed] [Google Scholar]
- 35.Eick A, Hu-Primmer J, Rowe T, Masseoud F, Fukuda K, Lim W. Seroprevalence of antibody to H9N2 viruses in poultry workers of Hong Kong. Poster 52. International Conference on Emerging Infectious Disease 2000; July 2000; Atlanta, GA, USA.
- 36.Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the internal genes of H5N1 viruses in Hong Kong. Proc Natl Acad Sci USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A. Influenza A viruses: relationship between H9N2 relationship between H9N2 and H5N1 human isolates. Proc Nat Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexander DJ. Ecology of avian influenza in domestic birds. In: Dodet B, Vicari M, editors. Emergence and control of zoonotic ortho- and paramyxovirus diseases. John Libbey Eurotext; Paris: 2001. pp. p25–p33. [Google Scholar]
- 39.Peiris JSM, Guan Y, Ghose P. Cocirculation of avian H9N2 and human H3N2 viruses in pigs in southern China. In: Osterhaus ADME, Cox NJ, Hampson A, editors. Proceedings of options for the control of influenza IV. Excerpta Medica; Amsterdam: 2001. pp. 195–200. [Google Scholar]
- 40.Ninomiya A, Takda A, Okazaki K, Shortridge K, Kida H. Seropidemiological evidence of avian H4, H5 and H9 influenza viruses transmission to pigs in south eastern China. Vet Microbio. 2002;88:107–114. doi: 10.1016/s0378-1135(02)00105-0. [DOI] [PubMed] [Google Scholar]
- 41.Saito T, Lim W, Suzuki T. Characterisation of a human H9N2 influenza virus isolated in Hong Kong. Vaccine. 2002;20:125–133. doi: 10.1016/s0264-410x(01)00279-1. [DOI] [PubMed] [Google Scholar]
- 42.Chin PS, Hoffmann E, Webby RJ. Molecular evolution of H6 influenza viruses from poultry in south eastern China: prevalence of H6N1 influenza viruses possessing seven A/HongKong/156/97 H5N1- like genes in poultry. J Virol. 2002;76:507–516. doi: 10.1128/JVI.76.2.507-516.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. J Gen Virol. 1999;80:3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- 44.Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the haemagglutinin polypeptide. Virology. 1975;68:440–454. doi: 10.1016/0042-6822(75)90285-8. [DOI] [PubMed] [Google Scholar]
- 45.Bosch FX, Orlich M, Klenk HD, Rott R. Proteolytic cleavage of influenza virus haemagglutinins: primary structure of the connecting peptide between HA1 and HA2 determines proteolytic cleavability and pathogenicity of avian influenza viruses. Virology. 1981;113:725–735. doi: 10.1016/0042-6822(81)90201-4. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Sastre A. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- 47.Goto H, Kawaoka Y. A novel mechanism for the acquisition of virulence by a human influenza A virus. Proc Nat Acad Sci USA. 1998;95:10224–10228. doi: 10.1073/pnas.95.17.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katz J, Lu X, Tumpey T, Smith C, Shaw M, Subbarao K. Molecular correlates of influenza A H5N1 virus pathogenesis. J Virol. 2000;74:10807–10810. doi: 10.1128/jvi.74.22.10807-10810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao P, Watanabe S, Ito T. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zitzow LA, Rowe T, Morken T. Pathogenesis of avian influenza A H5N1 viruses in ferrets. J Virol. 2002;76:4420–4429. doi: 10.1128/JVI.76.9.4420-4429.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hatta M, Gao P, Halfman P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- 53.Lu X, Cho D, Hall H. Pathogenicity and antigenicity of a new influenza A (H5N1) virus isolated from duck meat. J Med Virol. 2003;69:553–559. doi: 10.1002/jmv.10344. [DOI] [PubMed] [Google Scholar]
- 54.Cheung CY, Poon LLM, Lau AS. Induction of proinflammatory cytokines in human macrophages by influenza A H5N1 viruses: a mechanism for the unusual severity of human disease? Lancet. 2002;360:1831–1837. doi: 10.1016/s0140-6736(02)11772-7. [DOI] [PubMed] [Google Scholar]
- 55.Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–954. doi: 10.1038/nm757. [DOI] [PubMed] [Google Scholar]
- 56.Reid AR, Fanning TG, Hultin JV, Taubenberger JK. Origin and evolution of the 1918 Spanish influenza virus heamagglutinin gene. Proc Nat Acad Sci USA. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reid AR, Fanning TG, Janczewski TA, Taubenberger JK. Characterisation of the 1918 Spanish influenza virus neuraminidase gene. Proc Nat Acad Sci USA. 2000;97:6785–6790. doi: 10.1073/pnas.100140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furminger IGS. Vaccine production. In: Nicholson KG, Webster RG, Hay AJ, editors. The textbook of influenza. Blackwell; Oxford: 1998. pp. 325–332. [Google Scholar]
- 59.Meiklejohn G, Eickhoff TC, Graves P. Antigenic drift and efficacy of influenza virus vaccines. J Infect Dis. 1978;138:618–624. doi: 10.1093/infdis/138.5.618. [DOI] [PubMed] [Google Scholar]
- 60.Nichol KL. Efficacy/clinical effectiveness of inactivated influenza virus vaccines in adults. In: Nicholson KG, Webster RG, Hay AJ, editors. The textbook of influenza. Blackwell; Oxford: 1998. pp. 358–372. [Google Scholar]
- 61.Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull. 1979;35:69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
- 62.Delem A, Jovanovic D. Correlation between rate of infection and pre-existing titer of serum antibody as determined by single radial haemolysis during an epidemic of influenza A/Victoria/3/75. J Infect Dis. 1978;137:194–199. doi: 10.1093/infdis/137.2.194. [DOI] [PubMed] [Google Scholar]
- 63.Circulaire no 96–0661: 1–22. Committee for proprietary medicinal products; London: September 1996. Note for guidance on harmonisation of requirements for influenza vaccines, CPMP/BWP/214/96. [Google Scholar]
- 64.Kistner O, Barrett PN, Mundt W. Development of a Vero cell-derived influenza whole virus vaccine. Dev Biol Stand. 1999;98:101–110. [PubMed] [Google Scholar]
- 65.Medical Research Council Antibody responses and reactogenicity of graded doses of inactivated influenza A/New Jersey.76 whole virus vaccine in humans. J Infect Dis. 1977;136:S475–S483. doi: 10.1093/infdis/136.supplement_3.s475. [DOI] [PubMed] [Google Scholar]
- 66.Parkman PD, Hopps HE, Rastogi SC. Summary of influenza virus vaccine trials in adults. J Infect Dis. 1977;136:S722–S730. doi: 10.1093/infdis/136.supplement_3.s722. [DOI] [PubMed] [Google Scholar]
- 67.Wright PF, Thompson J, Vaughan WK. Trials of influenza A/New Jersey/76 virus vaccine in normal children: an overview of age related antigenicity and reactogenicity. J Infect Dis. 1977;136:S731–S741. doi: 10.1093/infdis/136.supplement_3.s731. [DOI] [PubMed] [Google Scholar]
- 68.Nicholson KG, Tyrrell DA, Harrison P. Clinical studies of monovalent inactivated whole virus and subunit A/USSR/77 (H1N1) vaccine: serological responses and clinical reactions. J Biol Stand. 1979;7:123–136. doi: 10.1016/s0092-1157(79)80044-x. [DOI] [PubMed] [Google Scholar]
- 69.Jennings R, Potter CW, Massey PMO. Responses of volunteers to inactivated influenza virus vaccines. J Hyg. 1981;86:1–16. doi: 10.1017/s0022172400068698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davenport FM, Henessey AV, Askin FB. Lack of adjuvant effect of AlPO4 on purified influenza virus haemagglutinin in man. J Immunol. 1968;100:1139–1140. [PubMed] [Google Scholar]
- 71.Hehme N, Engelmann H, Kunzel W. Pandemic preparedness: lessons learnt from H2N2 and H9N2 candidate vaccines. Med Microbiol Immunol. 2002;191:203–208. doi: 10.1007/s00430-002-0147-9. [DOI] [PubMed] [Google Scholar]
- 72.Takada A, Kuboki N, Okazaki K. Avirulent avian influenza virus as a vaccine strain against a potential human pandemic. J Virol. 1999;73:8303–8307. doi: 10.1128/jvi.73.10.8303-8307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wood JM, Major D, Daly J. Vaccine against H5N1 influenza. Vaccine. 2000;18:579–580. doi: 10.1016/s0264-410x(99)00268-6. [DOI] [PubMed] [Google Scholar]
- 74.Rimmelzwann GF, Class EJ, van Amerongen G, de Jong JC, Osterhaus ADME. ISCOM vaccine induced protection against a lethal challenge with a human H5N1 influenza virus. Vaccine. 1999;17:1355–1358. doi: 10.1016/s0264-410x(98)00390-9. [DOI] [PubMed] [Google Scholar]
- 75.Nicholson KG, Colegate AC, Podda A. Safety and antigenicity of nonadjuvanted and MF59- adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomised trial of two potential vaccines against H5N1 influenza. Lancet. 2001;357:1937–1943. doi: 10.1016/S0140-6736(00)05066-2. [DOI] [PubMed] [Google Scholar]
- 76.Stephenson I, Nicholson KG, Colegate A. Boosting immunity to influenza H5N1 with MF59- adjuvanted H5N3 A/Duck/Singapore/97 vaccine in a primed human population. Vaccine. 2003;21:1687–1693. doi: 10.1016/s0264-410x(02)00632-1. [DOI] [PubMed] [Google Scholar]
- 77.Leese J, Tamblyn SE. Pandemic planning. In: Nicholson KG, Webster RG, Hay AJ, editors. The textbook of influenza. Blackwell; Oxford: 1998. pp. 53–58. [Google Scholar]
- 78.Profeta ML, Palladino G. Serological evidence of human infections with avian influenza viruses. Arch Virol. 1986;90:355–360. doi: 10.1007/BF01317384. [DOI] [PubMed] [Google Scholar]
- 79.Rowe T, Abernathy RA, Primmer JH. Detection of antibody to avian influenza (H5N1) virus in human sera by using a combination of serological assays. J Clin Micro. 1999;37:937–943. doi: 10.1128/jcm.37.4.937-943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stephenson I, Zambon MC, Wood JM, Nicholson KG. Sialic acid receptor specificity of erythrocytes affects haemagglutinin-inhibition test detection of antibody to avian haemagglutinin. J Med Virol. 2003;70:391–398. doi: 10.1002/jmv.10408. [DOI] [PubMed] [Google Scholar]
- 81.Wood JM, Melzack D, Newman RW. A single radial haemolysis assay for antibody to H5 haemagglutinin. In: Osterhaus ADME, Cox NJ, Hampson A, editors. Proceedings of options for the control of influenza IV. Excerpta Medica; Amsterdam: 2001. pp. 461–466. [Google Scholar]
- 82.Stephenson I, Wood JM, Nicholson KG, Zambon MC, Charlett A. Detection of anti-H5 responses in human sera by HI using horse erythrocytes following MF59-adjuvanted influenza A/Duck/Singapore/97 vaccine. Virus Research. 2004;103:91–95. doi: 10.1016/j.virusres.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 83.Laky DL, Treanor JJ, Betts RF. Recombinant baculovirus influenza A haemagglutinin vaccines are well tolerated and immunogenic in healthy adults. J Infect Dis. 1996;174:838–841. doi: 10.1093/infdis/174.4.838. [DOI] [PubMed] [Google Scholar]
- 84.Treanor JJ, Betts RF, Smith GE. Evaluation of recombinant haemagglutinin expressed in insect cells as an influenza vaccine in young and elderly adults. J InfectDis. 1996;173:1467–1470. doi: 10.1093/infdis/173.6.1467. [DOI] [PubMed] [Google Scholar]
- 85.Crawford J, Wilkinson B, Vosnesensky A. Baculovirus-derived haemagglutinin vaccines protect against lethal infection by avian H7 and H5 subtypes. Vaccine. 1999;17:2265–2274. doi: 10.1016/s0264-410x(98)00494-0. [DOI] [PubMed] [Google Scholar]
- 86.Treanor JJ, Wilkinson BE, Masseoud M. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine. 2001;19:1732–1737. doi: 10.1016/s0264-410x(00)00395-9. [DOI] [PubMed] [Google Scholar]
- 87.Liu M, Wood JM, Ellis T. Preparation of a standardized, efficacious agricultural H5N3 vaccine by reverse genetics. Virology. 2003;314:580–590. doi: 10.1016/s0042-6822(03)00458-6. [DOI] [PubMed] [Google Scholar]
- 88.Subbarao K, Chen H, Swayne D. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid-based reverse genetics. Virology. 2003;305:192–200. doi: 10.1006/viro.2002.1742. [DOI] [PubMed] [Google Scholar]
- 89.Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. Cross-subtype protection in humans during sequential overlapping and/or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. J Infect Dis. 1985;151:81–88. doi: 10.1093/infdis/151.1.81. [DOI] [PubMed] [Google Scholar]
- 90.Ulmer J. Influenza DNA vaccines. Vaccine. 2002;20:S74–S76. doi: 10.1016/s0264-410x(02)00136-6. [DOI] [PubMed] [Google Scholar]
- 91.Kodihalli S, Kobasa DL, Webster RG. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine. 2000;18:2592–2599. doi: 10.1016/s0264-410x(99)00485-5. [DOI] [PubMed] [Google Scholar]
- 92.Bright RA, Ross TM, Subbarao K, Robinson HL, Katz JM. Impact of glycosylation on the immunogenicity of a DNA based influenza H5 vaccine. Virology. 2003;308:270–278. doi: 10.1016/s0042-6822(03)00008-4. [DOI] [PubMed] [Google Scholar]
- 93.Epstein SL, Tumpey T, Misplon JA. DNA vaccine expressing conserved influenza virus proteins protective against H5N1 challenge infection in mice. Emerg Infect Dis. 2002;8:796–801. doi: 10.3201/eid0808.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sambhara S, Kurichh A, Miranda R. Heterotypic immunity against human influenza A viruses including recently emerged avian H5 and H9 viruses induced by ISCOM vaccine in mice requires both cytotoxic T-lymphocyte and macrophage function. Cell Immunol. 2001;211:143–153. doi: 10.1006/cimm.2001.1835. [DOI] [PubMed] [Google Scholar]
- 95.Rimmelzwaan GF, Nieuwkoop N, Brandenburg A. A randomized, double blind study in young healthy adults comparing cell mediated and humoral immune responses induced by influenza ISCOM vaccines and conventional vaccines. Vaccine. 2000;191:180–187. doi: 10.1016/s0264-410x(00)00310-8. [DOI] [PubMed] [Google Scholar]
- 96.Tumpey T, Renshaw M, Clements JD, Katz JM. Mucosal delivery of inactivated influenza vaccine induces B cell dependent heterosubtypic crossprotection against lethal influenza A H5N1 virus infection. J Virol. 2001;75:5141–5150. doi: 10.1128/JVI.75.11.5141-5150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu X, Renshaw M, Tumpey T, Kelly G, Hu-Primmer J, Katz J. Immunity to influenza A H9N2 viruses induced by infection and vaccination. J Virol. 2001;75:4896–4901. doi: 10.1128/JVI.75.10.4896-4901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen H, Subbarao K, Swayne D. Generation and evaluation of a high-growth reassortant H9N2 influenza A virus as a pandemic vaccine candidate. Vaccine. 2003;21:1974–1979. doi: 10.1016/s0264-410x(02)00809-5. [DOI] [PubMed] [Google Scholar]
- 99.Stephenson I, Nicholson KG, Gluck R. Safety and antigenicity of whole virus and subunit influenza A/Hong Kong/1073/99 (H9N2) vaccine in healthy adults: phase I randomised trial. Lancet. 2003;13:1959–1966. doi: 10.1016/S0140-6736(03)15014-3. [DOI] [PubMed] [Google Scholar]
- 100.Beyer WEP, Palache AM, de Jong JC, Osterhaus ADME. Cold-adapted live influenza vaccine versus inactivated vaccine: systemic vaccine reactions, local and systemic antibody responses and vaccine efficacy: a meta-analysis. Vaccine. 2002;20:1340–1353. doi: 10.1016/s0264-410x(01)00471-6. [DOI] [PubMed] [Google Scholar]
- 101.Cha T, Kao K, Zhao J. Genotypic stability of coldadapted influenza vaccine in an efficacy clinical trial. J Clin Microbiol. 2000;38:839–845. doi: 10.1128/jcm.38.2.839-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li S, Liu C, Klimov A. Recombinant influenza A virus vaccines for the pathogenic human A/Hong Kong/97 (H5N1) viruses. J Infect Dis. 1999;179:1132–1138. doi: 10.1086/314713. [DOI] [PubMed] [Google Scholar]
- 103.Cox RJ, Brokstad, Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand J Immunol. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 104.Belshe RB, Mendelman PM, Treanor JJ. The efficacy of live attenuated cold adapted trivalent intranasal influenza virus vaccine in children. N Eng J Med. 1998;338:1405–1412. doi: 10.1056/NEJM199805143382002. [DOI] [PubMed] [Google Scholar]
- 105.Takada A, Matsushita S, Ninomiya A. Intranasal immunization with formalin-inactivated virus vaccine induces a broad spectrum of heterosubtypic immunity against influenza A virus infection in mice. Vaccine. 2003;21:3212–3218. doi: 10.1016/s0264-410x(03)00234-2. [DOI] [PubMed] [Google Scholar]
- 106.Stephenson I, Nicholson KG. Influenza: vaccination and treatment. Eur Resp J. 2001;17:1–12. doi: 10.1183/09031936.01.00084301. [DOI] [PubMed] [Google Scholar]