Abstract
Background
Severe acute respiratory syndrome (SARS) is a newly recognized infectious disease that caused an outbreak in south China in 2003. The cause of SARS was identified as a novel coronavirus (CoV). The existence of asymptomatic seroconvertors and the detection of the SARS-CoV RNA in plasma during the course of infection all suggest that SARS could, as least theoretically, be transmitted by transfusion. An estimate of the risk of SARS transmission through blood transfusion will contribute to decisions concerning blood safety monitoring and may be useful in the design of strategies to decrease the risk of transfusion-transmitted infections.
Study design and methods
Case onset dates from the 2003 Shenzhen SARS epidemic and investigational results from Taiwan on viremia in humans are used to estimate the number of cases that were viremic throughout the epidemic. Estimates of the asymptomatic-to-clinically confirmed SARS-CoV infection ratio, the proportion of asymptomatic infections reported in a seroprevalence survey in Hongkong, and the population size of Shenzhen are used to infer the SARS-CoV transfusion–transmission risk. Statistical resampling methods are used.
Results
Based on data from Shenzhen, Hongkong and Taiwan, the maximum and mean risk (per million) of SARS-CoV transmission from donors in Shenzhen were estimated as 23.57 (95% CI: 6.83–47.69) and 14.11 (95% CI: 11.00–17.22), respectively. The estimated risk peaked on April 02, 2003.
Conclusions
Although there are currently no confirmed reports of the transmission of SARS-CoV from asymptomatic individuals, recent research data indicate that transfusion-transmitted SARS-CoV is at least theoretically possible. Although the risk is low, with its rapid spread of the disease, appearance of alarmingly high infectivity and high fatality rate, public health authorities need to consider strategies for blood donor recruitment and virus inactivation during an epidemic to further ensure blood safety.
Keywords: Blood donors, SARS-CoV, Transfusion-transmitted infection
1. Introduction and background
New pathogens and antimicrobial-resistant forms of older pathogens continue to emerge, some with the potential for rapid, global spread, and high morbidity and mortality. Three examples of pathogens that are current causes for human health concern are avian influenza viruses, West Nile virus (WNV) and the severe acute respiratory syndrome (SARS) coronavirus [1]. SARS is a newly described respiratory infection with a potential threat to the health of people throughout the world. The etiological agent is a novel coronavirus (CoV), the SARS-associated CoV (SARS-CoV). The first SARS case was identified in Foshan municipality on November 16, 2002, in Guangdong (GD) Province, China. The patterns of spread of SARS suggest droplet and contact transmission. Close proximity of persons and handling of human secretions (respiratory secretions, faeces, and the like) enhance the risk for transmission [2]. Because of its relative high transmissibility and mortality, SARS raised a great concern over the public health. Epidemiological investigations showed that a total of 8422 probable cases, with 916 deaths were reported from 29 countries during the outbreak (data at August 7, 2003), while World Health Organization (WHO) announced that the last chain of human transfusion was broken on July 5, 2003 [3].
Almost every newly emerging human pathogen is of concern for the safety of the blood supply during and after an epidemic crisis. The potential for transmission of SARS-CoV through blood and blood products is unknown. The possibility of viremia before the onset of clinical symptoms and/or after symptom resolution remains an important concern regarding blood safety. Mildly symptomatic or asymptomatic infections can occur as documented by seroconversion in healthcare workers (HCWs) and animal traders [4], [5], [6]. In April 2003, Drosten et al. reported that viral RNA was detected at extremely low concentrations in plasma during the acute symptomatic phase of infection [7]. In 2004, Singapore researchers reported that SARS-CoV can be detected in the blood of infected patients [8], and, also in 2004, Woo et al. investigated the relative rates of non-pneumonic SARS-CoV infection (mildly symptomatic infections, not asymptomatic cases) and SARS-CoV pneumonia [9]. In 2005, Taiwan researchers first reported that the patterns of viremia differ among different SARS patients and that some patients have a more protracted viremia [10]. Therefore, although there are no confirmed reports of the transmission of SARS-CoV from asymptomatic individuals, and we are unaware that SARS-CoV has been transmitted by the transfusion of blood components, the possibility of such transmission during a donor’s viremic phase is at least theoretically possible.
Detection by nucleic acid amplification (NAT) of SARS-CoV in blood specimens from persons acutely infected with SARS-CoV has been reported in a number of patients [7]. Patients with a mild course of SARS recover approximately 10 days after the development of clinical symptoms [12]. Antibodies to SARS-CoV were measured in patients on day 16 of the disease [7], [13], and NAT may therefore be the only way to identify patients with mild symptoms or in the early phase of disease who might nevertheless be viremic [7], [14]. Therefore, persons with SARS could potentially be viremic before the onset of symptoms and/or after symptom resolution. Transmission of SARS-CoV via human cells, tissues, cellular and tissue-based products (HCT/Ps) recovered during these time periods may be possible [15]. Because SARS occurred in 2002 and 2003 suddenly and disappeared quickly, however, data on SARS viremia prior to symptom onset is very rare. Our risk estimate is theoretical and would overestimate the true risk if viremia does not develop before symptom onset.
Using seroepidemiologic data from the outbreak of SARS in Shenzhen, Guangdong Province, China, in 2003, we estimated the temporal trend in the proportion of viremic, asymptomatic individuals throughout this outbreak using a statistical resampling approach [16]. We then scaled these estimates to estimate the risk of transfusion-associated SARS-CoV transmission during the outbreak. We further compared this result with the estimated risk based on data from Taiwan [10], Singapore [6] and Guangdong Center for Disease Control (CDC) [2]. The supplemental data for the former estimation is mainly based on a large epidemiology investigation done by Woo et al. [9] and the latter estimation is based on a relatively small scale investigation (described further below).
2. Materials and methods
2.1. General approach
Our study population was the population of Shenzhen, Guangdong Province, China during the outbreak in 2003. Guangdong is where the first case of SARS was identified, followed by 1511 clinically confirmed cases, including 58 deaths; of the 1511 confirmed cases, 46 lived in Shenzhen [2]. We first estimated how many of the 46 infected individuals with known symptom onset dates were viremic at each time point throughout the outbreak period. This was done using a statistical resampling approach that incorporated these 46 symptom onset dates, an assumed distribution of the length of the time between the onset of viremia and the onset of symptoms, and an estimated distribution of the length of viremia. The assumed distribution of the duration between onset of viremia and symptom onset was derived from data on the incubation period of SARS and the timing of viremia onset relative to symptom onset during this incubation period [10]. The estimated distribution of the duration of viremia was derived using data from an extensive seroprevalence study and mass screening for detection of subclinical and non-pneumonic infection in Hongkong [9]. We assumed that the dates of infection of 46 individuals with known symptom onset dates were similar to those of all Shenzhen residents who became infected that year. We then used the estimate of the number of viremic cases over time to infer the number of asymptomatic, viremic SARS-CoV infections in the population over time by using seroepidemiologic survey results from Guangdong CDC and Hongkong that provided estimates of the proportion of SARS-CoV-infected individuals who are clinically confirmed and of the proportion of asymptomatic SARS-CoV infections. Finally, we used the population size of Shenzhen, Guangdong province (4,670,000, http://www.demographia.com) to scale the estimated number of viremic, asymptomatic SARS-CoV infections in the population to estimate the risk or probability of a SARS-CoV infected blood donation.
2.2. Study population and data collection
Onset dates for the 46 clinically confirmed cases in the 2003 outbreak were obtained from Shenzhen Municipal Hospital. Symptom onset times (in days) of the 46 cases are shown in the pin plot in Fig. 1 , with time t = 0 corresponding to February 9, 2003, the date of the first reported onset, and time t = 88 corresponding to the last reported onset on May 9. The duration of this epidemic was thus considered to be 88 days.
Fig. 1.
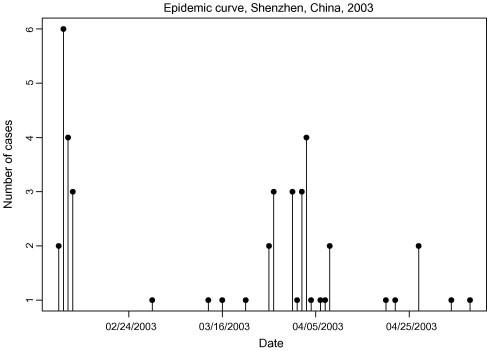
Symptom onset times of 46 individuals with SARS, Shenzhen, Guangdong, 2003.
2.3. Historic data on SARS viremia distribution in humans
Wang et al. reported on the course of SARS-CoV infection and viremia in humans, using results obtained from plasma [10]. The time from infection to symptom onset, the incubation period, is not precisely known but seems to be relatively short (approximately 2–14). WHO reported the incubation period as follows [3]: the maximum likelihood estimate of the mean and variance of the time from infection to onset were 6.37 days and 16.69 days, respectively (95% CI 5.29–7.75); therefore 95% of the patients would experience the onset of symptoms within 14.22 days of infection, based on the assumption that the incubation period followed a gamma distribution. There is approximately a 1–2-day lag between infection with the virus and the detection of virus in the blood [10], [14] so that the duration from onset of viremia to symptom onset is roughly 1–2 days shorter than the incubation period.
Efforts to detect SARS-CoV RNA in plasma during the course of infection found that the highest detection rate, 72%, was found between day 4 and day 11 of illness [10]. Analysis of sequential SARS-CoV load in plasma from six cases revealed different patterns of viremia, with the peak between day 4 and day 8. We calculated the infection dates from the day these patients were diagnosed with SARS using sequential plasma samples tested for SARS-CoV infection. Twenty-nine samples were tested, with a mean of 6.8 days and standard deviation of 3.9 days of viremia.
2.4. Statistical approach
We now describe the statistical method we used; a formal development along the lines of the method used here can be found in [16], where the methods were developed for West Nile virus.
Assume that the rate of blood donation is constant over the course of the epidemic, and assume that potential blood donors have the same risk of infection with SARS-CoV as the general population, which we anticipate may overestimate the risk. This latter assumption was based on the findings of a serosurvey of 400 healthy blood donors conducted in Hongkong [9], in which three seropositives were identified. Finally, we assumed that transfused blood components of SARS-CoV infected blood donors transmit the infection to recipients with 100% efficiency.
2.5. Estimating the proportion of asymptomatic, viremic infections throughout the outbreak
Our strategy was to view the symptom onset times of the cases (N = 46) as anchor times, then to use the information about how viremia relates to symptom onset to estimate the number of cases with viremia at any time t during the outbreak. Then, using this information and information on the asymptomatic or subclinical SARS-CoV infection-to-clinically confirmed SARS ratio (R), the proportion of infected individuals who remain asymptomatic (A), and the population size, we estimated the risk of SARS-CoV transmission by transfusion from a unit of blood donated at time t during the epidemic.
We used Monte Carlo simulation to estimate the number of asymptomatic, viremic individuals at a fixed time t as follows. Because individual case onset times are recorded to the day as discrete times, but the underlying infection process is instead continuous, we first smoothed the observed case onset times by adding a smoothing component. Next, simulated viremia time spans were computed for each case. To do this, the onset of viremia, relative to the case onset time, was chosen by taking a random sample from an assumed distribution for the duration from infection to symptom onset, based on the historic information provided above. The duration of viremia was then chosen by taking a random sample (with replacement) of the duration times from the data obtained in Taiwan [10]. We have assumed in this procedure that the relative timing and duration of viremia for a case is independent of the symptom onset time. Next, to reflect uncertainty in R, a random deviate r from the gamma distribution with mean R = 17.44 and variance Var[R] = 100.9 [9] was generated, and each case’s simulated viremia time was replicated [[r]] + 1 times to reflect the number of viremic individuals in the population over time. (Here, [[r]] denotes the largest integer in r.) Subsequently, to discount viremia times for symptomatic infections and account for uncertainty in A, a random deviate, a, from a beta distribution with mean A = 7.50 × 10−3 and variance Var[A] = 1.86 × 10−5 [10] was generated, and a random proportion a of the replicated viremia times were truncated to their onset times. At the end of each such run, the number of viremia times occurring at time t were counted over an equally spaced grid of 100 time points spanning the epidemic.
This Monte Carlo sampling process was repeated 5000 times, and the resulting counts of the number of asymptomatic, viremic individuals at each time t on the 100-point grid were averaged. Graphing these counts for all grid point times t yielded a curve representing the expected number of asymptomatic, viremic individuals over the course of the epidemic. Finally, this curve was divided by the population size of potential donors to yield the estimated risk or probability of SARS-CoV transfusion transmission. The population size of potential donors was estimated to be 80% of the total population size of Shenzhen, as approximately 80% of the population is at least 18 years of age (http://61.144.227/jingji/tongji/2003yxqk/200411230056.htm, accessed on December 21, 2006). We called the resulting curve the expected risk curve (ERC). We assumed that the possibility of SARS infection for the donors is the same as the whole eligible population.
We computed two summary measures of the ERC to aid interpretation, the maximum value and the mean value over the duration of the epidemic. The maximum is simply the point at which the curve is highest, and the timing of this maximum estimates when the expected transfusion-associated SARS-CoV risk was highest. The mean, computed by dividing the area under the ERC by the duration of the epidemic, provides a measure of the expected transmission risk over the whole course of the epidemic.
Confidence bands for the true risk curve (TRC) around the ERC can be computed in various ways. We used a simultaneous percentile-t approach, the details of which are outlined in [16]. A 95% CI for the true maximum is read from the confidence bands for the TRC, while the 95% CI for the true mean is computed using a percentile-t approach similar to that for the TRC [16].
2.6. Sensitivity analysis
Because of the relatively little available information on the epidemiology of SARS and SARS-CoV infection, we performed a second analyses using different sources of data for key parameters in our modelling approach. In addition to the analysis reported above, in the second analysis we used data drawn from Singapore [6] and Guangdong CDC [2], from which R = 0.22 (Var[R] = 0.0046), A = 0.012 (Var[A] = 0.00014). In the Singapore study, Wilder-Smith et al. [6] reported that of 80 hospital staff, 45 (56%) were positive by SARS-CoV serology. Of the 45 SARS-CoV-positive study participants, 37 (82%) were classified as having pneumonic SARS, 2 (4%) as having subclinical SARS, and 6 (13%) as having asymptomatic SARS-CoV infection. Guangdong CDC found one seropositive adult from 84 healthy adults at the study clinic [2].
All computations were performed with the statistical software S-Plus, version 7 (Insightful Corporation, Seattle, WA, USA) using either built-in functions or functions written by one of the authors (B.J.B.).
3. Results
Five thousand simulations combined to produce the ERC, shown as the dark, solid line in Fig. 2 . The dashed lines are 95th percentile-t simultaneous confidence bands. Also shown in Fig. 2 are 100 randomly selected realizations of the ERC from the 5000 generated. The scale on the left axis is the risk (per million) of SARS-CoV transmission from transfusion of a single unit of blood. For results displayed in Fig. 2, estimation was based on the estimates obtained from Woo et al. [9], R = 17.44 (Var[R] = 100.9), A = 0.75 (Var[A] = 1.8 × 10−5). Given the assumptions outlined in the methods, we estimated the maximum risk (per million) of transmission from transfusion of a single unit as 23.57 (95% CI, 6.83–47.69), occurring on about April 2, 2003, and the mean risk (per million) of transmission over the course of the outbreak as 14.11 (95% CI, 11.00–17.22).
Fig. 2.
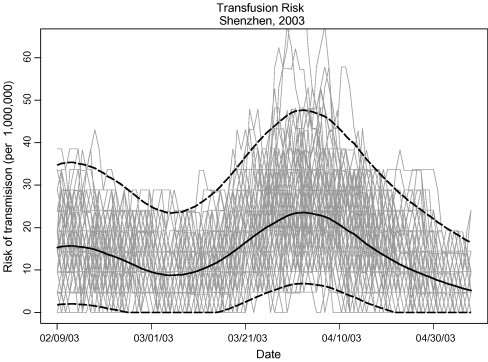
The ERC (solid, dark line) from 5000 simulation runs. Simultaneous 95th percentile-t confidence bands are shown as dashed lines. The left axis gives the scale as the risk of SARS-CoV transmission from transfusion of a single unit of blood, after inferring to the entire Shenzhen population and accounting for symptomatic individuals who would not donate or who would be deferred from donation. The light lines depict 100 sample realizations of the 5000 used to compute the ERC.
Results from the secondary analysis are reported in Table 1 , using the given values of R and A.
Table 1.
Comparison between estimations based on different data sources
| Data source | R | A (%) | Mean risk per million (95% Cl) | Maximum risk per million (95% Cl) | Peak time |
|---|---|---|---|---|---|
| Hongkong [9] and Taiwan [10] | 17.44 | 0.75 | 14.11 (11.00–17.22) | 23.57 (6.83–47.69) | April 2, 2003 |
| Singapore [6] and GD [2] | 0.22 | 1.19 | 0.59 (0.45–0.74) | 0.99 (0–2.05) | April 1, 2003 |
| CDC and Taiwan [10] |
R = ratio of the number of unapparent or subclinical infections to the number of apparent infections.
A = proportion of individuals who remain asymptomatic.
4. Discussion
Although donor screening and testing have nearly eliminated the risk of transfusion-acquired infections attributable to HIV and hepatitis viruses, the potential emergence and spread of other pathogens, particularly those associated with asymptomatic illness, could result in unrecognized transmission through blood transfusion [1], [17]. Our results indicate a small but non-zero risk of SARS-CoV transmission from transfusion of blood components during an epidemic, at least theoretically. Based on data from different published literature, we calculated that during an epidemic of SARS in Shenzhen in 2003, the risk peaked at approximately 23.57 per million donations in late March/early April, with a mean risk over the course of the outbreak of 14.11 per million donations. The calculated risk was highly limited in time, with the near-zero or zero risk among donations before February and after August, during times of no reported community SARS-CoV transmissions in Shenzhen, Guangdong, China. There were 10,766 donations in Shenzhen during the time of the epidemic, so based on our average risk estimate of 14.11 per million donations, the expected number of infected donations is 0.152 (1 in 71,000). Further, based on a simple binomial model, the probability that there was at least one infected donation in the 10,766 donations made is 1 − (1 − 14.11/1,000,000)10,766 = 0.14, or roughly 14%.
The ratio R is a crucial factor in our risk estimation approach. From Table 1, it is clear that our estimates are sensitive to the assumptions about this quantity. Good epidemiologic data strengthen inference, and improvements in the understanding of the epidemiology of SARS will provide opportunities to refine the estimates we report. For blood safety, surveillance should emphasize the existence of subclinical or non-pneumonic SARS-CoV infections, since persons with severe symptoms would be less likely to donate blood.
The comparison between the different estimates reported in Table 1 showed almost the same peak risk time, the 1st or 2nd of April, which is in line with the government report on the outbreak [19]. As the estimate of the timing of the peak in risk is driven by the case onset times, and not really affected by specification of R or A, this result is expected. This is further consistent with the SARS epidemic more broadly, as it was rampant in March, April and May 2003 in the mainland of China [19].
Our estimated mean transmission risk of 14.11 per million may be too high for several reasons. We assumed a rate of 100% transmission from viremic donors, and the true rate is probably lower. There are also uncertainties about the duration of viremia. On the other hand, we used 80% of the total population of Shenzhen (persons aged between 18 yr and 55 yr) as the denominator of our risk estimates, and the true number of potential donors is probably lower; this is expected to underestimate the true risk. Furthermore, there are no population-based seroepidemiological data available to inform specification of asymptomatic rates and the other, relevant parameters used in our modelling approach. While the estimates we used for the various parameters in our modelling are the best available, should there be biases in them, these biases would directly impact our results. As we took care in our analyses to use all available, relevant and published data, it is impossible without further (possibly population-based) estimates to adjust for such unknown biases.
Our sensitivity analysis showed that the values assumed for A and, particularly, for R are critical to the results we obtain. Since data from Singapore were based on a relatively small sample of healthcare workers, and data from Hongkong were based on an extensive seroprevalence study and mass screening for detection of subclinical and non-pneumonic infections, we consider our primary results to be those based on values for A and R obtained from Woo et al. [9].
Even with no confirmed cases of SARS-CoV transmission by blood transfusion, several precautionary measures were implemented by Chinese blood centers in spring 2003 to prevent the potential risk of SARS-CoV transmission through blood [20]. These measures included: (1) new questions were added to the donor questionnaire about predonation contact history with suspected SARS patients or people with contact history with SARS; (2) normal body temperature was added as a criterion for donor qualification; (3) all donors were required to notify the blood collection facilities if fever, cough, or other suspected SARS symptoms occurred within two weeks after donation; and (4) a SARS-CoV antibody testing research project was soon started using available tests on donor samples [20].
Kumar and Humar [21] reported that transplant patients are uniquely predisposed to emerging infections, and this applies to SARS. Lessons learned from West Nile virus [18] and SARS-CoV in transplantation should be applicable to future outbreaks of other emerging infectious diseases [21]. In the meantime, on the basis of current knowledge a return of SARS cannot be ruled out. Particularly, the origin of the virus is assumed to be the civet cat, which could not be quarantined [22]. Furthermore, the existence of asymptomatic SARS-CoV infections and the detection of SARS-CoV RNA in plasma during the course of infection suggest that the transmission of the SARS-CoV by transfusion is at least theoretically possible. Our analyses suggest, however, that this probability is very low during an outbreak similar in magnitude to the one in Shenzhen in 2003, with a maximum estimated risk of 23.57 per million donations. In case SARS does recur, preventive measures considered during the epidemic should include strategies for blood donor recruitment, and the estimates we provide herein may be used in a quantitative evaluation of such measures. The quantification of these effects will likely not be available unless and until SARS recurs.
References
- 1.AuBuchon J.P., Birkmeyer J.D., Busch M.P. Safety of the blood supply in the United States: opportunities and controversies. Ann Intern Med. 1997;127:904–909. doi: 10.7326/0003-4819-127-10-199711150-00009. [DOI] [PubMed] [Google Scholar]
- 2.Yu D., Li H., Xu R. Prevalence of IgG antibody to SARS-associated coronavirus in animal traders – Guangdong Province, China, 2003. MMWR Morb Mortal Wkly Rep. 2003;52:986–987. [PubMed] [Google Scholar]
- 3.Consensus Document on the Epidemiology of SARS, WHO/CDS/CSR/2003.11. <http://www.who.int/csr/sars/WHOconsensus.pdf>. [PubMed]
- 4.Ip M., Chan P.K.S., Lee N. Seroprevalence of antibody to severe acute respiratory syndrome (SARS)-associated coronavirus among health care workers in SARS and non-SARS medical wards. Clin Infect Dis. 2004;38(12):e116–e118. doi: 10.1086/421019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho K.Y., Singh K.S., Habib A.G. Mild illness associated with severe acute respiratory syndrome coronavirus infection: lessons from a prospective seroepidemiologic study of health-care workers in a teaching hospital in Singapore. J Infect Dis. 2004;189(4):642–647. doi: 10.1086/381558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilder-Smith A., Teleman M.D., Heng B.H. Asymptomatic SARS Coronavirus Infection among healthcare workers, Singapore. Emerg Infect Dis. 2005;11(7):1142–1145. doi: 10.3201/eid1107.041165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten C., Gunther S., Wolfgang Preiser. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Eng J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 8.Ng L.F.P., Wong M., Koh S. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J Clin Microbiol. 2004;42(1):347–350. doi: 10.1128/JCM.42.1.347-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo P.C.Y., Lau S.K.P., Tsio H. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet. 2004;363:841–845. doi: 10.1016/S0140-6736(04)15729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Wei-Kung, Fang Chi-Tai, Chen Hui-Ling. Detection of severe acute respiratory syndrome coronavirus RNA in plasma during the course of infection. J Clin Microbiol. 2005;43(2):962–965. doi: 10.1128/JCM.43.2.962-965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avendano M., Derkach P., Swan S. Clinical course and management of SARS in health care workers in Toronto: a case series. CMAJ. 2003;168:1649–1660. [PMC free article] [PubMed] [Google Scholar]
- 13.Enserink M., Vogel G. Infectious diseases: deferring competition, global net closes in on SARS. Science. 2003;300:224–225. doi: 10.1126/science.300.5617.224. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. Case definitions for surveillance of SARS. Geneva, 1 May 2003 (Online). <http://www.who.int/csr/sars/casedefinition/en/>; 2003 [accessed 29 March 2007].
- 15.Guidance for Industry. Eligibility determination for donors of human cells, tissues, and cellular and tissue-based products (HCT/Ps). US DHS, FDS, CBER; May 2004. [PubMed]
- 16.Biggerstaff B.J., Petersen L.R. Estimated risk of West Nile virus transmission through blood transfusion during an epidemic in Queens, New York City. Transfusion. 2002:1019–1026. doi: 10.1046/j.1537-2995.2002.00167.x. [DOI] [PubMed] [Google Scholar]
- 17.Schreiber G.B., Busch M.P., Kleinman S.H. The risk of transfusion-transmitted viral infections. The retrovirus epidemiology donor study. N Engl J Med. 1996;334:1685–1690. doi: 10.1056/NEJM199606273342601. [DOI] [PubMed] [Google Scholar]
- 18.Iwamoto M., Jeringan D.B., Guasch A. Transmission of West Nile Virus from an organ donor to four transplant recipients. N Engl J Med. 2003;348:2196–2203. doi: 10.1056/NEJMoa022987. [DOI] [PubMed] [Google Scholar]
- 19.Nie Q.H., Luo X.D., Zhang J.Z. Current status of severe acute respiratory syndrome in China. World J Gastroenterol. 2003;9(8):1635–1645. doi: 10.3748/wjg.v9.i8.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G.J., Qiu Y., Zhang P. Epidemiological study of SARS CoV among volunteer blood donors in Beijing. Chin J Transfus. 2003;16:223–225. [Google Scholar]
- 21.Kumar D., Humar A. Emerging viral infections in transplant recipients. Curr Opin Infect Dis. 2005;18(4):337–341. doi: 10.1097/01.qco.0000172697.44784.ff. [DOI] [PubMed] [Google Scholar]
- 22.Abbott A. Pet theory comes to the fore in fight against SARS. Nature. 2003;423:576. doi: 10.1038/423576b. [DOI] [PMC free article] [PubMed] [Google Scholar]


