Abstract
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 strain EDL933 encodes the single chromosomal 9-O-acetylesterase NanS, and several copies of prophage-encoded 9-O-acetylesterases (NanS-p). These enzymes have recently been shown to cleave 5-N-acetyl-9-O-acetyl neuraminic acid (Neu5,9Ac2) to yield de-O-acetylated Neu5Ac, the latter of which may serve as a carbon and/or nitrogen source.
In the current study, we investigated the NanS- and NanS-p-mediated digestion of synthetic O-acetylated neuraminic acids and bovine submaxillary glands mucin (BSM)-derived O-acetylneuraminic acids by high-performance thin-layer chromatography (HPTLC) and nano electrospray ionization mass spectrometry (nanoESI MS). Initial HPTLC analyses showed the expected activity of NanS and NanS-p variants for Neu5,9Ac2. However, all tested enzymes were unable to de-O-acetylate 5-N-acetyl-4-O-acetylneuraminic acid (Neu5,4 Ac2) in our test system. The nanoESI MS analysis of neuraminic acids after treatment of BSM with NanS-p gave evidence that NanS-p variants of EHEC O157:H7 strain EDL933 cleave off O-acetyl groups from mono-, di-, and tri-O-acetylated Neu5Ac and N-glycolylneuraminic acid (Neu5Gc), regardless of the carbon positions C7, C8 or C9 of the acetate esters. This enzyme activity leads to neuraminidase-accessible Neu5Ac and Neu5Gc on mucin glycans.
Moreover, we could demonstrate by HPTLC analyses that recombinant Bacteroides thetaiotaomicron sialidase (BTSA-His) was able to cleave Neu5Ac and Neu5,9Ac2 from BSM and that the combination of BTSA-His with both NanS-His and NanS-p-His derivatives enhanced the release of de-O-acetylated core Neu5Ac and Neu5Gc from mammalian mucin O-glycans.
Growth experiments with EHEC wildtype strain EDL933, its nanS and nanS/nanS-p1a-p7 mutant and exogenous BTSA-His in BSM demonstrated that the presence of BTSA-His enhanced growth of EDL933 and the nanS deletion mutant but not the nanS/nanS-p1a-p7 mutant.
Thus, we hypothesize that the expression of sialic acid O-acetylesterases with a broad specificity could be an advantage in competition with the gut microbiota for nutrients and facilitate EHEC colonization in the human large intestine.
Keywords: O-acety-l-esterase, Escherichia coli O157:H7, Bovine submaxillary gland mucin, De-O-acetylation, Neu5A, Neu5Ac2, NanS-p, NanS
1. Introduction
Infections with enterohemorrhagic Escherichia coli (EHEC) can cause hemorrhagic colitis (HC) and the life-threatening hemolytic-uremic syndrome (HUS) (Nataro and Kaper, 1998). One of the natural reservoirs of EHEC strains is the intestinal tract of ruminants, especially cattle, where they can survive in the presence of mucus-derived carbohydrates (Aperce et al., 2014; Bertin et al., 2013; Fox et al., 2009). EHEC O157:H7 strains have caused numerous cases and outbreaks during the last decades (Page and Liles, 2013) Most strains of this serotype express Shiga toxin (Stx) 1 and/or Stx2 with their different variants and the locus of enterocyte effacement (LEE), encoding a type III secretion system (Scheutz et al., 2011; Stevens and Frankel, 2014). The first reported EHEC O157:H7 outbreak occurred in 1982 where at least 47 people developed diarrhea in Michigan and Oregon after consuming contaminated meat in a fast-food restaurant (Riley et al., 1983). One of the strains that have been isolated during this outbreak was EDL933 (O’Brien et al., 1993). Up to now, this is one of the best-characterized EHEC O157:H7 strains worldwide and was therefore used in the current study.
The primary site of infection in the human body is the large intestine, where EHEC cells adhere to the intestinal epithelium, damage this cellular barrier by the action of type III effector proteins and translocate Stx into the blood system (Nguyen and Sperandio, 2012). Infecting EHEC strains may evade host defence, gain access to growth substrates and overcome colonization resistance mechanisms (Stecher and Hardt, 2011). They replicate in the colon during the course of infection, but up to now, the mechanisms of competition with the microbiota and carbon nutrition are only poorly understood. An important carbon source for EHEC are breakdown products of the gastrointestinal mucus that are produced by the host in high amounts each day (Conway et al., 2004). Many commensal gut microorganisms attain required C- and N-sources from mucin glycoproteins (Johansson et al., 2009, 2011; Marcobal et al., 2013). Endowed with various glycoside hydrolases and exploiting synergistic mechanisms between them they are well adapted to grow in the mucus environment (Marcobal et al., 2013).
A number of EHEC O157:H7 virulence factors seems to play primary roles in penetration of mucus or adhesion to the mucus-coated colonic epithelium such as the mucinase StcE (Hews et al., 2017) or the H7 flagella (Erdem et al., 2007). To get access to the limited nutrients in large intestine, replication of EHEC strains depends on mechanisms to compete at least for one of the mucus-derived carbohydrates or to be otherwise metabolically flexible (Conway and Cohen, 2015). Commensal non-pathogenic E. coli represent presumably the major competitors in the gut, because they exhibit a similar genetic background as EHEC do. Thus, the expression of additional genes involved in sugar catabolism, which are located on genetic mobile elements in EHEC, is imperative for the assertiveness of EHEC towards commensal E. coli. Mucus-derived sugars as fucose, galactose, N-acetylglucosamine, N-acetylgalactosamine or N-acetylneuraminic acid (Neu5Ac) can be utilized by EHEC as well as by commensal E. coli (Aperce et al., 2014; Chang et al., 2004; Fabich et al., 2008).
Neu5Ac is an α-keto sugar with a nine-carbon backbone, typically found terminally linked to glycan chains of mammalian mucins. Neu5Ac is highly abundant in glycans of the large intestine (Robbe et al., 2003), where E. coli preferentially colonizes, compared to the small intestine (Barnett Foster, 2013; Torres et al., 2005). N-glycolylneuraminic acid (Neu5Gc) is the hydroxylated derivative of Neu5Ac carrying an OH-group in the N-acetyl chain at carbon C5. Neu5Gc is generated by the enzyme CMP-Neu5Ac-hydroxylase, which is expressed as functional enzyme in all mammals except humans, who produce an inactive enzyme due a deletion in the corresponding chromosome (Peri et al., 2018). Although humans lack a functioning CMP-Neu5Ac-hydroxylase, Neu5Gc has been found in low quantities in human mucins most likely due to catabolic incorporation from nutritional Neu5Gc derived from red meat (Varki and Schauer, 2009). Furthermore, neuraminic acids exhibit a large heterogeneity in the gut with respect to structural modifications of their hydroxyl groups. A very common modification is the O-acetylation with up to four O-acetyl residues at carbon atoms C4, C7, C8 and C9 (Varki and Schauer, 2009; Vimr, 2013). Neu5Ac is the major sialic acid present in mucin, followed by the mono-O-acetylated Neu5Ac-variants Neu5,9Ac2, Neu5,7Ac2 and Neu5,8Ac2. Less common are the di-O-acetylated Neu5Ac-species Neu5,8,9Ac3 and Neu5,7,9Ac3 (Robbe et al., 2003).
Nonpathogenic and pathogenic E. coli strains can encode the three chromosomal operons nanATEK-yhcH, nanCMS (formerly yjhATS), and yjhBC, which are involved in sialic acid metabolism. Neu5Ac inactivates the repressor NanR, whereby the nan-operons are activated and sialic acids are catabolized (Rangarajan et al., 2011). While Neu5Ac is directly taken up by the transporter NanT, O-acetylated sialic acids have to be deacetylated first (Steenbergen et al., 2009). NanS is an N-acetyl-9-O-acetylneuraminic acid esterase, which deacetylates mono-O-acetylated Neu5,9Ac2 to Neu5Ac, which is then transported by NanT and utilized as an energy source. It has been shown that NanS is also able to deacetylate mono-O-acetylated Neu5,8Ac2, but not the Neu5,7Ac2 or Neu5,4 Ac2 isomeric structures (Steenbergen et al., 2009).
Expressing NanS, commensal non-pathogenic E. coli are able to utilize O-acetylated sialic acids as carbon sources. NanS belongs to the large family of SGNH serine proteases, which are not only found in commensal bacteria, but also in a number of pathogens that produce sialate O-acetylesterases (Corfield et al., 1992, 1993; Phansopa et al., 2015). Vimr (2013) have extensively reviewed the bacterial sialometabolism and reported that many enteropathogenic E. coli strains harbor further nanS-homologs.
The pathogenic E. coli O157:H7 strain EDL933 encodes several nanS-homologous alleles (nanS-p), which are localized in prophage genomes in the region of late gene expression (Saile et al., 2016). Recently, the enzymatic characteristics of recombinantly expressed NanS-p1, NanS-p2, and NanS-p4 of strain EDL933 were determined and an N-acetyl-9-O-acetylneuraminic acid esterase activity was detected for the three enzymes. The three variants were investigated as representatives of the EDL933 NanS-p and were selected due to phylogenetic reasons (Saile et al., 2016). For analysis of the role of NanS-p, several nanS and nanS-p deletion mutants have been constructed up to a supermutant that did not contain any nanS or nanS-p genes, and their role in substrate utilization has been demonstrated (Saile et al., 2016). Compared to nanS, the nanS-p genes harbour additional nucleotides downstream of the SGNH hydrolase-like domain and a DUF1737 domain. The function of the DUF1737 domain is still unknown (Rangel et al., 2016; Saile et al., 2016).
In the human large intestine, commensal mucin-degrading species of the phyla Bacteroidetes, Firmicutes, Actinobacteria and Verrucomicrobia can cleave bound sialic acids from glycans by intrinsic sialidases, providing free nutritional sialic acids for sialidase-negative E. coli (Robinson et al., 2017; Tailford et al., 2015; Vimr et al., 2004). Sialidases are substrate-specific and the decoration of sialic acids with one or more O-acetyl residues prevents sialidases from enzymatic cleavage. The activity of sialidases increases upon removal of O-acetyl groups from sialic acids by sialate O-acetylesterases (Phansopa et al., 2015). B. thetaiotaomicron, for example, represents a commensal of the gut, that produces a number of glycoside hydrolases such as the B. thetaiotaomicron sialidase BTSA (Park et al., 2013), enabling this microorganism to release terminally bound sialic acids from mucin.
The aim of the current study was to further characterize the enzymatic properties of recombinant NanS and NanS-p derivatives to cleave O-acetyl residues from O-acetylated neuraminic acids and BSM, also in combination with recombinant BTSA. Moreover, growth experiments with EHEC wildtype strains and nanS-p mutants were performed to study the influence of the enzymes in vitro.
2. Materials and methods
2.1. Bacterial strains
Bacterial strains used in this study are listed in Table 1 .
Table 1.
Characteristics of bacterial strains used in this study.
| Bacterial strains | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli O157:H7 EDL933 | nanS+,nanS-p1a+ to nanS-p7+ | (O’Brien et al., 1993) |
| E. coli O157:H7 EDL933ΔnanS | Deletion of nanS | (Saile et al., 2016) |
| E. coli O157:H7 EDL933ΔnanSΔnanS-p1a-p7 | Deletion of nanS, nanS-p1a, nanS-p2, nanS-p3, nanS-p4, nanS-p5, nanS-p6, nanS-p7 | (Saile et al., 2016) |
| E. coli BL21(DE3) | F– ompT hsdSB(rB−,mB−) gal dcm (DE3) | (Studier and Moffatt, 1986) |
| E. coli BL21(DE3)/pET-22b(+) | contains plasmid pET-22b(+) (Novagene, Merck) | (Nübling et al., 2014) |
| E. coli BL21(DE3)/pET-Z1466-his | Recombinant 933 Wp42 (syn. NanS-p1) | (Nübling et al., 2014) |
| E. coli BL21(DE3)/pET-nanS-p2-his | Recombinant NanS-p2 | (Saile et al., 2016) |
| E. coli BL21(DE3)/pET-nanS-p4-his | Recombinant NanS-p4 | (Saile et al., 2016) |
| E. coli BL21(DE3)/pET-BT_0455-his | Recombinant BTSA (without signal sequence) | (Saile et al., 2018) |
| E. coli BL21(DE3)/pET-nanS-his | Recombinant NanS |
This study |
2.2. Cloning and expression techniques
E. coli O157:H7 strain EDL933 was grown on LB agar (1% (w/v) tryptone, 0.5% (w/v) yeast extract, 1% (w/v) NaCl, pH 7) at 37 °C with shaking at 180 rpm. Amplification of nanS from EDL933 was performed by designing primers Z5905_NdeI_for (5’-cccCATATGAACGCAATAATATCGCCTG-3’) and Z5905_XhoI_rev (5’-aaaCTCGAGCCTTTCGCGCCAAAACTGC-3’) using the NCBI reference sequence NZ_CP015855 (Saile et al., 2016). PCR conditions were as follows: after initial DNA denaturation for 5 min at 95 °C, 30 cycles of each 30 s 94 °C, 30 s 66 °C, and 120 s 72 °C were carried out. After a final extension step of 5 min at 72 °C, the PCR products were cooled down to 4 °C and further processed. The PCR product was then digested with the restriction enzymes NdeI and XhoI (Thermo Scientific, USA), ligated into expression vector pET-22b(+), transformed into laboratory E. coli strain BL21(DE3), and correct insertion of the PCR product was confirmed by DNA sequencing as described previously (Nübling et al., 2014).
Expression, verification of protein purity and determination of the concentrations of BTSA-His, NanS-His, NanS-p1-His, NanS-p2-His and NanS-p4-His by SDS-PAGE and Bradford staining were performed as recently described (Nübling et al., 2014; Saile et al., 2016, 2018).
2.3. Growth of EDL933 and its O-acetyl esterase knockout derivatives in bovine submaxillary gland mucin supplemented with BTSA-His
E. coli O157:H7 strain EDL933, EDL933ΔnanS and EDL933ΔnanSΔnanS-p1a-p7 were incubated for 24 h at 37 °C and 180 rpm in M9 minimal medium (Sambrook and Russell, 2001) containing 0.8% (w/v) mucin from bovine submaxillary gland (BSM, cat. 84195-52-8, Merck Millipore, Germany). The initial optical density (OD600) was 0.1 for each culture. Liquid culture of EDL933ΔnanSΔnanS-p1a-p7, supplemented with 4 μg/ml NanS-p4-His, was used as a control.
To compare the effect of recombinant BTSA-His on growth of EHEC strains, the cultures were grown in parallel, whereby one of the parallel culture each contained 3.86 μg/ml BTSA-His. All experiments were performed in triplicate with two technical replicates, respectively. Statistical significance was determined using Student’s t test.
2.4. Enzymatic digestion of BSM
One mg/ml of BSM dissolved in 50 mM Tris-HCl pH 7.5 was incubated with 5 μg NanS-His, NanS-p1-His, NanS-p2-His or NanS-p4-His for 2 h at 25 °C. Then another 5 μg of enzyme was added (10 μg of NanS-p equates to 0.15 μM; 10 μg of NanS equates to 0.27 μM) and incubation was prolonged for 2 h. In addition, 1 mg/ml BSM was incubated with 0.96 μg/mg (0.02 μM) BTSA-His for 4 h at 25 °C. Digestions of BSM with NanS-His/NanS-p-His and BTSA-His were carried out similar to the BSM digestion with NanS-His/NanS-p-His but contained in addition 0.96 μg/mg BTSA-His. BSM was incubated for 4 h at 25 °C without enzyme and used as negative control. All reactions were stopped after the incubation time by heat inactivation at 95 °C for 10 min.
2.5. Enzymatic de-O-acetylation of Neu5,9Ac2 and Neu5,4 Ac2
One hundred μg of mono-O-acetylated Neu5,9Ac2 (provided by Wolfgang Fessner and Ning He, Technical University Darmstadt, Darmstadt, Germany) or mono-O-acetylated Neu5,4 Ac2 (Applied Biotech GmbH, Austria, no. S-4-S-10) were dissolved in 100 μl of 50 mM Tris-HCl pH 7.5 and incubated each with 5 μg NanS-His, NanS-p1-His, NanS-p2-His or NanS-p4-His at 25 °C for 1 h. Another 5 μg of enzyme (in total 1.5 μM of NanS-p and 2.7 μM of NanS) were added to each reaction and incubation was prolonged for 1 h. After incubation the enzymes were heat inactivated at 95 °C for 10 min. As a negative control 100 μg of sialic acids (Neu5Ac, Neu5,9Ac2, Neu5,4 Ac2) were incubated for 2 h at 25 °C without enzymes.
2.6. High-performance thin-layer chromatography (HPTLC) analysis of sialic acids
HPTLC analysis was performed with glass-backed HPTLC plates (20 cm x 10 cm) coated with silica gel 60 F254 (Merck, Germany). Three to 6 μl of heat-inactivated enzymatic digestions and Neu5Ac, Neu5,9Ac2 and Neu5,4 Ac2 standards were applied onto plates as a fine 8 mm wide streak using the Automatic TLC Sampler 4 (ATS4, CAMAG, Switzerland) equipped with a 100 μl Hamilton® syringe. After sample application the plates were developed in a twin trough developing chamber (CAMAG) filled with n-butanol:H2O:acetic acid (5:2.5:2.5, each by vol.) as a mobile phase. Plates were first air-dried and then dipped in diphenylamine-aniline-phosphoric acid (DAPA, Carl Roth, Germany) reagent for one second and kept on a TLC plate heater (CAMAG, Switzerland) at 110 °C until colored bands became apparent. Visualization was done under UV light (366 nm) using a TLC visualizer (CAMAG). The chromatograms were processed with winCATS software (CAMAG).
2.7. Sample preparation for nanoESI MS
BSM samples were incubated with NanS-His, NanS-p1-His, NanS-p2-His, NanS-p4-His, or without enzyme followed by treatment with 12.5% (v/v) acetic acid for 2 h at 95 °C. After chilling to ambient temperature the samples were dried in vacuo and dissolved in 40% methanol containing 0.5% formic acid for MS analysis. Samples treated with BTSA-His (vide supra) were spiked with an equal volume of methanol an incubated on ice for 30 min. After centrifugation at 15,800 x g the supernatants were analyzed by nanoESI MS.
2.8. nanoESI mass spectrometry analysis
Mass spectrometry (MS) analyses were performed using a SYNAPT G2-S mass spectrometer (Waters, Manchester, UK) equipped with a Z-spray source in the positive ion sensitivity mode. Typical source settings were: temperature 80 °C, capillary voltage 0.8 kV, sampling cone voltage 20 V, and offset voltage 50 V (Steil et al., 2018).
3. Results
3.1. Recombinant EHEC O-acetylesterases NanS, NanS-p1, NanS-p2 and NanS-p4 deacetylate mono-O-acetylated Neu5,9Ac2, but not Neu5,4 Ac2
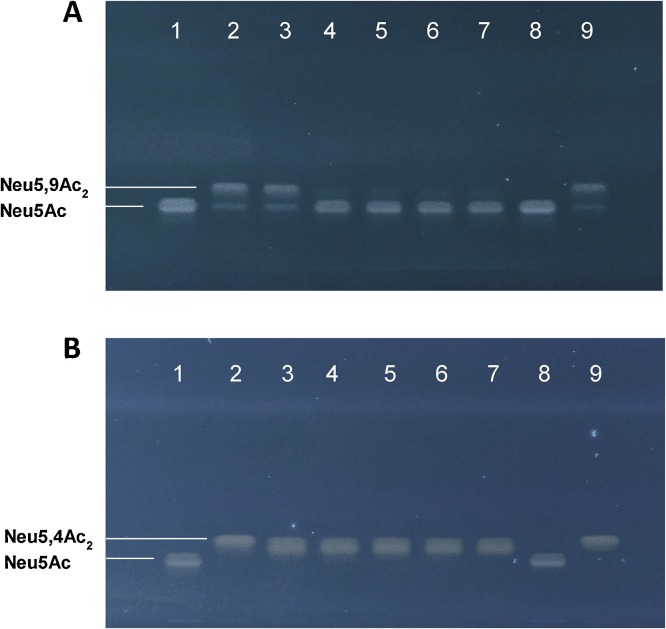
The native O-acetylesterases NanS and NanS-p have been shown to remove the 9-O-acetyl group from Neu5,9Ac2 (Nübling et al., 2014; Saile et al., 2016). In order to explore the activity and specificity of purified NanS and NanS-p, we cloned His-tagged nanS, nanS-p1, nanS-p2, and nanS-p4 and expressed the enzymes in laboratory E. coli strain BL21 (DE3) following IPTG induction as described above. Cells were lysed and recombinant enzymes were purified with Ni-NTA-columns, followed by determining their purity and concentration by SDS-PAGE and Bradford staining. NanS-His, NanS-p1-His, NanS-p2-His and NanS-p4-His were incubated for 2 h with mono-O-acetylated Neu5,9Ac2 or Neu5,4 Ac2; samples of Neu5,9Ac2 and Neu5,4 Ac2 without enzymes served as controls. After enzyme treatment, we qualitatively analysed the extent of O-acetylation by means of HPTLC (see above). The results of the exposure of Neu5,9Ac2 and Neu5,4 Ac2 with the EHEC O-acetylesterases are shown in Fig. 1 A and Fig. 1B, respectively.
Fig. 1.
(A) HPTLC analysis of mono-O-acetylated Neu5,9Ac2 and (B) Neu5,4 Ac2 incubated with NanS-p EHEC O-acetylesterases. Three μl of 1 mg/mL Neu5Ac (Fig. 1A,B, lanes 1,8), Neu5,9Ac2 (Fig. 1A, lanes 2,9) and Neu5,4 Ac2 (Fig. 1B, lanes 2,9) preparations each, were applied onto HPTLC plates as controls. In addition, three μl of 1 mg/ml Neu5,9Ac2 (Fig. 1A, lane 3), and Neu5,4 Ac2 (Fig. 1B, lane 3) containing reaction buffer without enzymes were applied as buffer controls. Neu5,9Ac2 was digested with NanS-His (Fig.1A, lane 4), NanS-p1-His (Fig.1A, lane 5), NanS-p2-His (Fig.1A, lane 6) and NanS-p4-His (Fig.1A, lane 7). Neu5,4 Ac2 was digested with NanS-His (Fig.1B, lane 4), NanS-p1-His (Fig. 1B, lane 5), NanS-p2-His (Fig. 1B, lane 6), and NanS-p4-His (Fig. 1B, lane 7).
Incubation of Neu5,9Ac2 with NanS-His (Fig. 1A, lane 4), NanS-p1-His (Fig. 1A, lane 5), NanS-p2-His (Fig. 1A, lane 6) and NanS-p4-His (Fig. 1A, lane 7) resulted in a HPTLC band that separated at the position of Neu5Ac reference in the chromatogram (Fig. 1A), indicating the de-O-acetylation of Neu5,9Ac2 to Neu5Ac. Thus, all tested enzymes were capable to cleave the O-acetyl group from carbon atom C9, while Neu5,9Ac2 remained intact in the buffer controls without enzyme (Fig. 1A, lane 3) when compared to the applied Neu5,9Ac2 standard (Fig. 1A, lane 2). It should be mentioned that the Neu5,9Ac2 preparation used in this study contained a small amount of Neu5Ac, which did not affect complete de-O-acetylation of the Neu5,9Ac2 substrate to the Neu5Ac end product by the four EHEC O-acetylesterases. On the other hand, incubation of Neu5,4 Ac2 with NanS-His (Fig. 1B, lane 4), NanS-p1-His (Fig. 1B, lane 5), NanS-p2-His (Fig. 1B, lane 6) and NanS-p4-His (Fig. 1B, lane 7) revealed bands at the position of Neu5,4 Ac2 (Fig. 1B, lane 2), indicating that none of the recombinant O-acetylesterases was able to cleave the O-acetyl group from carbon C4.
3.2. NanoESI MS analysis of O-acetylesterase-treated BSM revealed enzymatic de-O-acetylation of mucin-bound mono-, di-, and tri-O-acetylated sialic acids
In a next step, we wanted to determine the specificity of recombinant NanS and NanS-p O-acetylesterases more precisely, since O-acetyl groups do not only occur at positions C4 or C9, but also at C7 or C8 of sialic acids. Additionally, di- and tri-O-acetylated derivatives exist and unravelling NanS- or NanS-p-mediated de-O-acetylation of such sialic acids was also in the focus of our investigation. Consequently, we used BSM as a source for mucin-bound Neu5Ac, Neu5Gc and their diverse O-acetylated derivates, which are components of highly sialylated O-glycans of BSM as proposed in the manufacturer’s data sheet (Merck, Darmstadt). Moreover, in recent work we could already demonstrate NanS-p activity in BSM (Saile et al., 2016).
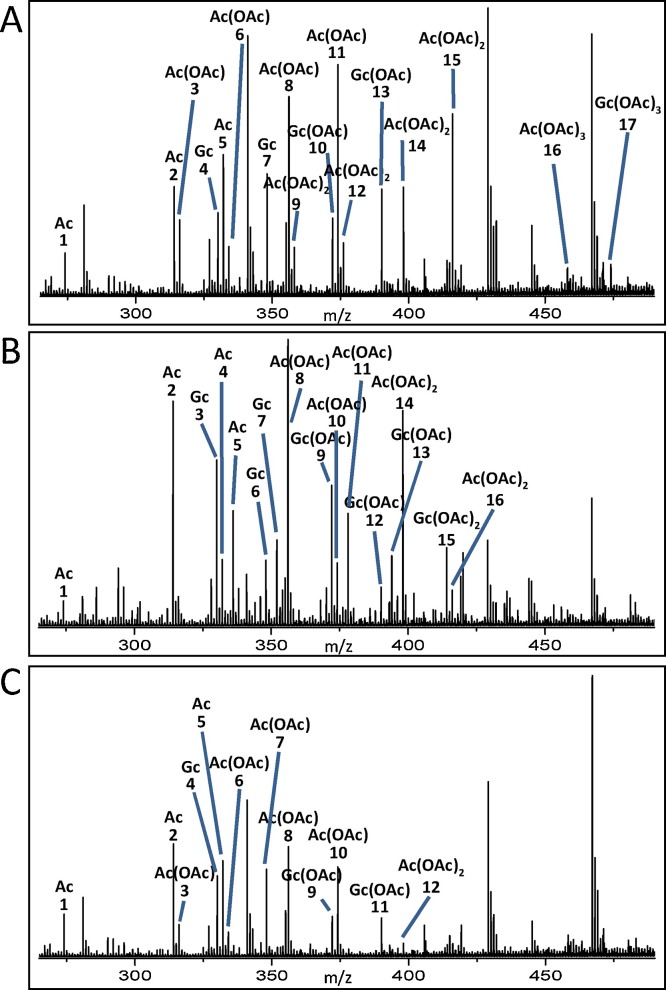
In the initial experiment, BSM-derived sialic acids, obtained by mild acid treatment, were analyzed by nanoESI MS. The obtained mass spectrum demonstrated the presence of Neu5Ac as well as the corresponding mono-, di-, and tri-O-acetylated Neu5Ac derivatives (Fig. 2 A). In addition, mono-, di-, and tri-O-acetylated Neu5Gc variants were detected. All detected sialic acid derivatives, which were chemically released from BSM O-glycans, are listed together with their m/z values in Table 2 A.
Fig. 2.
nanoESI mass spectra of chemically released sialic acids from BSM (A), and of sialic acids obtained by chemical release upon pretreatment of BSM with NanS-His (B) and NanS-p1-His (C). (A) The overview mass spectrum of BSM-derived sialic acids indicates the presence of Neu5Ac, Neu5Gc, Neu5Ac(OAc), Neu5Gc(OAc), Neu5,9(OAc)2, Neu5Ac(OAc)3 and Neu5Gc(OAc)3 in the O-glycans of BSM. (B) Ions corresponding to Neu5Ac, Neu5Gc, Neu5Ac(OAc), Neu5Gc(OAc), Neu5Ac(OAc)2 and Neu5Gc(OAc)2 were detected in the spectrum BSM-derived sialic acids after treatment with NanS-His. (C) The spectrum obtained from BSM-derived sialic acids after treatment with NanS-p1-His gives rise to ions corresponding to Neu5Ac, Neu5Gc, Neu5Ac(OAc), Neu5Gc(OAc) and Neu5Ac(OAc)2. Ac, Neu5Ac; Ac(OAc), Neu5Ac(OAc); Ac(OAc)2, Neu5Ac(OAc)2; Ac(OAc)3, Neu5Ac(OAc)3. Gc, Neu5Gc; Gc(OAc), Neu5Gc(OAc); Gc(OAc)2, Neu5Gc(OAc)2; Gc(OAc)3, Neu5Gc(OAc)3. The numbered ions in the mass spectra depicted in panel A, B, and C, are listed in Table 2A–C, respectively.
Table 2.
Detected ions derived from chemically released sialic acids of BSM without enzyme (A), NanS-His treated BSM (B), and NanS-p1-His treated BSM (C), and corresponding m/z values.
| No. | Sialic acid species | m/z | No. | Sialic acid species | m/z |
|---|---|---|---|---|---|
| A | |||||
| 1 | Neu5Ac+ -H2O | 274.10 | 10 | Neu5Gc(OAc)+ -H + Na | 372.09 |
| 2 | Neu5Ac+ -H + Na | 314.09 | 11 | Neu5Ac(OAc) [M + Na]+ | 374.11 |
| 3 | Neu5Ac(OAc)+ -H2O | 316.11 | 12 | Neu5Ac(OAc)2+ | 376.13 |
| 4 | Neu5Gc+ -H + Na | 330.08 | 13 | Neu5Gc(OAc) [M + Na]+ | 390.10 |
| 5 | Neu5Ac [M + Na]+ | 332.10 | 14 | Neu5Ac(OAc)2+ -H + Na | 398.11 |
| 6 | Neu5Ac(OAc)+ | 334.12 | 15 | Neu5Ac(OAc)2 [M + Na]+ | 416.12 |
| 7 | Neu5Gc [M + Na]+ | 348.09 | 16 | Neu5Ac(OAc)3 [M + Na]+ | 458.13 |
| 8 | Neu5Ac(OAc)+ -H + Na | 356.10 | 17 | Neu5Gc(OAc)3 [M + Na]+ | 474.13 |
| 9 | Neu5Ac(OAc)2+ -H2O | 358.11 | |||
| B | |||||
| 1 | Neu5Ac+ -H2O | 274.10 | 9 | Neu5Gc(OAc)+ -H + Na | 372.09 |
| 2 | Neu5Ac+ -H + Na | 314.09 | 10 | Neu5Ac(OAc) [M + Na]+ | 374.11 |
| 3 | Neu5Gc+ -H + Na | 330.08 | 11 | Neu5Ac(OAc)+ -2H+2Na | 378.08 |
| 4 | Neu5Ac [M + Na]+ | 332.10 | 12 | Neu5Gc(OAc) [M + Na]+ | 390.10 |
| 5 | Neu5Ac+ -2H+2Na | 336.07 | 13 | Neu5Gc(OAc)+ -2H+2Na | 394.07 |
| 6 | Neu5Gc [M + Na]+ | 348.09 | 14 | Neu5Ac(OAc)2+ -H + Na | 398.11 |
| 7 | Neu5Gc+ -2H+2Na | 352.06 | 15 | Neu5Gc(OAc)2+ -H + Na | 414.09 |
| 8 | Neu5Ac(OAc)+ -H + Na | 356.10 | 16 | Neu5Ac(OAc)2 [M + Na]+ | 416.12 |
| C | |||||
| 1 | Neu5Ac+ -H2O | 274.10 | 7 | Neu5Gc [M + Na]+ | 348.09 |
| 2 | Neu5Ac+ -H + Na | 314.09 | 8 | Neu5Ac(OAc)+ -H + Na | 356.10 |
| 3 | Neu5Ac(OAc)+ -H2O | 316.11 | 9 | Neu5Gc(OAc)+ -H + Na | 372.09 |
| 4 | Neu5Gc+ -H + Na | 330.08 | 10 | Neu5Ac(OAc) [M + Na]+ | 374.11 |
| 5 | Neu5Ac [M + Na]+ | 332.10 | 11 | Neu5Gc(OAc) [M + Na]+ | 390.10 |
| 6 | Neu5Ac(OAc)+ | 334.12 | 12 | Neu5Ac(OAc)2+ -H + Na | 398.11 |
In order to clarify, whether NanS-His and NanS-p-His O-acetylesterases are able to deacetylate mono-, di-, and tri-O-acetylated Neu5Ac or Neu5Gc species detected in BSM, we incubated BSM with either NanS-His, NanS-p1-His, NanS-p2-His or NanS-p4-His. Afterwards, the chemically released sialic acids were identified by nanoESI MS. Mass spectra of sialic acids obtained after exposure of BSM to the recombinant O-acetylesterases are exemplarily shown for NanS-His and NanS-p1-His in Fig. 2B and 2C, respectively. Identified sialic acids with the corresponding ion species and their m/z values are listed in Table 2B and C, respectively. Spectra of BSM-derived sialic acids upon exposure to NanS-His or NanS-p1-His evidenced predominant Neu5Ac and Neu5Gc core structures, accompanied by weaker signals, which could be assigned to mono-O-acetylated Neu5Ac(OAc) and Neu5Gc(OAc) species and the di-O-acetylated Neu5Ac(OAc)2 derivative. Importantly, Neu5Ac(OAc)3 and Neu5Gc(OAc)3 variants, which appear in the spectra at m/z 458.13 and m/z 474.13, respectively, were undetectable after O-acetylesterase treatment. Similar results were obtained with recombinant NanS-p2-His and NanS-p4-His (data not shown). Our results show that NanS as well as NanS-p are able to release acetyl residues not only from mono-O-acetylated sialic acids, but also from di- and tri-O-acetylated sialic acids as demonstrated for Neu5Ac(OAc)2, Neu5Ac(OAc)3, and their di- and tri-O-acetylated Neu5Gc counterparts. Furthermore, we could show that NanS and NanS-p have the ability to deacetylate O-acetylated sialic acids in soluble form and linked to O-glycans of mammalian mucin.
3.3. De-O-acetylation of mucin-bound sialic acids by NanS-His/NanS-p-His results in Neu5Ac and Neu5Gc substrates accessible to BTSA-His
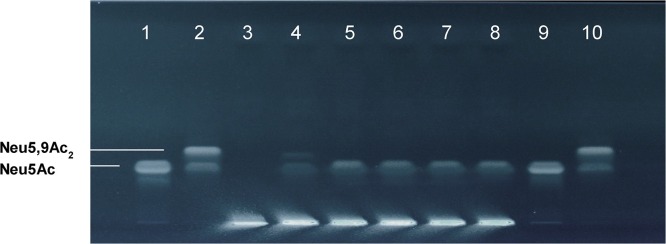
To explore the general accessibility of mucin-derived Neu5Ac or Neu5Gc as a substrate for commensal sialidases like BTSA from B. thetaiotaomicron VPI-5482, we exposed BTSA-His and NanS-His or its homologs simultaneously to BSM, suggesting that de-O-acetylated sialic acids are convenient substrates for the sialidase. For this purpose, we incubated BSM with either sialidase BTSA-His, or coincubated BSM with sialidase BTSA-His and either NanS-His, NanS-p1-His, NanS-p2-His or NanS-p4-His. After an exposure time of 4 h, we determined the sialic acids released from BSM first by means of HPTLC. The results of the enzymatic digestions of BSM using BTSA-His alone and combined with NanS-His, NanS-p1-His, NanS-p2-His or NanS-p4-His are shown in Fig. 3 . One mg/mL Neu5Ac (Fig. 3, lanes 1, 9) or Neu5,9Ac2 (Fig. 3, lanes 2, 10) were used as standards. Due to its higher polarity, Neu5,9Ac2 clearly separates beyond Neu5Ac allowing for unequivocal distinction of the two compounds. BSM incubated in buffer only without enzymes served as negative control (Fig. 3, lane 3).
Fig. 3.
Enzymatic cleavage of BSM-bound sialic acids by BTSA-His alone and combined with EHEC O-acetylesterases. Aliquots from enzymatic digests and standards were applied and sialic acids were stained with the DAPA reagent after chromatography. Approaches shown are BSM without enzymes (lane 3) and BSM with BTSA-His alone (lane 4), BTSA-His + NanS-His (lane 5), BTSA-His + NanS-p1-His (lane 6), BTSA-His + NanS-p2-His (lane 7), and BTSA-His + NanS-p4-His (lane 8). Neu5Ac (lanes 1, 9), and Neu5,9Ac2 (lanes 2, 10) served as standards.
The HPTLC analysis of enzymatically released sialic acids from BSM after incubation with BTSA-His alone (Fig. 3, lane 4) revealed two clearly DAPA-stained bands indicating the presence of enzymatically cleaved de-O-acetylated and mono-O-acetylated sialic acids. On the other hand, the combination of BTSA-His with NanS-His (Fig. 3, lane 5), NanS-p1-His (Fig. 3, lane 6), NanS-p2-His (Fig. 3, lane 7) or NanS-p4-His (Fig. 3, lane 8) revealed a single strong band, which separates at the position of Neu5Ac in the chromatogram as deduced from standard Neu5Ac (Fig. 3, lanes 1, 9).
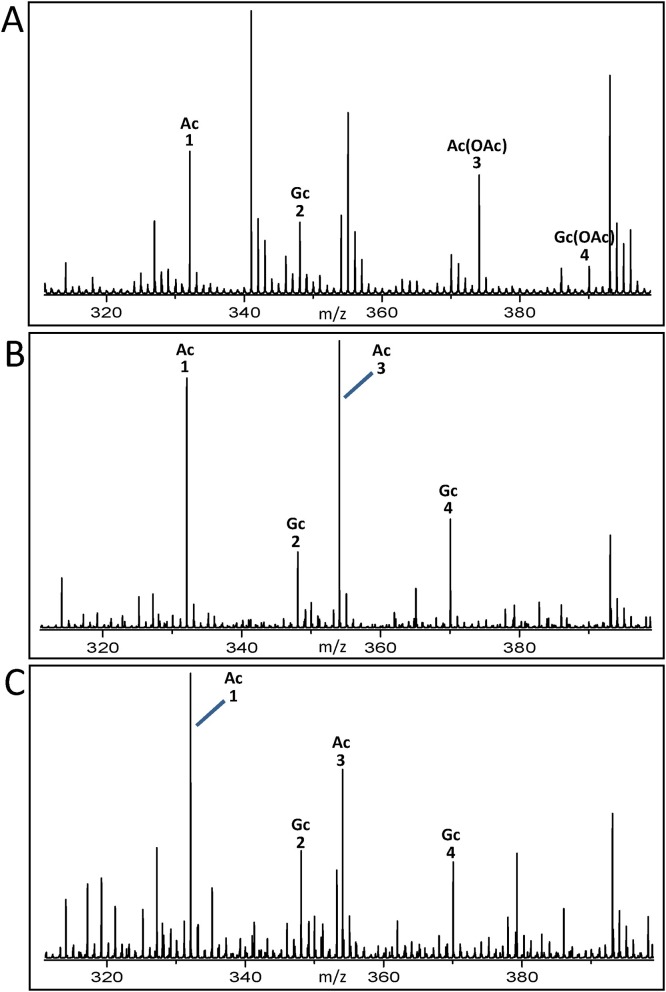
Collectively, our experimental data gave evidence that BTSA-His is able to cleave Neu5Ac and Neu5,9Ac2 from BSM. The enzyme seems not to exhibit any specificity for Neu5Ac or the O-acetylated derivative. Co-incubation of BSM with BTSA-His and O-acetylesterases revealed Neu5Ac only, while Neu5,9Ac2 could not be detected. Due to the suggested capability of NanS-His and its homologs to deacetylate mono-, di-, and tri-O-acetylated sialic acids, the above described digests were repeated and the released sialic acids were detected using mass spectrometry. For this purpose, proteins of the digests were precipitated with methanol and the supernatants were directly applied to nanoESI MS. Mass spectra of sialic acids obtained from digests of BSM using BTSA-His alone, BTSA-His combined with NanS-His and BTSA-His combined with NanS-p1-His are shown in Fig. 4 A, 4B and 4C, respectively. Identified sialic acids with the m/z values of the corresponding ions are listed in Table 3A–C, respectively. The mass spectrum obtained from BSM exposed to BTSA-His alone (Fig. 4A) shows ions derived from Neu5Ac and Neu5Gc basic structures and Neu5Ac(OAc) and Neu5Gc(OAc) (listed in Table 3 A). Thus, BTSA-His was capable to cleave off Neu5Ac and Neu5Gc as well as the mono-O-acetylated counterparts from glycans. Di- and tri-O-acetylated sialic acids were not detected and hence not released by BTSA-His. The combination of BTSA-His with the O-acetylesterases NanS-His or NanS-p1-His revealed ions indicating liberated Neu5Ac and Neu5Gc as shown in the mass spectra of Fig. 4B (listed in Table 3B) and Fig. 4C (listed in Table 3C), respectively. Both combined approaches revealed Neu5Ac and Neu5Gc but no O-acetylated sialic acids. The combination of BTSA-His and NanS-p2-His or NanS-p4-His gave the same results (data not shown).
Fig. 4.
nanoESI mass spectra of sialic acids obtained from digests of BSM treated with BTSA-His alone (A), BTSA-His combined with NanS-His (B), and BTSA-His combined with NanS-p1-His (C). The nanoESI MS analysis revealed ions which could be assigned to Neu5Ac(OAc) and Neu5Gc(OAc) only in the digests when using BTSA-His alone (A), whereas the combined digests (B and C) solely revealed the Neu5Ac and Neu5Gc cores. Ac, Neu5Ac; Ac(OAc), Neu5Ac(OAc); Gc, Neu5Gc; Gc(OAc), Neu5Gc(OAc). The numbered ions in the mass spectra depicted in panel A, B, and C, are listed in Table 3A–C, respectively.
Table 3.
Detected ions assigned to sialic acids obtained by treatment of BSM with BTSA-His alone (A), BTSA-His combined with NanS-His (B), and BTSA-His combined with NanS-p1-His (C) and corresponding m/z values.
| No. | Sialic acid species | m/z |
|---|---|---|
| A | ||
| 1 | Neu5Ac [M + Na]+ | 332.10 |
| 2 | Neu5Gc [M + Na]+ | 348.09 |
| 3 | Neu5Ac(OAc) [M + Na]+ | 374.11 |
| 4 | Neu5Gc(OAc) [M + Na]+ | 390.10 |
| B | ||
| 1 | Neu5Ac [M + Na]+ | 332.10 |
| 2 | Neu5Gc [M + Na]+ | 348.09 |
| 3 | Neu5Ac [M-H+2Na]+ | 354.08 |
| 4 | Neu5Gc [M-H+2Na]+ | 370.07 |
| C | ||
| 1 | Neu5Ac [M + Na]+ | 332.10 |
| 2 | Neu5Gc [M + Na]+ | 348.09 |
| 3 | Neu5Ac [M-H+2Na]+ | 354.08 |
| 4 | Neu5Gc [M-H+2Na]+ | 370.07 |
In conclusion, the results demonstrate that the combination of both BTSA-His with NanS-His or NanS-p1-His, respectively, enhances the release of de-O-acetylated core Neu5Ac and Neu5Gc from mammalian mucin O-glycans, which might serve as a carbon and/or nitrogen source for EHEC.
4. Recombinant BTSA supports the growth of EHEC O157:H7 strain EDL933 on BSM dependent on the presence of NanS-p
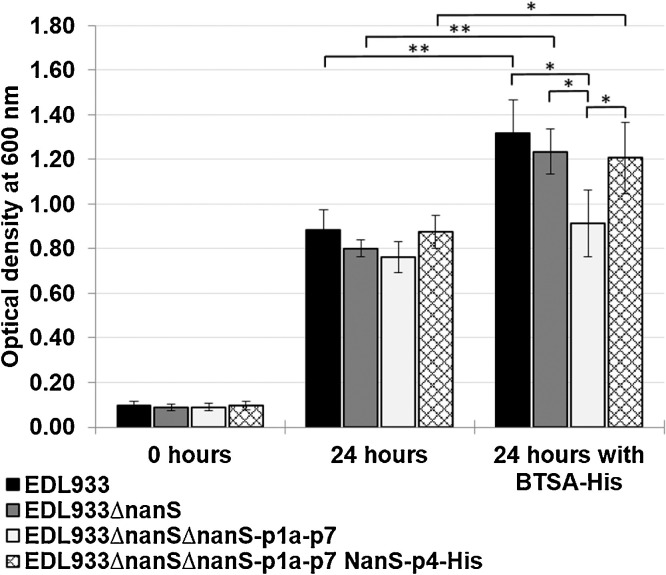
We compared the growth behaviour (OD600 values) of EHEC O157:H7 strain EDL933 (NZ_CP015855) (Saile et al., 2016), EDL933ΔnanS, and EDL933ΔnanSΔnanS-p1a-p7 for 24 h in growth medium containing BSM with or without BTSA-His. Despite an expected trend of the OD600 values, which decreased from 0.88, 0.8 to 0.76 for EDL933, EDL933ΔnanS and EDL933ΔnanSΔnanS-p1a-p7, respectively, the differences were not statistically significant (Fig. 7). However, if BTSA-His was added to the medium, significant differences in the OD600 between EDL933ΔnanSΔnanS-p1a-p7 and EDL933 or EDL933ΔnanS was detected with values of 0.91, 1.32 or 1.24, respectively (Fig. 5 ). For all strains, an increased OD600 was observed when BTSA-His was added, compared to the cultivation without BTSA-His. An explanation for this phenomenon is that both, Neu5Ac and Neu5,9Ac2, terminally bound to the mucin, were released by BTSA-His, whereby the degradation of Neu5Ac was unaffected by the deletions. Thus, Neu5,9Ac2 may be additionally utilized by EDL933 or EDL933ΔnanS. As a control a culture of EDL933ΔnanSΔnanS-p1a-p7 was supplemented with NanS-p4-His, resulting in an OD600 comparable to that of a wildtype EDL933 culture.
Fig. 5.
Growth behaviour (OD600) of E. coli O157:H7 strain EDL933 (black), EDL933ΔnanS (dark grey), EDL933ΔnanSΔnanS-p1a-p7 (light grey) and EDL933ΔnanSΔnanS-p1a-p7 supplemented with NanS-p4-His (hachured) in M9 minimal medium supplemented with 0.8% BSM at time points 0 and 24 h with or without BTSA-His. Error bars represent the standard deviations of 3 independent biological replicates. Significance was assessed with the paired Student t test (* p < 0.05; ** p < 0.01).
The results of this study showed for the first time, that NanS-p O-acetylesterases are active in a mucin environment and can de-O-acetylate mucin-derived sialic acids in vitro, in combination with sialidase treatment.
5. Discussion
According to current concepts, EHEC have to outcompete the gut microbiota for access to energy sources for a successful infection process. Sialic acids are highly abundant in the large intestine, where they decorate the mucus layer(s) in terminally exposed positions on the mucin O-glycans. Sialic acids can be modified by up to three acetic acid esters at the positions of the hydroxyl groups of the sialic acid core including Neu5Gc species, whereby Neu5Gc and its derivatives do exist in all mammals except humans (Robbe et al., 2003; Varki and Schauer, 2009; Vimr, 2013). While pure Neu5Ac and Neu5Gc are readily cleavable by bacterial sialidases, O-acetylation protects them from enzymatic hydrolysis (Juge et al., 2016). The intestinal E. coli strains are generally considered competitors for EHEC strains owing to their similar genetic background. Both groups harbor genes for sialic acid metabolism and are able to utilize free sialic acids as carbon and nitrogen sources. Compared to commensal nonpathogenic E. coli, EHEC harbor several phage-encoded NanS homologs (NanS-p) (Saile et al., 2016, 2018). These enzymes catalyze de-O-acetylation of Neu5,9Ac2 to Neu5Ac, which after sialidase action can be taken up and metabolized by both, commensal and pathogenic E. coli (Saile et al., 2016, 2018). Currently, it is not known, why EHEC strains contain a chromosomal nanS gene and multiple prophage-associated nanS-p genes. One hypothesis is that multiple nanS-p genes confer a gene dose effect to ensure substrate utilization and to outcompete commensal E. coli in the gut (Saile et al., 2016).
In this study, we demonstrated that recombinant NanS and NanS-p O-acetylesterases of EHEC O157:H7 strain EDL933 were able to de-O-acetylate sialic acids using BSM as a source for glycan-bound sialic acids. NanoESI MS spectra of chemically released sialic acids from BSM revealed ion signals of mono-, di-, and tri-O-acetylated Neu5Ac and Neu5Gc derivatives. NanS and NanS-p were capable to de-O-acetylate such O-acetylated sialic acids of BSM as determined by nanoESI MS analysis. Since various of the known microbial 9-O-acetylesterases are unable to de-O-acetylate Neu5,4 Ac2, we probed the specificity of NanS and the NanS-p esterases with respect to cleavage of the acetyl ester at C4 of Neu5Ac. HPTLC analysis revealed that neither NanS nor the NanS-p esterases are able to split off the O-acetyl group from this position. Mass spectra of NanS- or NanS-p-treated mucin still exhibited ion signals of O-acetylated sialic acids. Since O-acetyl groups are mostly located at position C4 and/or C9, it is tempting to speculate that the detected mono-O-acetylated Neu5Ac-species represent the Neu5,4 Ac2 isomer. Otherwise, an incomplete digestion or inactivation of the enzymes due to long-lasting incubation at elevated temperature could be the reason.
Furthermore, we could show that NanS and the NanS-p esterases deacetylate not only free sialic acids, but also glycosidically bound sialic acid of BSM. Although EHEC does not encode a sialidase, de-O-acetylation of bound sialic acids leads to liberation of acetate, which in turn can be readily used as an additional carbon source. However, de-O-acetylation results in sialidase-accessible Neu5Ac or Neu5Gc basic structures, which are common substrates of commensal sialidases, e.g., BTSA of B. thetaiotaomicron. In order to verify this hypothesis, we probed the removal of BSM-bound sialic acids with BTSA-His. This sialidase hydrolyzes pure Neu5Ac and Neu5Gc and also the mono-O-acetylated counterparts Neu5Ac(OAc) and Neu5Gc(OAc). Di- and tri-O-acetylated sialic acids were resistant towards BTSA giving evidence for a protective effect of O-acetylated sialic acids from degradation.
Combined exposure of BTSA with NanS or NanS-p resulted in release of Neu5Ac and Neu5Gc from BSM, which can then be metabolized in soluble form by EHEC. Even though B. thetaiotaomicron encodes sialidases, this bacterium lacks the nan operon and is unable to utilize released sialic acids as a nutrient. Therefore, the complementary action of both enzymes may result in a higher content of free sialic acids in the gut. In order to assess the growth-promoting effect of a mixed enzyme treatment, we cultivated the EHEC O157:H7 wildtype strain EDL933, and its mutant strains EDL933ΔnanS and EDL933ΔnanSΔnanS-p1a-p7 on mucin, mucin supplemented with BTSA-His and mucin exposed to BTSA-His combined with NanS-p-His. The combination revealed a significantly enhanced bacterial growth suggesting that EHEC exploits sialidases of commensal gut bacteria, and is supported by own O-acetylesterases to get access to O-glycan-derived sialic acids of intestinal mucus. On principle, sialic acids per se and particularly the O-acetylated species prevent from subsequent digest of desialylated glycans by glycosidases (Phansopa et al., 2015). Collectively, the sialidase-mediated release of sialic acids combined with complementary saponification of acetic acid side chains allows for ensuing stepwise digest of desialylated glycans by glycosidases such as galactosidases, N-acetylhexosaminidases or fucosidases. It can be speculated that the primary role of NanS-p esterases during infection could be the perturbation of the mucus layer integrity and initiation of commensal degradation, which may enable or at least facilitate EHEC strains crossing of the mucus barrier and getting access to the epithelial cells in the colon.
Interestingly, a number of viruses do exist, e.g., human coronavirus OC43 or mouse hepatitis virus, encoding for O-acetylesterases that may play a comparative role like the bacterial esterases during infection. These enzymes are mostly specific for Neu5,9Ac2 (human coronavirus OC43) or Neu5,4 Ac2 (mouse hepatitis virus) (Strasser et al., 2004). The influenza C virus encodes for an O-acetylesterase being capable to hydrolyze glycosidically bound Neu5,9Ac2, Neu5,7(8),9Ac3 and Neu5Gc9Ac of BSM in a similar manner like NanS-p-His (Strasser et al., 2004). This O-acetylesterase enables the virus to attach to the host membrane, destroys receptors accompanied with altered signal transduction in the host cell and enables viral entry (Ayora-Talavera, 2018; Matrosovich et al., 2015; Strasser et al., 2004).
We propose that NanS-p esterases might have an important complementary role during mixed viral and bacterial infections due to their broad specificity compared to that of the influenza C virus esterase. Thus, EHEC NanS-p esterases, which have the potency to de-O-acetylate mono-, di-, and tri-O-acetylated receptor-bound sialic acids, might have a yet underestimated functional role throughout initial infection and immune evasion of EHEC. Further work is necessary to clarify the role of NanS-p O-acetylesterases with regard to their involvement in maintenance and successful colonization of EHEC strains.
Acknowledgements
This work was supported by grant Schm1360/6-1 from the Deutsche Forschungsgemeinschaft (DFG). J. M. further acknowledges financial support by the German Center for Infection Research (DZIF, TTU 06.801). We thank Anja Voigt for experimental support in cloning of nanS.
References
- Aperce C.C., Heidenreich J.M., Drouillard J.S. Capacity of the bovine intestinal mucus and its components to support growth of Escherichia coli O157:H7. Animal. 2014;8:731–737. doi: 10.1017/S1751731114000147. [DOI] [PubMed] [Google Scholar]
- Ayora-Talavera G. Sialic acid receptors: focus on their role in influenza infection. JRLCR. 2018;10:1–11. [Google Scholar]
- Barnett Foster D. Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence. 2013;4:315–323. doi: 10.4161/viru.24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin Y., Chaucheyras-Durand F., Robbe-Masselot C., Durand A., de La Foye A., Harel J., Cohen P.S., Conway T., Forano E., Martin C. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ. Microbiol. 2013;15:610–622. doi: 10.1111/1462-2920.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D.-E., Smalley D.J., Tucker D.L., Leatham M.P., Norris W.E., Stevenson S.J., Anderson A.B., Grissom J.E., Laux D.C., Cohen P.S., Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U S A. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Cohen P.S. Commensal and pathogenic Escherichia coli metabolism in the gut. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MBP-0006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Krogfelt K.A., Cohen P.S. The kife of commensal Escherichia coli in the mammalian intestine. EcoSal Plus. 2004;1 doi: 10.1128/ecosalplus.8.3.1.2. [DOI] [PubMed] [Google Scholar]
- Corfield A.P., Wagner S.A., Clamp J.R., Kriaris M.S., Hoskins L.C. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 1992;60:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfield A.P., Wagner S.A., O’Donnell L.J., Durdey P., Mountford R.A., Clamp J.R. The roles of enteric bacterial sialidase, sialate O-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj. J. 1993;10:72–81. doi: 10.1007/BF00731190. [DOI] [PubMed] [Google Scholar]
- Erdem A.L., Avelino F., Xicohtencatl-Cortes J., Girón J.A. Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J. Bacteriol. 2007;189:7426–7435. doi: 10.1128/JB.00464-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabich A.J., Jones S.A., Chowdhury F.Z., Cernosek A., Anderson A., Smalley D., McHargue J.W., Hightower G.A., Smith J.T., Autieri S.M., Leatham M.P., Lins J.J., Allen R.L., Laux D.C., Cohen P.S., Conway T. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.T., Drouillard J.S., Shi X., Nagaraja T.G. Effects of mucin and its carbohydrate constituents on Escherichia coli O157 growth in batch culture fermentations with ruminal or fecal microbial inoculum. J. Anim. Sci. 2009;87:1304–1313. doi: 10.2527/jas.2008-1166. [DOI] [PubMed] [Google Scholar]
- Hews C.L., Tran S.-L., Wegmann U., Brett B., Walsham A.D.S., Kavanaugh D., Ward N.J., Juge N., Schüller S. The StcE metalloprotease of enterohaemorrhagic Escherichia coli reduces the inner mucus layer and promotes adherence to human colonic epithelium ex vivo. Cell Microbiol. 2017;19 doi: 10.1111/cmi.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.E.V., Thomsson K.A., Hansson G.C. Proteomic analyses of the two mucus layers of the colon barrier reveal that their main component, the Muc2 mucin, is strongly bound to the Fcgbp protein. J. Proteome Res. 2009;8:3549–3557. doi: 10.1021/pr9002504. [DOI] [PubMed] [Google Scholar]
- Johansson M.E.V., Larsson J.M.H., Hansson G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juge N., Tailford L., Owen C.D. Sialidases from gut bacteria: a mini-review. Biochem. Soc. Trans. 2016;44:166–175. doi: 10.1042/BST20150226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcobal A., Southwick A.M., Earle K.A., Sonnenburg J.L. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology. 2013;23:1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M., Herrler G., Klenk H.D. Sialic acid receptors of viruses. Top. Curr. Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nataro J.P., Kaper J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Y., Sperandio V. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect. Microbiol. 2012;2:90. doi: 10.3389/fcimb.2012.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübling S., Eisele T., Stöber H., Funk J., Polzin S., Fischer L., Schmidt H. Bacteriophage 933W encodes a functional esterase downstream of the Shiga toxin 2a operon. Int. J. Med. Microbiol. 2014;304:269–274. doi: 10.1016/j.ijmm.2013.10.008. [DOI] [PubMed] [Google Scholar]
- O’Brien A.D., Melton A.R., Schmitt C.K., McKee M.L., Batts M.L., Griffin D.E. Profile of Escherichia coli O157:H7 pathogen responsible for hamburger-borne outbreak of hemorrhagic colitis and hemolytic uremic syndrome in Washington. J. Clin. Microbiol. 1993;31:2799–2801. doi: 10.1128/jcm.31.10.2799-2801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A.V., Liles W.C. Enterohemorrhagic Escherichia coli infections and the hemolytic-uremic syndrome. Med. Clin. North Am. 2013;97:681–695. doi: 10.1016/j.mcna.2013.04.001. xi. [DOI] [PubMed] [Google Scholar]
- Park K.-H., Kim M.-G., Ahn H.-J., Lee D.-H., Kim J.-H., Kim Y.-W., Woo E.-J. Structural and biochemical characterization of the broad substrate specificity of Bacteroides thetaiotaomicron commensal sialidase. Biochim. Biophys. Acta. 2013;1834:1510–1519. doi: 10.1016/j.bbapap.2013.04.028. [DOI] [PubMed] [Google Scholar]
- Peri S., Kulkarni A., Feyertag F., Berninsone P.M., Alvarez-Ponce D. Phylogenetic distribution of CMP-Neu5Ac hydroxylase (CMAH), the enzyme synthetizing the proinflammatory human xenoantigen Neu5Gc. Genome Biol. Evol. 2018;10:207–219. doi: 10.1093/gbe/evx251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phansopa C., Kozak R.P., Liew L.P., Frey A.M., Farmilo T., Parker J.L., Kelly D.J., Emery R.J., Thomson R.I., Royle L., Gardner R.A., Spencer D.I.R., Stafford G.P. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. Biochem. J. 2015;472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- Rangarajan E.S., Ruane K.M., Proteau A., Schrag J.D., Valladares R., Gonzalez C.F., Gilbert M., Yakunin A.F., Cygler M. Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7. Protein Sci. 2011;20:1208–1219. doi: 10.1002/pro.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel A., Steenbergen S.M., Vimr E.R. Unexpected diversity of Escherichia coli sialate O-Acetyl esterase NanS. J. Bacteriol. 2016;198:2803–2809. doi: 10.1128/JB.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley L.W., Remis R.S., Helgerson S.D., McGee H.B., Wells J.G., Davis B.R., Hebert R.J., Olcott E.S., Johnson L.M., Hargrett N.T., Blake P.A., Cohen M.L. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- Robbe C., Capon C., Maes E., Rousset M., Zweibaum A., Zanetta J.-P., Michalski J.-C. Evidence of regio-specific glycosylation in human intestinal mucins: presence of an acidic gradient along the intestinal tract. J. Biol. Chem. 2003;278:46337–46348. doi: 10.1074/jbc.M302529200. [DOI] [PubMed] [Google Scholar]
- Robinson L.S., Lewis W.G., Lewis A.L. The sialate O-acetylesterase EstA from gut Bacteroidetes species enables sialidase-mediated cross-species foraging of 9-O-acetylated sialoglycans. J. Biol. Chem. 2017;292:11861–11872. doi: 10.1074/jbc.M116.769232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile N., Voigt A., Kessler S., Stressler T., Klumpp J., Fischer L., Schmidt H. Escherichia coli O157:H7 strain EDL933 harbors multiple functional prophage-associated genes necessary for the utilization of 5-N-acetyl-9-O-acetyl neuraminic acid as a growth substrate. Appl. Environ. Microbiol. 2016;82:5940–5950. doi: 10.1128/AEM.01671-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile N., Schwarz L., Eißenberger K., Klumpp J., Fricke F.W., Schmidt H. Growth advantage of Escherichia coli O104:H4 strains on 5-N-acetyl-9-O-acetyl neuraminic acid as a carbon source is dependent on heterogeneous phage-borne nanS-p esterases. Int. J. Med. Microbiol. 2018;308:459–468. doi: 10.1016/j.ijmm.2018.03.006. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (N.Y.): 2001. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- Scheutz F., Nielsen E.M., Frimodt-Møller J., Boisen N., Morabito S., Tozzoli R., Nataro J.P., Caprioli A. Characteristics of the enteroaggregative shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Euro Surveill. 2011;16 doi: 10.2807/ese.16.24.19889-en. [DOI] [PubMed] [Google Scholar]
- Stecher B., Hardt W.-D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011;14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Steenbergen S.M., Jirik J.L., Vimr E.R. YjhS (NanS) is required for Escherichia coli to grow on 9-O-acetylated N-acetylneuraminic acid. J. Bacteriol. 2009;191:7134–7139. doi: 10.1128/JB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steil D., Pohlentz G., Legros N., Mormann M., Mellmann A., Karch H., Müthing J. Combining mass spectrometry, surface acoustic wave interaction analysis, and cell viability assays for characterization of Shiga toxin subtypes of pathogenic Escherichia coli bacteria. Anal. Chem. 2018;90:8989–8997. doi: 10.1021/acs.analchem.8b01189. [DOI] [PubMed] [Google Scholar]
- Stevens M.P., Frankel G.M. The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.EHEC-0007-2013. EHEC-0007-2013. [DOI] [PubMed] [Google Scholar]
- Strasser P., Unger U., Strobl B., Vilas U., Vlasak R. Recombinant viral sialate-O-acetylesterases. Glycoconj. J. 2004;20:551–561. doi: 10.1023/B:GLYC.0000043292.64358.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F.W., Moffatt B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tailford L.E., Owen C.D., Walshaw J., Crost E.H., Hardy-Goddard J., Le Gall G., de Vos W.M., Taylor G.L., Juge N. Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation. Nat. Commun. 2015;6:7624. doi: 10.1038/ncomms8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A.G., Zhou X., Kaper J.B. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 2005;73:18–29. doi: 10.1128/IAI.73.1.18-29.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Schauer R. 2nd ed. 2009. Essentials of Glycobiology: Sialic Acids. Cold Spring Harbor (NY) [Google Scholar]
- Vimr E.R. Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013;(2013):816713. doi: 10.1155/2013/816713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E.R., Kalivoda K.A., Deszo E.L., Steenbergen S.M. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]