Abstract
An 9.4-kDa antifungal peptide designated as campesin was isolated from seeds of the cabbage Brassica campestris. The isolation procedure involved affinity chromatography on Affi-gel blue gel, ion exchange chromatography on Q-Sepharose and Mono S, and gel filtration on Superdex 75 and Superdex Peptide. The peptide was adsorbed on the first three chromatographic media. It exerted an inhibitory action on mycelial growth including Fusarium oxysporum and Mycosphaerella arachidicola, with an IC50 of 5.1 μM and 4.4 μM, respectively. The peptide was characterized by remarkable thermostability and pH stability. It inhibited proliferation of HepG2 and MCF cancer cells with an IC50 of 6.4 μM and 1.8 μM, and the activity of HIV-1 reverse transcriptase with an IC50 of 3.2 μM. It demonstrated lysolecithin binding activity.
Key words: Antifungal peptide, Isolation, Chinese cabbage, Brassica campestris, Brassicaceae
Living organisms produce a diversity of defense proteins to protect themselves from invading microbes, pathogens, and other noxious organisms. The defense proteins comprise antifungal proteins (1), antimicrobial proteins (2), ribosome inactivating proteins 3., 4., lectins 1., 5., ribonucleases (6) and protease inhibitors 7., 8..
Antifungal proteins are produced by a diversity of organisms including animals (9), plants 1., 2., 3., 4., 5., 6., 7., 8. and fungi (10). In plants, various tissues including bulbs (11), fruits (12), leaves (13), roots (14), seeds 1., 2., 3., 7., and tubers (5) produce antifungal proteins and peptides. Plant antifungal proteins constitute a repertoire of different proteins (15). They include chitinases and chitinase-like proteins 5., 14., 16., chitin-binding proteins 1., 13., lipid transfer proteins 2., 17., protease inhibitors 7., 8. ribosome inactivating proteins 3., 4., thaumatin-like proteins 12., 18., glucanases (16), embryo abundant protein-like proteins (19), and defensin-like peptides (20).
From plants belonging to the genus Brassica, various proteins including napins (21) and a number of enzymes including amine oxidase (22), pyruvate kinase (23), phosphoenolpyruvate carboxylase (24), myrosinase (25) and glyoxalase (26) have been purified. However, antifungal proteins have been isolated from only several Brassica species 17., 27., 28., 29.. Recently, a lipid transfer protein with antifungal activity, referred to hereinafter as BCLTP, has been reported from Brassica campestris seeds (17). Another antifungal peptide with N-terminal sequence similar to B. napus trypsin inhibitor but without trypsin inhibitory activity has been isolated from the seeds of B. parachinensis (27). Alboglabrin is an antifungal peptide from B. alboglabra seeds with an N-terminal sequence different from those of B. campestris and B. parachinensis (28). It is known that the same seed may produce more than one type of antifungal proteins (30). The intent of the present investigation was to isolate and characterize an additional antifungal protein from B. campestris seeds and to ascertain if the antifungal protein is different from the previously isolated lipid transfer peptide (29). It was found that the isolated peptide displayed various kinds of antipathogenic activities including antifungal, antitumor and anti-HIV reverse transcriptase which are of potential therapeutic value. The peptide, which could be synthesized, is of special interest because the aforementioned activities are thermostable and its antifungal activity is pH-stable.
Materials and methods
Materials
Seeds of B. campestris L. var. purpurea Bailey were purchased from a local vendor, Beijing Seed Company, in Mainland China. The seeds have been authenticated by Prof. Shiuying Hu, Honorary Professor of Chinese medicine, CUHK. They were deposited in laboratory 302, Department of Biochemistry, CUHK, under the voucher number BC883. The fungi were provided by Department of Microbiology, China Agricultural University, China. SP-Sepharose, Mono S column and Superdex Peptide column were from GE Healthcare (Sweden), Affi-gel blue gel was from Bio-Rad (USA). All chemicals were of the highest purity available.
Isolation of antifungal peptides
The seeds of B. campestris L. var. purpurea Bailey were purchased from a local vendor. The crude seed extract was subjected to ion exchange chromatography on a 5 × 20 cm column of Q-Sepharose (GE healthcare) which had been equilibrated with and was then eluted with 10 mM Tris–HCl buffer (pH 7.8). After unadsorbed proteins (fraction Q1) had come off the column, the column was eluted with 10 mM Tris–HCl buffer (pH 7.8) containing 1 M NaCl to yield fraction Q2. Fraction Q1 was then chromatographed on a 2.5 × 20 cm column of Affi-gel blue gel (Bio-Rad) in 10 mM Tris–HCl buffer (pH 7.8). Unadsorbed proteins (fraction B1) were eluted with the same buffer while adsorbed proteins (fraction B2) were eluted with 10 mM Tris–HCl buffer (pH 7.8) containing 1 M NaCl. Fraction B2 was taken for purification on a Mono S column by FPLC in 10 mM NH4OAc buffer (pH 4.5). After removal of the unadsorbed proteins, adsorbed proteins were eluted by three successive linear NaCl concentration gradients: 0–0.2 M, 0.2–0.4 M and 0.4–1 M. Fraction S5 eluted by the first gradient was subjected to purification on a Superdex 75 HR 10/30 column (GE healthcare) in 10 mM NH4OAc buffer (pH 4.5). Fraction SU3 from the column was purified on a Superdex Peptide column (GE Healthcare). The main resulting peak constituted purified antifungal peptide.
Protein determination
Protein concentration was determined by the dye-binding method (Bio-Rad) using bovine serum albumin as a standard.
Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis [Tricine-SDS-PAGE]
It was conducted according to the method of Schagger and von Jagow (31). After electrophoresis using 18% acrylamide gel, the gel was stained with Coomassie Brilliant Blue. The molecular mass of the isolated antifungal peptide was determined by comparison of its electrophoretic mobility with those of Kaleidoscope prestained standards (Bio-Rad) including carbonic anhydrase, soybean trypsin inhibitor, lysozyme, aprotinin and insulin with molecular mass of 38.8 kDa, 25 kDa, 16.3 kDa, 7.8 kDa and 3.4 kDa, respectively.
N-terminal amino acid sequence analysis
The N-terminal amino acid sequence of the purified peptide was determined by Edman degradation using an amino acid sequencer.
Assay of antifungal activity
The assay for antifungal activity was performed using 100 × 15 mm petri dishes containing 10 ml of potato dextrose agar. After the mycelial colony had developed, sterile blank paper disks (0.625 cm in diameter) were placed around and at a distance of 1 cm away from the periphery of the mycelial colony. An aliquot (8 μl containing 60 μg or 300 μg) of the purified peptide in 20 mM PBS buffer (pH 6.0) was added to a disk. The plates were incubated at 23 °C for 72 h until mycelial growth had surrounded peripheral disks containing the control (buffer) and had produced crescents of inhibition around disks containing samples with antifungal activity. The fungal species included Fusarium oxysporum and Mycosphaerella arachidicola (19).
To determine the IC50 value for the antifungal activity of the isolated antifungal peptide, four doses of the peptide were added separately to four aliquots each containing 4 ml potato dextrose agar at 45 °C, mixed quickly and poured into four separate small Petri dishes. After the agar had cooled down the same amount of mycelia was added to each plate. Buffer only without antifungal peptide served as a control. After incubation at 23 °C for 72 h, the area of the mycelial colony was measured and the inhibition of fungal growth determined. The concentration of the isolated antifungal peptide that brought about 50% reduction in the area of mycelial colony is the IC50 (19).
To investigate the thermal [0–100 °C] stability and pH [pH 0–14] stability of the antifungal activity, the isolated antifungal peptide was pretreated for 30 min accordingly and the antifungal assay was then conducted as mentioned above. After thermal treatment, the antifungal peptide solution in 10 mM Tris–HCl buffer (pH 7.8) was cooled down to room temperature before the assay for antifungal activity. After pH treatment, the low-molecular-weight components in the mixture were removed by centricon (Millipore) and pH adjusted to 7.8 using Tris–HCl buffer.
A solution of the isolated antifungal peptide [1 mg/ml] was incubated with an equal volume of trypsin or pepsin [1 mg/ml] at 37 °C for 1 h. At the end of the incubation, the reaction mixture was examined for antifungal activity.
Assay of lipid binding
Binding of lysolecithin was conducted at 25 °C with a Carywin-100 spectrofluorimeter (Varian Ltd, USA). The excitation wavelength was at 229 nm and emission spectra were recorded from 300 to 400 nm with 4-nm bandwidths and were corrected for the buffer contribution. Small amounts of concentrated lyso-C12 (lyso-α-lauroyl-phosphatidylcholine) solution in water (5 mg/ml) were added stepwise to a cuvet containing 1 ml of a solution of the isolated antifungal peptide (0.25 mg/ml) in 20 mM Tris–HCl buffer (pH 7.8). For each lipid–protein ratio, the maximum fluorescence intensity at 329 nm was used for constructing lipid titration curves. This maximum intensity was determined by averaging the intensity values obtained at 328 nm, 329 nm and 330 nm (29). To investigate the thermal [0–100°C] stability of the lipid binding activity, the isolated antifungal peptide was pretreated for 30 min accordingly and the assay was then conducted as mentioned above.
Assay of antiproliferative activity on tumor cell lines
This assay was performed in view of reports that some antifungal proteins have this activity 20., 29.. Breast cancer MCF-7 cells, heptoma HepG2 cells and normal human embryonic liver cells WRL68 were suspended in RPMI medium and the cell density was adjusted to 2 × 104 cells/ml. A 100-μl aliquot of the cell suspension was seeded to a well of a 96-well plate, followed by incubation for 24 h. Different concentrations of the antifungal peptide in 100 μl complete RPMI medium were then added to the wells and incubated for 72 h. After 72 h, 20 μl of 5 mg/ml solution of [3-[4,5-of imethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide] [MTT] in phosphate buffered saline was spiked into each well. The plates were incubated for another 4 h. The plates were then centrifuged at 324 ×g for 5 min. The supernatant was carefully removed and 150 μl of dimethyl sulfoxide was added to each well to dissolve the MTT-formazan formed at the bottom of the wells. Ten minutes later, the absorbance at 590 nm was determined by using a microplate reader (20). Blank was set by adding 100 μl of dimethyl sulfoxide. Negative control was set as 100% of cell survival. To investigate the thermal [0–100 °C] stability of the antiproliferative activity, the isolated antifungal peptide was pretreated for 30 min accordingly and the assay was then conducted as mentioned above.
Assay for HIV-1 reverse transcriptase inhibitory activity
The assay for HIV reverse transcriptase inhibitory activity was conducted in accordance with procedures supplied with the assay kit from Boehringer Mannheim (Germany) was carried out since some antifungal proteins possess this activity 8., 11., 20., 29.. The assay makes use of the ability of reverse transcriptase to synthesize DNA, starting from the template/primer hybrid poly (A) oligo (dT) 15. The digoxigenin- and biotin-labeled nucleotides in an optimized ratio are incorporated into one of the same DNA molecule, which is freshly synthesized by the reverse transcriptase (RT). The detection and quantification of synthesized DNA as a parameter for RT activity follows a sandwich ELISA protocol. Biotin-labeled DNA binds to the surface of streptavidin-precoated microtiter plate modules. In the next step, a precoated antibody to digoxigenin, conjugated to peroxidase, binds to the digoxigenin-labeled DNA. In the final step, the peroxidase substrate is added. The peroxidase enzyme catalyzes the cleavage of the substrate, forming a colored reaction product. The absorbance of the sample at 405 nm can be measured using a microtiter plate (ELISA) reader and is directly correlated to the level of RT activity. A fixed amount (4–6 ng) of recombinant HIV-1 reverse transcriptase was used. The inhibitory activity of the antifungal peptide was calculated as percent inhibition as compared to a control without the antifungal peptide (11). To investigate the thermal [0–100 °C] stability of HIV-1 reverse transcriptase inhibitory activity, the isolated antifungal peptide was pretreated for 30 min accordingly and the assay was then conducted as mentioned above.
Assay of ability to inhibit HIV-1 integrase
The assay was carried out since some antifungal proteins possess this activity (32).
Expression and purification of recombinant HiV-1 integrase
The plasmid that expressed His-tagged wild-type HIV-1 integrase, pT7-7-His (Y|TX)-HIV-1-IN, was a generous gift from Dr. S.A. Chow (School of Medicine, UCLA). To express the protein, a 1-liter culture of E. coli BL21 (DE3) cells containing the expression plasmid was grown at 37 °C until OD600 reached 0.7–0.8. Cells were induced by addition of 0.8 mM IPTG and harvested after 4 h incubation by centrifugation at 6000 ×g for 10 min at 4 °C. Cells were suspended at a concentration of 10 ml/g wet cell paste in 20 mM Tris–HCl (pH 8.0), containing 0.1 mM EDTA, 2 mM β-mercaptoethanol, 0.5 M NaCl and 5 mM imidazole. Lysozyme was added to a concentration of 0.2 mg/ml. After 1 h incubation at 4 °C, the lysate was sonicated and centrifuged at 40,000 ×g at 4 °C for 20 min. The pellet was homogenized in 50 ml buffer A (20 mM Tris–HCl, pH 8.0, 2 M NaCl, 2 mM β-mercaptoethanol) containing 5 mM imidazole. The suspension was rotated at 4 °C for 1 h and cleared by centrifugation at 40,000 ×g at 4 °C for 20 min. The supernatant was loaded onto a 1 ml chelating Sepharose column charged with 50 mM imidazole. The column was washed with five column volumes of buffer A containing 5 mM imidazole and the protein was eluted with three column volumes of buffer A containing 200 mM and 400 mM imidazole, respectively. Protein-containing fractions were pooled and EDTA was added to a final concentration of 5 mM. The protein was dialyzed against buffer B (20 mM HEPES, pH 7.5, 1 mM EDTA, 1 M NaCl, 20% glycerol) containing 2 mM β-mercaptoethanol and then against buffer B containing 1 mM dithiothreitol. Aliquots of the protein were stored at − 70 °C.
HIV-1 integrase assay
A non-radioactive ELISA-based HIV-1 integrase assay was performed according to the DNA-coated plates method. In this study, 1 μg of Smal-linearized p Bluescript SK was coated onto each well in the presence of 2 M NaCl as target DNA. The donor DNA was prepared by annealing VU5BR (5′-biotin-GTGTGGAAAATCTCTAGCAGT-3′) and VU5 (5′-ACTGCTAGAGATTTTCCACAC-3′) in 10 mM Tris-HC1, pH 8.0, 1 mM EDTA and 0.1 M NaCl at 80 °C followed by 30 min at room temperature. Integrase reaction was performed in 20 mM HEPES (pH 7.5), containing 10 mM MnCl2, 30 mM NaCl, 10 mM dithiothreitol and 0.05% Nonidet-P40 (Sigma). After the integrase reaction, the biotinylated DNA immobilized on the wells was detected by incubation with streptavidin-conjugated alkaline phosphatase (Boehringer Mannheim) followed by colorimetric detection with 1 mg/ml p-nitrophenyl phosphate in 10% diethanolamine buffer, pH 9.8, containing 0.5 mM MgCl2. The absorbance due to the alkaline phosphatase reaction was measured at 415 nm. The ribosome inactivating protein trichosanthin was used as a positive control 32., 33..
Screening for inhibitory effect on SARS Coronavirus (CoV) protease
Some antifungal proteins have been tested for this activity (33). The activity of SARS CoV protease was indicated by a designed substrate which was composed of two proteins linked by a cleavage site for SARS CoV protease. The substrate for assay is a recombinant protein that has a cleavable linker sequence (TSAVLQ↓SGFRK) between two fluorescent proteins, viz cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP). The former one is the donor protein of which the emission spectrum overlaps with the excitation spectrum of the latter acceptor protein. The reaction was performed in a mixture containing 5 μM SARS CoV protease, 5 μM sample, 20 μM substrate and buffer (20 mM Tris–HCl (pH7.5), 20 mM NaCl and 10 mM beta-mercaptoethanol) for 40 min at 37 °C. After 40 min, the reaction was stopped by heating at 100 °C for 2 min. Then the reaction mixture was analyzed by SDS-PAGE. If SARS CoV protease is inhibited by the test sample, there is only one band, which is the intact substrate, shown in the SDS-PAGE.
Assay for trypsin-inhibitory activity
This assay was conducted in view of the finding that some antifungal proteins have trypsin inhibitory activity (8) and that B. parachinensis antifungal peptide has an N-terminal sequence similar to a trypsin inhibitor (27). Trypsin activity was determined by using N-α-benzoyl-l-arginine ethyl ester hydrochloride (BAEE) as the substrate. Ten microliters of a trypsin solution (250 μg/ml) in assay buffer (50 mM Tris–HCl, pH 8, containing 20 mM CaCl2) were added to 980 μl of assay buffer and then 10 μl of BAEE in assay buffer was added to give a final concentration of 0.6 mM. The reaction rate was determined by monitoring the absorbance change at 253 nm for 1 min.
To assay for trypsin-inhibitory activity, 10 μl test sample in assay buffer was added to trypsin, and incubated at 25 °C for 15 min before addition of substrate (BAEE) to initiate the reaction. Trypsin-inhibitory activity was calculated as follows:
where Abs control is absorbance change in absence of sample, Abs sample is absorbance change in presence of sample, trypsin (mg) is the amount of trypsin in assay mixture. One unit of trypsin-inhibitory activity refers to the activity capable of inhibiting 1 mg trypsin. A similar assay was conducted using casein as substrate instead of BAEE (8).
Assay of mitogenic activity
This assay was carried out since some antifungal proteins possess this activity (20). Four C57BL/6 mice [20–25 g] were sacrificed by cervical dislocation. The spleens were removed aseptically and spleen cells were isolated by pressing the tissue through a sterilized 100-mesh stainless steel sieve and resuspended to 5 × 106 cells/ml in RPMI 1640 culture medium supplemented with 10% fetal bovine serum, 100 U penicillin/ml, and 100 μg streptomycin/ml. The cells [7 × 105 cells/100 μl/well] were seeded into a 96-well culture plate. Serial dilutions of a solution of the isolated peptide in 100 μl medium were added. After incubation of the cells at 37 °C in a humidified atmosphere of 5% CO2 for 24 h, 10 μl [methyl-3H]-thymidine [0.25 μCi, GE Healthcare] was added. The cells were incubated for a further 6 h under the same conditions. The cells were then harvested with an automated cell harvester onto a glass fiber filter. The radioactivity was measured with a Beckman model LS 6000SC scintillation counter. All reported values are the means of triplicate samples. Con A was used as positive control and bovine serum albumin as a negative control (20).
Results
Isolation of campesin and determination of molecular mass and N-terminal sequence
The crude seed extract was fractionated by ion exchange chromatography on Q-Sepharose to produce a large unadsorbed fraction Q1 with antifungal activity and a sharp adsorbed fraction Q2 devoid of antifungal activity. Affinity chromatography of fraction Q1 on Affi-gel blue gel yielded a broad unadsorbed fraction B1 without antifungal activity and a smaller adsorbed fraction B2 with antifungal activity. The results have been shown in a previous publication [17]. FPLC-ion exchange chromatography of fraction B2 on Mono S gave rise to several fractions (Fig. 1A). Purification of fraction S5 with antifungal activity was performed by gel filtration on a Superdex 75 column to yield a large peak (SU3) two small peaks (SU1 and SU2) (Fig. 1B). Fraction SU3 with antifungal activity was purified on a Superdex peptide column to yield a large absorbance peak with antifungal activity (P1) and a tiny inactive peak (Fig. 1C). The yields of crude extract, fractions Q1, B2, S5, SU3 and P1 from 400 g seeds were 19430, 7310, 850, 40, 25 and 20 mg, respectively (Table 1 ). Fraction P1, the purified peptide, showed a molecular mass of 9.4 Da in Tricine-SDS-PAGE (Fig. 2 ) and 9.4 kDa in gel filtration on Superdex peptide (Fig. 1C). It resembled the previously reported B. campestris lipid transfer peptide (BCLTP) in N-terminal sequence (Table 2 ).
Fig. 1.
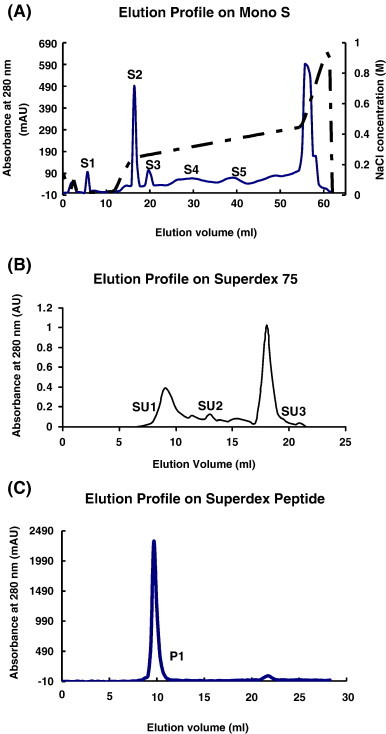
(A) FPLC-ion exchange chromatography on Mono S. Sample: fraction of seed extract previously adsorbed on Q-Sepharose and subsequently adsorbed on Affi-gel blue gel. Fraction S5 with antifungal activity was collected. (B) Gel filtration on a Superdex 75 HR 10/30 column. Fraction SU3 with antifungal activity was collected. (C) Gel filtration on a Superdex Peptide column. Fraction P1 with antifungal activity was collected.
Table 1.
Summary of purification of campesin from the seed Brassica campestris.
| Content | Active chromatographic fraction | Yield (mg) from 400 g seeds |
|---|---|---|
| – | Crude extract | 19430 |
| Q-Sepharose | Q1 | 7310 |
| Affi-gel Blue gel | B2 | 850 |
| Mono S | S5 | 40 |
| Superdex 75 | SU1 | 25 |
| Superdex Peptide | P1 | 20 |
Fig. 2.
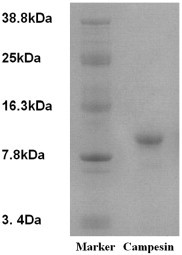
Tricine-SDS-PAGE of campesin. Left lane: molecular Kaleidoscope prestained molecular standards including carbonic anhydrase, soybean trypsin inhibitor, lysozyme, aprotinin and insulin with molecular mass of 38.8 kDa, 25 kDa, 16.3 kDa, 7.8 kDa and 3.4 kDa, respectively. Right lane: campesin.
Table 2.
N-terminal sequence of campesin in comparison with other Brassica antifungal peptides.
Biological activities of campesin
The peptide exerted antifungal activity against various fungal species including M. arachidicola (A) and F. oxysporum (B) with the IC50 values being respectively 4.4 μM (A) and 5.1 μM (B) (Fig. 3 ). Its lipid transfer activity is shown in Fig. 4 . The antifungal activity of the peptide was retained after exposure to trypsin, chymotrypsin and pepsin, and to various temperatures from 0 °C to 100 °C, and to the pH range 0–14. It inhibited proliferation of HepG2 cells and MCF7 cells (Fig. 5A) with an IC50 of 6.4 μM and 1.8 μM, respectively, and reduced the activity of HIV-1 reverse transcriptase with an IC50 of 3.2 μM (Fig. 5B). However, there was no antiproliferative activity toward normal embryonic liver WRL68 cells (Fig. 5A). The antiproliferative and HIV-1 reverse transcriptase inhibitory activities were fully preserved after exposure to various temperatures from 0 °C to 100 °C for 30 min. For the purpose of comparison, the thermostability of the various activities of BCLTP which had not previously been examined, was tested and the activities were found to be completely preserved at various temperatures from 0 °C to 100 °C. Table 3 shows a comparison of the antifungal peptides from various Brassica species. It lacked trypsin inhibitory activity toward casein and BAEE when tested up to 10 μM and 50 μM, respectively (not shown). It was devoid of mitogenic activity (Fig. 5C). However, it had no inhibitory effect on HIV-1 integrase and SARS proteinase (not shown).
Fig. 3.
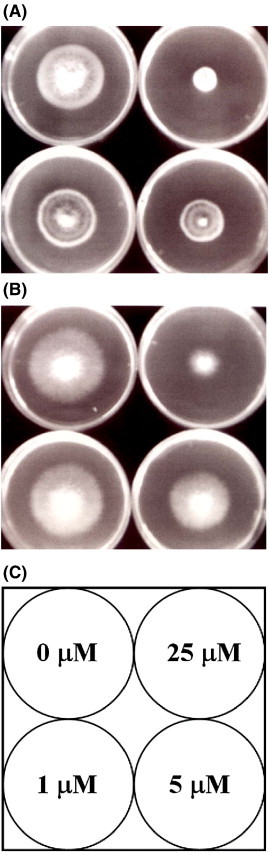
Determination of IC50 of antifungal activity of campesin. The fungi tested were: plate (A) Mycosphaerella arachidicola and (B) Fusarium oxysporum. (C) shows concentration of campesin in each plate.
Fig. 4.
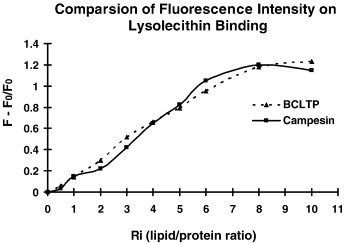
Comparison of increase in fluorescence intensity at 329 nm on lysolecithin binding due to campesin and BCLTP. F–F0 represents the fluorescence intensity of campesin. F is the intensity obtained at each lipid–protein ratio.
Fig. 5.
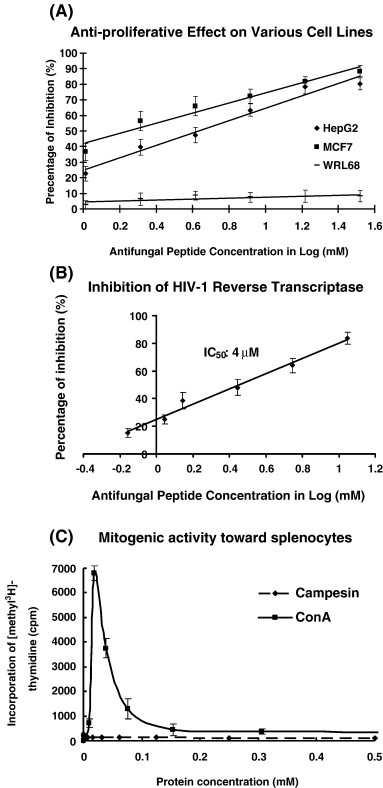
Campesin demonstrated antiproliferative activity (A), and HIV-1 reverse transcriptase inhibitory activity (B), but lacked mitogenic activity (C). Results represent mean ± SD (n = 3).
Table 3.
Comparison with other Brassica antifungal peptides.
| Campesin, (B. campestris (this study) | BCLTP (B. campestris) (29) | Antifungal peptide (B. alboglabra)(28) | Antifungal peptide (B. parachinensis) (27)a | |
|---|---|---|---|---|
| Molecular weight | 9.4 kDa | 9414 Da | 5907 Da | 5716 Da |
| Lipid transfer activity | Present | Present | Absent | Absent |
| Antifungal activity (IC50) | Mycosphaerella arachidicola: 4.4 μM | M. arachidicola: 8.3 μM | M. arachidicola: 2.4 μM | M. arachidicola: 2.6 μM |
| Fusarium oxysporum: 5.1 μM | F. oxysporum: 4.5 μM | F. oxysporum: 4.3 μM | F. oxysporum: 3.9 μM | |
| Antitumor activity (IC50) | HepG2: 6.4 μM | HepG2: 5.8 μM | HepG2: 2.7 μM | HepG2: 19.2 μM |
| MCF7: 1.8 μM | MCF7: 1.6 μM | MCF7: 3.4 μM | MCF7: 4.8 μM | |
| Antiproliferative activity on normal human embryonic liver cells (WRL68) | Devoid of activity | Not tested | Not tested | Not tested |
| Anti-HIV reverse transcriptase activity (IC50) | 3.2 μM | 4 μM | 4.9 μM | 17.8 μM |
| pH stability of antifungal activity | Stable from pH 0–14 | Stable from pH 0–14 | Stable from pH 2–11 | Stable from pH 1–13 |
| Thermostability | Up to 100 °C (shown for all aforementioned activities) | Up to 100 °C (shown for all aforementioned activities) | Up to 80 °C (shown for antifungal activity only) | Up to 100 °C (shown for antifungal activity only) |
Discussion
An antifungal peptide (BCLTP) with a molecular mass of 9414 Da and an N-terminal sequence similar to nonspecific lipid transfer proteins has previously been isolated from B. campestris seeds (29). The present study constitutes another report on the isolation of an antifungal peptide from seeds of the same species. Nevertheless, the N-terminal sequence of the antifungal peptide isolated in the present study, designated as campesin, is similar to that of BCLTP except for the replacement of some amino acids in BCLTP by more basic residues but different from those of antifungal peptides from B. parachinensis and B. alloglabra although all of them demonstrate pronounced thermostability and pH stability, HIV-1 reverse transcriptase inhibitory activity and antiproliferative activity toward tumor cells. They exert antifungal activity toward a variety of fungal species including F. oxysporum, M. arachidicola and a Helminthosporium species. Brassica parachinensis napin shows N-terminal sequence similarity to B. napus trypsin inhibitor as well as trypsin inhibitory activity (21). B. parachinensis antifungal peptide also exhibits N-terminal sequence to trypsin inhibitor but it is devoid of trypsin inhibitory activity (27). The absence of trypsin inhibitory activity in B. parachinensis antifungal peptide in spite of N-terminal sequence similarity to trypsin inhibitors is reminiscent of the lack of antifungal activity of thaumatin despite the fact that thaumatin-like proteins have antifungal activity (18). However, campesin does not resemble napin in N-terminal sequence and manifests no trypsin inhibitory activity. This is noteworthy in view of the fact that some of the trypsin inhibitors demonstrate antifungal activity (8). The two B. campestris antifungal peptides, campesin and BCLTP were isolated by using similar protocols. Ion exchange chromatography on Q-Sepharose, affinity chromatography on Affi-gel blue gel, ion exchange chromatography on Mono S and gel filtration on Superdex Peptide were used for purification of B. campestris lipid transfer peptide (29), the only difference in the present protocol being an additional gel filtration on Superdex 75.
The yield of campesin (50 mg/kg) is lower than that of BCLTP (175 mg/kg). The HIV-1 reverse transcriptase inhibitory activity of the former (IC50 = 3.2 μM) is also lower than that of the latter (IC50 = 4 μM) but more potent than many anti-HIV-1 natural products (32). Similarly, the antiproliferative activity of campesin toward HepG2 cells and MCF-7 cells (IC50 = 6.4 μM and 1.8 μM, respectively) is lower than that of BCLTP (IC50 = 5.8 μM and 1.6 μM, respectively). Both antifungal peptides are devoid of mitogenic activity toward mouse splenocytes. It has previously been shown that some 8., 20. but not other (29) antifungal proteins/peptides exhibit mitogenic activity. BCLTP and campesin have similar lipid transfer activity. They have no inhibitory effect on HIV-1 integrase, unlike some other antifungal proteins (32). They are also inactive toward SARS proteinase. French bean defensin-like antifungal peptides resemble BCLTP and campesin in that all of them do not have any suppressive effect on HIV-1 integrase and SARS CoV proteinase (33).
Lysolecithin is employed as a model phospholipid for lipid transfer activity assay in this study since it manifests in aqueous solution a simple and quick equilibrium between dispersed monomeric molecules and micellar aggregates. Lipid binding by nsLTPs, in aqueous solution by fluorescence spectroscopy with substrate lyso-C12, was first studied using maize nsLTP and wheat nsLTP (34). The relative enhancement fluorescence intensity of campesin shows similarity to that of wheat nsLTP, while maize nsLTP evokes a considerably smaller relative increase (35). The presence of a stable lysolecithin–protein complex in aqueous solution is analogous to a condition in which nsLTP serves as a carrier that can extract one lipid molecule from a membrane and transfer it to another (36). The results disclose that the relative increase of fluorescence intensity continues as lipid/protein ratio rises until a plateau is attained, indicating that the purified peptide possesses good lipid transfer activity. To date, it has been impossible to isolate a stable complex involving lysolecithin and to establish the model of binding of such a molecule to the nsLTP. Recent monolayer experiments (34) provide addition information on the interaction between the wheat nsLTP and diacylphospholipids. A model is proposed involving a collisional complex-shuttle mechanism. In this model, only one acyl chain of the lipid is inserted into the protein hydrophobic tunnel and the transfer implies an intimate contact between the donor and acceptor membranes to prevent transfer of the second acyl chain in the aqueous solvent. Ongoing studies aim to establish more precisely the binding mode of multiacyl phospholipids by nsLTPs.
The physiological importance of lipid transfer protein in B. campestris seeds remains to be clarified although different functions of these proteins have been shown. These comprise surface wax synthesis in leaves (37), lipid reserves mobilization in cotyledons (38), fatty acyl CoA binding (39), calmodulin binding (40), defense reaction (41), protection of thylakoids of nonacclimated plants from freeze–thaw damage (42), and food IgE-mediated allergy (43). The promoter of a lipid transfer gene is upregulated by blue and red light and viral infection in transgenic Arabidopsis (44). The physiologic significance of nsLTP in Brassica seeds is likely to comprise mobilization of lipid reserves during germination (38) and resistance to pathogenic organisms (44).
Lipid transfer proteins with antifungal activity have been demonstrated in only a small number of plants including maize (45), wheat (46), onion (47), sunflower (48), pepper (49), mungbean (50), rice (51), cheeseweed (52), sugar beet (53), and radish (54). Antifungal activity has not been demonstrated in lipid transfer protein from Ginkgo biloba (55) and some lipid transfer protein isoforms from grape (56). The presence of lipid transfer protein isoforms in B. campestris seeds revealed by the present study is in line with the findings on grape (56) although both isoforms in B. campestris have antifungal activity whereas only one isoform in grape manifests antifungal activity (56).
Most of the aforementioned lipid transfer proteins have been tested only for antifungal and/or antibacterial activity and the other activities tested in the present investigation were not examined. The antiproliferative activity toward tumor cells and inhibitory activity toward HIV-1 reverse transcriptase observed in both BCLTP and campesin are in line with their role as defense proteins. It is shown only in the present study that campesin is devoid of antiproliferative activity on normal embryonic liver cells. Thus, it appears that its antiproliferative activity is confined to tumor cells. It is also demonstrated that all the biological activities of both BCLTP and campesin are thermostable, while in previous studies only the antifungal activity of the plant lipid transfer proteins has been shown to be thermostable. The peptide nature, and highly potent as well as thermostable antifungal, antitumor and anti-HIV reverse transcriptase activities of both BCLTP and campesin make them candidates for development into therapeutic agents.
In summary, the two antifungal peptides isolated from B. campestris seeds (i.e. campesin and BCLTP) have potentially exploitable activities. In contrast, some antifungal proteins like mungbean chitinase lack HIV-1 reverse transcriptase inhibitory activity and antiproliferative activity toward tumor cells (29). The finding in the present study of two closely similar antifungal peptides is reminiscent of the report of two highly homologous ribosome inactivating proteins with antifungal activity from bitter melon seeds (4). Other previous investigations revealed the presence of two structurally disparate antifungal proteins in the same plant tissue (30).
References
- 1.Broekaert W.F., Van Parijs J., Leyns F., Joos H., Peumans W.J. A chitin-binding lectin from stinging nettle rhizomes with antifungal properties. Science. 1989;245:1100–1102. doi: 10.1126/science.245.4922.1100. [DOI] [PubMed] [Google Scholar]
- 2.Cammue B.P.A., Thevissen K., Hendriks M., Eggermont K., Goderis I.J., Proost P., Van Damme J., Osborn R.W., Guerbette F., Kader J.C., Broekaert W.F. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer protein. Plant. Physiol. 1995;109:445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leah R., Tommerup H., Svendsen I., Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J. Biol. Chem. 1991;246:1564–1573. [PubMed] [Google Scholar]
- 4.Wang B., Shi X., Guo C., Ye X., Wang Z., Rao P. Isolation and purification of ribosome-inactivating proteins from bitter melon seeds by ion exchange chromatographic columns in series. Se Pu. 2004;22:543–546. [PubMed] [Google Scholar]
- 5.Gozia O., Ciopraga J., Bentia T., Lungu M., Zamfirescu I., Tudor R., Roseanu A., Nitu F. Antifungal properties of lectin and new chitinases from potato tuber. FEBS Lett. 1995;370:245–249. [PubMed] [Google Scholar]
- 6.Wang H., Ng T.B. Isolation of a novel deoxyribonuclease with antifungal activity from Asparagus officinalis seeds. Biochem. Biophys. Res. Commun. 2001;289:102–104. doi: 10.1006/bbrc.2001.5963. [DOI] [PubMed] [Google Scholar]
- 7.Joshi B.N., Sainani M.N., Bastawade K.B., Gupta V.S., Ranjekar P.K. Cysteine protease inhibitor from pearl millet: a new class of antifungal protein. Biochem. Biophys. Res. Commun. 1998;246:382–387. doi: 10.1006/bbrc.1998.8625. [DOI] [PubMed] [Google Scholar]
- 8.Ye X.Y., Ng T.B., Rao P.F. A Bowman–Birk-type trypsin–chymotrypsin inhibitor from broad beans. Biochem. Biophys. Res. Commun. 2001;289:91–96. doi: 10.1006/bbrc.2001.5965. [DOI] [PubMed] [Google Scholar]
- 9.Bulet P., Hetru C., Dimarcq J.L., Hoffmann D. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 1999;23:329–344. doi: 10.1016/s0145-305x(99)00015-4. [DOI] [PubMed] [Google Scholar]
- 10.Wang H., Ng T.B. Eryngin, a novel antifungal peptide from fruiting bodies of the edible mushroom Pleurotus eryngii. Peptides. 2004;25:1–5. doi: 10.1016/j.peptides.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Wang H., Ng T.B. Ascalin, a new antifungal peptide with human immunodeficiency virus type 1 reverse transcriptase inhibitory activity from shallot bulbs. Peptides. 2002;23:1025–1029. doi: 10.1016/s0196-9781(02)00032-3. [DOI] [PubMed] [Google Scholar]
- 12.Pressey R. Two isoforms of NP24: a thaumatin-like protein in tomato fruit. Phytochem. 1997;44:1241–1245. doi: 10.1016/s0031-9422(96)00667-x. [DOI] [PubMed] [Google Scholar]
- 13.Huang X., Xie W., Gong Z. Characteristics and antifungal activity of a chitin binding protein from Ginkgo biloba. FEBS Lett. 2000;478:123–126. doi: 10.1016/s0014-5793(00)01834-2. [DOI] [PubMed] [Google Scholar]
- 14.Lam S.K., Ng T.B. Isolation of a small chitinase-like antifungal protein from Panax notoginseng (sanchi ginseng) roots. Int. J. Biochem. Cell Biol. 2001;33:287–292. doi: 10.1016/s1357-2725(01)00002-4. [DOI] [PubMed] [Google Scholar]
- 15.Selitrennikoff C.P. Antifungal proteins. Appl. Environ. Microbiol. 2001;67:2883–2894. doi: 10.1128/AEM.67.7.2883-2894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogelsang R., Barz W. Purification, characterization and differential hormonal regulation of a β-1, 3-glucanase and two chitinases from chickpea (Cicer arietinum L.) Planta. 1993;189:60–69. doi: 10.1007/BF00201344. [DOI] [PubMed] [Google Scholar]
- 17.Lin P., Xia L., Ng T.B. First isolation of an antifungal lipid transfer peptide from seeds of a Brassica species. Peptides. 2007;28:1514–1519. doi: 10.1016/j.peptides.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 18.Chu K.T., Ng T.B. Isolation of a large thaumatin-like antifungal protein from seeds of the Kweilin chestnut Castanopsis chinensis. Biochem. Biophys. Res. Commun. 2003;301:364–370. doi: 10.1016/s0006-291x(02)02998-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang H., Ng T.B. Ginkbilobin, a novel antifungal protein from Ginkgo biloba seeds with sequence similarity to embryo-abundant protein. Biochem. Biophys. Res. Commun. 2000;279:407–411. doi: 10.1006/bbrc.2000.3929. [DOI] [PubMed] [Google Scholar]
- 20.Wong J.H., Ng T.B. Gymnin, a potent defensin-like antifungal peptide from the Yunnan bean Gymnocladus chinensis Baill. Peptides. 2003;24:963–968. doi: 10.1016/s0196-9781(03)00192-x. [DOI] [PubMed] [Google Scholar]
- 21.Ngai P.H.K., Ng T.B. Isolation of a napin-like polypeptide with potent translation-inhibitory activity from Chinese cabbage (Brassica parachinensis cv green-stalked) seeds. J. Peptide. Sci. 2003;9:442–449. doi: 10.1002/psc.460. [DOI] [PubMed] [Google Scholar]
- 22.Lim T.S., Chitra T.R., Han P., Pua E.C., Yu H. Cloning and characterization of Arabidopsis and Brassica juncea flavin-containing amine oxidases. J. Exp. Bot. 2006;57:4155–4169. doi: 10.1093/jxb/erl193. [DOI] [PubMed] [Google Scholar]
- 23.Smith C.R., Knowles V.L., Plaxton W.C. Purification and characterization of cytosolic pyruvate kinase from Brassica napus (rapeseed) suspension cell cultures: implications for the integration of glycolysis with nitrogen assimilation. Eur. J. Biochem. 2000;267:4477–4485. doi: 10.1046/j.1432-1327.2000.01494.x. [DOI] [PubMed] [Google Scholar]
- 24.Moraes T.F., Plaxton W.C. Purification and characterization of phosphoenolpyruvate carboxylase from Brassica napus (rapeseed) suspension cell cultures: implications for phosphoenolpyruvate carboxylase regulation during phosphate starvation, and the integration of glycolysis with nitrogen assimilation. Eur. J. Biochem. 2000;267:4465–4476. doi: 10.1046/j.1432-1327.2000.01495.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen S., Halkier B.A. Functional expression and characterization of the myrosinase MYR1 from Brassica napus in Saccharomyces cerevisiae. Protein Expr. Purif. 1999;17:414–420. doi: 10.1006/prep.1999.1158. [DOI] [PubMed] [Google Scholar]
- 26.Deswal R., Sopory S.K. Glyoxalase I from Brassica juncea is a calmodulin stimulated protein. Biochim. Biophys. Acta. 1999;1450:460–467. doi: 10.1016/s0167-4889(99)00047-6. [DOI] [PubMed] [Google Scholar]
- 27.Lin P., Ng T.B. A novel and exploitable antifungal peptide from kale (Brassica alboglabra) seeds. Peptides. 2008;29:1664–1671. doi: 10.1016/j.peptides.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin P., Ng T.B. Brassiparin, an antifungal peptide from Brassica parachinensis seeds. J. Appl. Microbiol. 2009;106:554–563. doi: 10.1111/j.1365-2672.2008.04025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin P., Xia L., Wong J.H., Ng T.B., Ye X.Y., Wang S.Y., Shi X.Z. Lipid transfer proteins from Brassica campestris and mung bean surpass mung bean chitinase in exploitability. J. Peptide. Sci. 2007;13:642–648. doi: 10.1002/psc.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye X.Y., Ng T.B., Rao P.F. Cicerin and arietin, novel chickpea peptides with different antifungal potencies. Peptides. 2002;23:817–822. doi: 10.1016/s0196-9781(02)00005-0. [DOI] [PubMed] [Google Scholar]
- 31.Schagger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of protein in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Ng T.B., Au T.K., Lam T.L., Ye X.Y., Wang C.C. Inhibitory effects of antifungal proteins on human immunodeficiency virus type 1 reverse transcriptase, protease and integrase. Life Sci. 2002;70:927–936. doi: 10.1016/s0024-3205(01)01458-8. [DOI] [PubMed] [Google Scholar]
- 33.Leung E.H.W., Wong J.H., Ng T.B. Concurrent purification of two defense proteins from French bean seeds: a defensin-like antifungal peptide and a hemagglutinin. J. Peptide Sci. 2008;14:349–353. doi: 10.1002/psc.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subirade M., Salesse C., Marion D., Pezolet M. Interaction of a non-specific wheat lipid transfer protein with phospholipid monolayers imaged by fluorescence microscopy and studied by infrared spectroscopy. J. Biophys. 1995;69:974–988. doi: 10.1016/S0006-3495(95)79971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomar J., Petit M.C., Sodano P., Sy D., Marion D., Kader J.C., Vovelle F., Ptak M. Solution structure and lipid binding of a nonspecific lipid transfer protein extracted from maize seeds. Protein Sci. 1996;5:565–577. doi: 10.1002/pro.5560050402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kader J.C. Lipid-transfer proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:627–654. doi: 10.1146/annurev.arplant.47.1.627. [DOI] [PubMed] [Google Scholar]
- 37.Pyee J., Yu H., Kolattukudy P.E. Identification of a lipid transfer protein as the major protein in the surface wax of broccoli (Brassica oleracea) leaves. Arch. Biochem. Biophys. 1994;311:460–468. doi: 10.1006/abbi.1994.1263. [DOI] [PubMed] [Google Scholar]
- 38.Soufleri I.A., Vergnolle C., Miginiac E., Kader J.C. Germination-specific lipid transfer protein cDNAs in Brassica napus L. Planta. 1996;199:229–237. doi: 10.1007/BF00196563. [DOI] [PubMed] [Google Scholar]
- 39.Wirtz K.W., Wouters F.S., Bastiaens P.H., Wanders R.J., Seedorf U., Jovin T.M. The non-specific lipid transfer protein (sterol carrier protein 2) acts as a peroxisomal fatty acyl-CoA binding protein. Biochem. Soc. Trans. 1998;26:374–378. doi: 10.1042/bst0260374. [DOI] [PubMed] [Google Scholar]
- 40.Liu H., Xue L., Li C., Zhang R., Ling Q. Calmodulin-binding protein BP-10, a probable new member of plant nonspecific lipid transfer protein superfamily. Biochem. Biophys. Res. Commun. 2001;285:633–638. doi: 10.1006/bbrc.2001.5219. [DOI] [PubMed] [Google Scholar]
- 41.Garcý'a-Olmedo F., Molina A., Segura A., Moreno M. The defensive role of non specific lipid-transfer proteins in plants. Trends Microbiol. 1995;3:72–74. doi: 10.1016/s0966-842x(00)88879-4. [DOI] [PubMed] [Google Scholar]
- 42.Hincha D.K., Neukamm B., Sror H.A., Sieg F., Weckwarth W., Ruckels M., Lullien-Pellerin V., Schroder W., Schmitt J.M. Cabbage cryoprotectin is a member of the nonspecific plant lipid transfer protein gene family. Plant Physiol. 2001;125:835–846. doi: 10.1104/pp.125.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palacin A., Cumplido J., Figueroa J., Ahrazem O., Sanchez-Monge R., Carrillo T., Salcedo G., Blanco C. Cabbage lipid transfer protein Bra o 3 is a major allergen responsible for cross-reactivity between plant foods and pollens. J. Allergy Clin. Immunol. 2006;117:1423–1429. doi: 10.1016/j.jaci.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 44.Sohal A.K., Pallas J.A., Jenkins G.I. The promoter of a Brassica napus lipid transfer protein gene is active in a range of tissues and stimulated by light and viral infection in transgenic Arabidopsis. Plant Mol. Biol. 1999;41:75–87. doi: 10.1023/a:1006232700835. [DOI] [PubMed] [Google Scholar]
- 45.Perri F., Della Penna S., Rufini F., Patamia M., Bonito M., Angiolella L., Vitali A. Antifungal proteins production in Maize suspension cultures. Biotechnol. Appl. Biochem. 2008 doi: 10.1042/BA20080060. Elecronic publication. [DOI] [PubMed] [Google Scholar]
- 46.Kirubakaran S.I., Begum S.M., Ulaganathan K., Sakthivel N. Characterization of a new antifungal lipid transfer protein from wheat. Plant Physiol. Biochem. 2008;46:918–927. doi: 10.1016/j.plaphy.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 47.Edqvist J., Rönnberg E., Rosenquist S., Blomqvist K., Viitanen L., Salminen T.A., Nylund M., Tuuf J., Mattjus P. Plants express a lipid transfer protein with high similarity to mammalian sterol carrier protein-2. J. Biol. Chem. 2004;279:53544–53553. doi: 10.1074/jbc.M405099200. [DOI] [PubMed] [Google Scholar]
- 48.Gonorazky A.G., Regente M.C., de la Canal L. Stress induction and antimicrobial properties of a lipid transfer protein in germinating sunflower seeds. J. Plant Physiol. 2005;162:618–624. doi: 10.1016/j.jplph.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Jung H.W., Kim K.D., Hwang B.K. Identification of pathogen-responsive regions in the promoter of a pepper lipid transfer protein gene (CALTPI) and the enhanced resistance of the CALTPI transgenic Arabidopsis against pathogen and environmental stresses. Planta. 2005;221:361–373. doi: 10.1007/s00425-004-1461-9. [DOI] [PubMed] [Google Scholar]
- 50.Wang S.Y., Wu J.H., Ng T.B., Ye X.Y., Rao P.F. A non-specific lipid transfer protein with antifungal and antibacterial activities from the mung bean. Peptides. 2004;25:1235–1242. doi: 10.1016/j.peptides.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 51.Ge X., Chen J., Li N., Lin Y., Sun C., Cao K. Resistance function of rice lipid transfer protein LTP110. J. Biochem. Mol. Biol. 2003;36:603–607. doi: 10.5483/bmbrep.2003.36.6.603. [DOI] [PubMed] [Google Scholar]
- 52.Wang X., Bunkers G.J., Walters M.R., Thoma R.S. Purification and characterization of three antifungal proteins from cheeseweed (Malva parviflora) Biochem. Biophys. Res. Commun. 2001;282:1224–1228. doi: 10.1006/bbrc.2001.4716. [DOI] [PubMed] [Google Scholar]
- 53.Kristensen A.K., Brunstedt J., Nielsen K.K., Roepstorff P., Mikkelsen J.D. Characterization of a new antifungal non-specific lipid transfer protein (nsLTP) from sugar beet leaves. Plant Sci. 2000;155:31–40. doi: 10.1016/s0168-9452(00)00190-4. [DOI] [PubMed] [Google Scholar]
- 54.Terras F.R., Goderis I.J., Van Leuven F., Vanderleyden J., Cammue B.P., Broekaert W.F. In vitro antifungal activity of a radish (Raphanus sativus L.) seed protein homologous to nonspecific lipid transfer proteins. Plant Physiol. 1992;100:1055–1058. doi: 10.1104/pp.100.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawano Y., Hatano K., Miyakawa T., Komagata H., Miyauchi Y., Yamazaki H., Tanokura M. Proteinase inhibitor from ginkgo seeds is a member of the plant nonspecific lipid transfer protein gene family. Plant Physiol. 2008;146:1909–1919. doi: 10.1104/pp.107.111500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomès E., Sagot E., Gaillard C., Laquitaine L., Poinssot B., Sanejouand Y.H., Delrot S., Coutos-Thévenot P. Nonspecific lipid-transfer protein genes expression in grape (Vitis sp.) cells in response to fungal elicitor treatments. Mol. Plant Microbe. Interact. 2003;16:456–464. doi: 10.1094/MPMI.2003.16.5.456. [DOI] [PubMed] [Google Scholar]


