Highlights
-
•
Widespread changes in lncRNA expresssion are associated with the immune response.
-
•
lncRNAs regulate the inflammatory response following activation of innate immunity.
-
•
lncRNAs regulate T cell differentiation and migration.
-
•
The action of long non-coding RNAs is mediated via diverse mechanisms.
Keywords: long noncoding RNA; antisense; lincRNA, inflammation; innate immune response; adaptive immune response
Abstract
It is increasingly clear that long non-coding RNAs (lncRNAs) regulate a variety biological responses, and that they do so by a diverse range of mechanisms. In the field of immunology, recent publications have shown widespread changes in the expression of lncRNAs during the activation of the innate immune response and T cell development, differentiation, and activation. These lncRNAs control important aspects of immunity such as production of inflammatory mediators, differentiation, and cell migration through regulating protein–protein interactions or via their ability to basepair with RNA and DNA. We review the current understanding of the mechanism of action of these immune-related lncRNAs, discuss their impact on physiological and pathological processes, and highlight important areas of inquiry at the intersection between immunology and lncRNA biology.
What are lncRNAs?
The first draft of the human genome uncovered several surprises including the observation that exonic regions of protein-coding genes represented <2% of the genome. Although some of the remaining DNA plays a crucial role in the maintenance of DNA structure and regulation of mRNA expression (i.e., transcription binding sites, promoter and enhancer regions), subsequent studies have shown that a significant proportion is transcribed into ‘non-coding RNAs’ (ncRNAs). Indeed, the recent release of the Encyclopedia of DNA Elements (the ‘ENCODE’ project aims to catalogue all the functional elements in human DNA) has concluded that ∼80% of DNA is functional and, importantly, that the majority (∼62%) is transcribed into non-coding RNA (ncRNA) 1, 2. However, the broad definition employed to identify functional/transcribed regions of DNA, which included an association with modified histones, methylated CpG dinucleotides or open chromatin, means this is likely to be an overestimate 3, 4. Indeed, examination of evolutionary conservation gives a more conservative estimate of ∼10% functional DNA, although this still allows for large regions that might be transcribed into ncRNA 3, 4.
ncRNAs are broadly classified as either short ncRNAs (<200 nucleotides) or long ncRNAs (>200 nucleotides). The microRNA (miRNA) family of short ncRNAs are best characterised and are known to induce mRNA degradation or block mRNA translation via the RNA interference pathway. By contrast, much less in known about lncRNAs although, in comparison with mRNAs, these are generally shorter in length, contain fewer albeit longer exons, and are expressed at lower levels (median is ∼10% of mRNAs) 1, 5, 6. lncRNAs also demonstrate low evolutionary sequence conservation, with estimates that only ∼15% of mouse protein-coding genes have homologues in humans, leading to speculation that the majority of lncRNAs are non-functional 6, 7, 8, 9. The most recent release from GenCode (version 19) has annotated ∼14 000 lncRNA genes in humans [5] although, given that many lncRNAs are expressed in a cell-, tissue-, and developmental stage-specific manner, this is likely to be a significant underestimate 1, 10. Presently, lncRNAs are classified by their position relative to protein-coding mRNAs and comprise the long intergenic ncRNA (lincRNA), intronic lncRNA, antisense lncRNA, transcribed pseudogene lncRNAs, and enhancer RNA (eRNA) (Box 1 ). However, these arbitrary definitions will need refining in the light of the increasing volume of sequencing data and the accumulating information on lncRNA function and mechanism.
Box 1. Classification of lncRNAs.
With limited information on their function and mechanism of action, lncRNAs are currently classified by their positions relative to protein-coding genes (Figure I ). In general, most lncRNAs are transcribed by RNA polymerase II (RNAP II) and are therefore capped, polyadenylated and commonly spliced. Antisense lncRNAs are transcribed across the exons of protein-coding genes from the opposite strand [76] whereas intronic lncRNAs are transcribed from intronic regions in either the sense or antisense orientation. The largest group are the intergenic lncRNAs (lincRNAs), that are located between protein-coding genes 32, 77. Two additional groups that could arguably be classified as lincRNAs are transcribed pseudogenes and enhancer RNAs (eRNAs). Transcribed pseudogenes arise when a gene loses the ability to produce a protein, either through mutation or inaccurate duplication [78]. eRNAs are produced by either mono- and bidirectional transcription at intergenic enhancer regions and can be differentiated from other lincRNAs by the presence of high H3K4me1 marks in their promoter regions 61, 79, 80. This contrasts with the members of the antisense, intronic, intergenic, and pseudogene lncRNA families that are associated with epigenetic marks characteristic of protein-coding genes (i.e., high H3Kme3 in the promoter region).
Figure I.
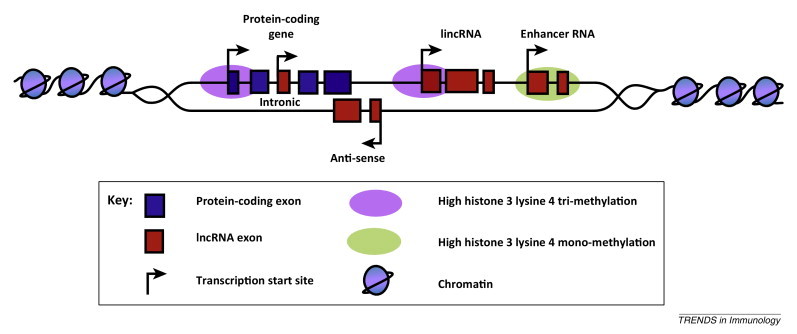
Locations of lncRNAs relative to protein-coding genes.
There is now accumulating evidence that lncRNAs are important regulators of physiological and pathological responses 11, 12. However, their potential importance in the immune response is only now emerging and it is this question that represents the topic of this review. As described in the forthcoming sections, immune-related lncRNAs are generally identified through examination of differential expression in response to activation of immune cells. This provides the basis for the subsequent functional and mechanistic analysis of individual lncRNAs. For clarity, these individual lncRNAs will be reviewed under the headings of innate immunity, acquired immunity, host–pathogen interaction, and enhancer RNAs. However, before proceeding it is worth highlighting emerging trends surrounding the mechanism of action of these immune-related lncRNAs.
Mechanism of action of immune-related lncRNAs
An interesting observation from the sequencing data is that many of the immune-related lncRNAs are located close to, or partially overlapping, the 5′ end (upstream) or 3′ end (downstream) of protein-coding genes implicated (and differentially expressed) in the immune response 13, 14. These are commonly transcribed in the antisense direction (relative to the protein-coding gene) which, in the case of upstream lncRNAs, can produce bidirectional transcription of the lncRNA and mRNA from a shared promoter region. Confusingly, these are often termed antisense lncRNAs despite there being only partial or no overlap with protein-coding genes and, as a result, we have recently proposed that these should instead be classified as mRNA-flanking lncRNAs (mf-lncRNAs) [13]. Given their locations, this group of lncRNAs which include THRIL (TNFα and hnRNPL related immunoregulatory lincRNA) [15], PACER (p50-associated COX-2 extragenic RNA) [16], lincR-Ccr2-5′AS [14], lnc-IL7R [17], and IL1β-RBT46 [13], have been shown to regulate the expression of their adjacent protein-coding gene in cis. eRNAs have also been demonstrated to control expression of protein-coding genes in cis, although chromatin looping is thought to be responsible for bringing the distal enhancer regions into close proximity with the promoter regions. The majority of the remaining immune-related lncRNAs, which are located in intergenic regions, regulate the immune response in trans. In addition to their cis actions, the four mRNA-flanking lncRNAs THRIL [15], lincR-Ccr2-5′AS [14], lnc-IL7R [17], and IL1β-RBT46 [13] have also been shown regulate the expression of multiple additional genes in trans.
As with proteins, it is speculated that lncRNAs are composed of domains that permit either protein binding and/or base-pairing with RNA or DNA sequences 18, 19, 20, 21, 22. Presently, it would appear that the action of most immune-related lncRNAs is mediated through protein binding. Targets include splicing factor proline/glutamine rich (SFPQ) [23], importin-β family [24], and the transcription factors, nuclear factor-κB (NF-κB) 16, 25, signal transducer and activator of transcription 3 (STAT3) [26], and the glucocorticoid receptor (GR) [27]. In these known examples, lncRNAs have been shown to act as ‘decoys’ to prevent protein–DNA binding (SFQR, NF-κB, and GR) or as antagonists of protein–protein interactions (importin-β and STAT3). Immune-related lncRNAs have also been shown to interact with members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family 15, 28 and components of chromatin-modifying complexes including the polycomb repressor complex 2 (PRC2) [29], WD repeat domain 5 (WDR5), a core subunit of the MLL methyltransferase complex [30], and the UTX/JMJD3 demethylases [31]. Although the exact mechanism is undefined, it has been speculated that they might act as scaffolds to bring together proteins and/or target these to the DNA through base-pairing, a situation that has been described with other chromatin-associated lncRNAs [12].
LncRNAs regulate the innate immune response
The first evidence of a potential role for lncRNAs in the innate immune response was a report by Guttman et al. [32] who used the intergenic deposition of epigenetic marks to identify 20 lincRNAs induced in lipopolysaccharide (LPS)-stimulated mouse bone marrow-derived dendritic cells (BMDD). Microarray and RNA sequencing (RNA-Seq) have now demonstrated differential lncRNA expression following activation of monocytes, macrophages, dendritic cells, and fibroblasts, as well as following viral infection in mouse lungs 15, 25, 26, 28, 33, 34, 35 (Table 1 ). As outlined in the following sections, investigators have then examined the role of individual lncRNAs in the innate immune response.
Table 1.
Long non-coding RNAs implicated in the immune response.
| LncRNA | Model system | Observation | Refs |
|---|---|---|---|
|
Innate immune response | |||
| Multiple | Coronavirus infection in mouse lung | RNA-seq demonstrated widespread differential expression of lncRNAs following lung infection with severe acute respiratory syndrome coronavirus in four mouse strains (129/S1, CAST, PWK, and WSB) | [33] |
| Multiple | LPS-stimulated mouse macrophages | Identification of multiple lincRNAs and eRNAs using pol II and H3K36me3 epigenetic marks. Eight of 11 lincRNAs were validated by qRT-PCR | [35] |
| LincRNA-Cox2 | LPS-stimulated mouse bone marrow-derived dendritic cells | Identification of 20 lincRNAs including lincRNA-Cox2 using deposition of epigenetic marks of active transcription (H3K4me3 at their promoters and H3K36me3 within the transcribed region) | [32] |
| LincRNA-Cox2 | Pam3CSK4-stimulated mouse bone marrow-derived macrophages | Revealed that lincRNA-Cox2 repressed the expression of 787 genes in non-stimulated cells and the increased expression of 713 genes following exposure to Pam3CSK4. The actions of lincRNA-Cox2 were mediated through interaction with hnRNP-A/B and hnRNP-A2/B1 | [28] |
| THRIL | Pam3CSK4-stimulated human monocytic THP-1 cells | Microarray analysis identified 159 differentially expressed lincRNAs including down-regulation of antisense lncRNA THRIL (TNFα and hnRNPL related immunoregulatory lincRNA). THRIL was shown to regulate both basal and Pam3CSK4-stimulated gene expression through an interaction with hnRNPL | [15] |
| Lethe | TNFα-stimulated mouse embryonic fibroblasts | RNA-seq identified 112 lncRNAs and 54 transcribed pseudogenes that were differentially expressed including Rps15a-ps4 (renamed Lethe). Lethe was induced in response to IL1β and dexamethasone and shown to interact and block the binding of the RelA (p65) subunit of NF-κB | [25] |
| NEAT1 | Poly(IC)- or influenza-stimulated HeLa and human epithelial A549 cells | Increased NEAT1 expression induced the formation of paraspeckle formation. Redistribution of SFPQ from the CXCL8 promoter to the paraspeckles following NEAT1 binding leads to increased CXCL8 expression | [23] |
| Ptprj-as1 | LPS-stimulated mouse bone marrow-derived macrophages | Induced in response to LPS | [34] |
| IL1β-RBT46 and IL1β-eRNA | LPS-stimulated human monocytes and monocytic THP-1 cells | RNA-seq identified 76 eRNAs, 40 lincRNAs, 65 antisense RNAs, and 35 regions of bidirectional transcription (RBTs) that are differentially expressed. IL1β-RBT46 and IL1β-eRNA were shown to regulate LPS-induced IL1β and CXCL8 expression | [13] |
| Unnamed | LPS-stimulated K562 leukemias cells | Multiple lncRNAs were located upstream of TNF and shown to negatively regulate TNF expression, possibly through binding to the transcriptional repressor, LRRFIP1 [leucine rich repeat (in FLII) interacting protein 1] | [81] |
| Lnc-IL7R | LPS-stimulated monocytic THP1 cells | Lnc-IL7R is transcribed from the 3′-UTR of IL7R in the sense orientation. Induced following LPS stimulation and negatively regulates IL7R, IL8, IL-6, VCAM-1, and E-selectin expression, a process associated with diminished H3K27me3 levels | [17] |
| PACER | PMA- and LPS-stimulated human U937 monocytic cell line | PACER (p50-associated COX-2 extragenic RNA) is expressed upstream of the Cox2 promoter and positively regulates COX2 production. PACER binds to, and drives the release of, the repressive p50 dimer of NF-κB from the Cox2 promoter | [16] |
| Lnc-DC | Differentiation of human and mouse dendritic cells | Lnc-DC (LOC645638) is required for monocyte differentiation into dendritic cells (DC). Lnc-DC promotes phosphorylation and activation of STAT3, a transcription involved in DC differentiation, by blocking its dephosphorylation by SHP1 | [26] |
|
Adaptive immune response | |||
| Multiple | Human CD8+ T cells | Microarray studies identified 100s of lymphoid-specific lncRNAs and showed differential expression during CD8+ T cell activation and following differentiation into CD8+ memory and effector T cells | [44] |
| NTT | Human T cell lines | NTT (noncoding transcript in CD4+ T cells) was identified in activated T cells | [41] |
| Gas5 | Human primary T cells and T cell lines (CEM-C7 and Jurkat) | Gas5 (growth arrest specific transcript-5) levels increase upon growth arrest and inhibit cell-cycle progression and promote apoptosis | [42] |
| Gas5 | Human primary T cells | Inhibition of T cell proliferation through the mTOR antagonist rapamycin is mediated by upregulation of Gas5 | [43] |
| NRON | Human Jurkat T cell | NRON (noncoding repressor of NFAT) blocked the nucleocytoplasmic transport and therefore the transcriptional activity of NFAT through interaction with multiple proteins including members of the importin-β superfamily | [24] |
| NRON | Human Jurkat T cells and mouse T cells | NRON shown to attenuate NFAT dephosphorylation and thereby block NFAT nuclear translocation, activation, and induction of IL-2 | [45] |
| NeST | Transgenic mouse infected with Salmonella and Theiler's virus | Overexpression of NeST (Nettoie Salmonella pas Theilers's) was shown to increase clearance of bacterial Salmonella infection but reduce resistance to the mouse Theiler's picornavirus. NeST induced the expression of IFN-γ through an interaction with WD repeat domain 5 (WDR5), a core subunit of the MLL histone H3 lysine 4 (H3K4) methyltransferase complex | [30] |
| LincR-Ccr2-5′AS | Mouse CD4+ TH2 cells | RNA-seq studies identified 1524 lincRNAs in 42 mouse T cell subsets. LincR-Ccr2-5′AS was located at the 5′-end of Ccr2 in CD4+ TH2 cells and was shown to regulate both the induction and suppression of gene expression during TH2 differentiation. LincR-Ccr2-5′AS is also implicated in chemokine-mediated signalling including cell migration | [14] |
| Multiple | Mouse T and B cells | LncRNAs shown to regulate chromatin remodelling associated with variable, diversity, and joining (V(D)J) recombination required to produce antigen receptors (Ig or TCR) | [48] |
| Multiple | Mouse B cells | Transcription of antisense and sense lncRNAs is associated with looping of VH regions into close proximity with the DJH region during recombination in pro-B cells, a process that occurs within transcription factories | [49] |
|
Pathogen-associated | |||
| PAN | KSHV-infected B cell lines | PAN (polyadenylated nuclear) RNA expression from KSHV was shown to modulate host cell response including downregulation of IFNγ, IL18, and α-interferon 16 | [52] |
| PAN | KSHV-infected B- and T cell lines | PAN RNA-mediated suppression of host genes is mediated through polycomb repression complex 2 (PRC2)-mediated histone methylation | [29] |
LincRNA-Cox2 mediates the induction and repression of gene expression in mouse macrophages
Initially identified by Guttman et al. [32] and located 50 kb downstream from the Cox2 (Ptgs2) gene, lincRNA-Cox2 was demonstrated to repress expression of 787 genes in non-stimulated bone marrow-derived macrophages (BMDM) and induce an increase in 713 genes following exposure to palmitoyl-3-cysteinyl-seryl-(lysyl)4 (Pam3CSK4; a TLR1/2 agonist) [28] (Figure 1A). Gene Ontology (GO) analysis showed these groups were enriched for genes involved in the immune response and included Ccl5 and Il6. Although the precise mechanism is unknown, the repressive action of lincRNA-Cox2 was mediated through interaction with hnRNP-A/B and hnRNP-A2/B1 [28]. These hnRNPs are members of a family of multifunctional RNA-binding proteins that are known to have a role in the processing of precursor mRNA, as well as in regulating gene expression [36]. Of relevance, Sauvageau et al. [37] have recently reported the production of a lincRNA-Cox2 knockout mouse which will provide an invaluable resource for the in vivo studies. Limited preliminary analyses have shown that lincRNA-Cox2 is selectively expressed in the lung, and that knockout does not impact upon the development of these animals [37].
Figure 1.
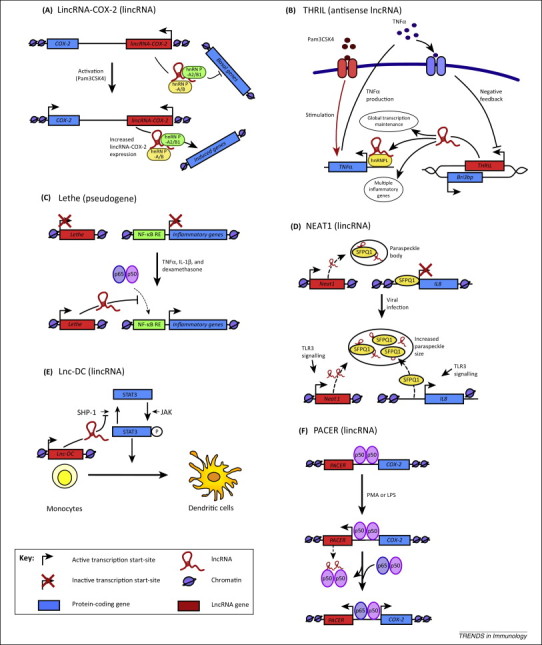
LncRNAs in the innate immune response. (A)LincRNA-COX-2 is located 3′ of the COX2 gene and is expressed in response to Pam3Csk4 stimulation of mouse bone marrow-derived macrophages. It has widespread effects on inflammatory gene expression, repressing the transcription of anti-inflammatory genes in non-stimulated cells and promoting the expression of proinflammatory genes following Pam3Csk4 exposure via an interaction with hnRNP-A2/B1 and hnRNP-A/B [28]. (B) Using human THP1 macrophages, THRIL was identified as an antisense lncRNA (overlapping BRI3BP) that promotes TNF transcription by forming a complex with hnRNPL and binding to the promoter of TNF[15]. THRIL is expressed basally; however, this is decreased in a negative feedback loop following Pam3Csk4-induced TNFα release [15]. THRIL has also been shown to regulate basal and Pam3Csk4-stimulated gene expression. (C)Lethe expression is induced in mouse embryonic fibroblasts following treatment with TNFα, IL1β, and dexamethasone, and prevents NF-κB binding to NF-κB response elements [25]. (D) In basal cells, SFPQ1 is bound to the CXCL8 promoter to repress its transcription, as well as to NEAT1 lincRNA within the paraspeckle bodies. NEAT1 expression is upregulated upon viral infection, leading to an increase in the size of NEAT1-containing paraspeckle bodies, the relocation of SFPQ1 from the CXCL8 promoter and derepression of CXCL8 transcription [23]. (E)Lnc-DC expression is required for differentiation of human monocytes into dendritic cells. Lnc-DC promotes STAT3 phosphorylation through inhibiting the action of Src homology region 2 domain-containing phosphatase-1 (SHP-1) [26]. (F)PACER is located upstream of the Cox2 transcriptional start site and is expressed in the antisense direction. PACER induced COX2 expression by removing the repressive action of the p50 homodimer (of NF-κB) bound at the Cox2 promoter [16].
THRIL regulates TNFα release and global gene expression in human monocytic THP-1 cells
THRIL is located downstream of the gene encoding the BRI3 binding protein (Bri3 bp), is transcribed from the opposite strand, and partially overlaps the 3′-end of Bri3 bp. Expression of THRIL in human monocytic THP-1 cells is reduced in response to Pam3CSK4, although this effect was indirect and mediated through autocrine/paracrine TNFα release. THRIL knockdown reduced expression of 444 genes (only 10 genes were increased) in non-stimulated cells and blocked differential expression of 317 of the 618 genes seen in response to Pam3CSK4, including multiple inflammatory genes such as IL6, CXCL8, CXCL10, CCL1, and CSF1 [15]. As with lincRNA-Cox2, THRIL was shown to interact with hnRNPL, with the resultant complex binding to the TNFα promoter and driving transcription in both control and Pam3CSK4-stimulated THP1 macrophages (Figure 1B).
Lethe blocks NF-κB-driven inflammatory responses in mouse fibroblasts
Rapicavoli et al. [25] identified a large increase in expression of the Rps15a-ps4 pseudogene following TNFα-induced activation of mouse embryonic fibroblasts (MEF). This pseudogene, renamed Lethe, was also induced following exposure to IL-1β and the anti-inflammatory GR agonist, dexamethasone [25]. Lethe was shown to block NF-κB (p65 or relA)-DNA binding and by this mechanism to inhibit both the inflammatory response to TNFα and IL-1β, and promote the anti-inflammatory actions of dexamethasone [25] (Figure 1C).
NEAT1-mediated SFPQ relocation into nuclear paraspeckles promotes CXCL8 expression in response to viral infection
The lincRNA NEAT1 (nuclear paraspeckle assembly transcript 1) has a key role in regulating nuclear paraspeckle body formation 38, 39. Under basal conditions, paraspeckles contain several proteins including SFPQ, which are bound to NEAT1. However, in addition to NEAT1, SFPQ is also known to repress the transcription of CXCL8 through binding within its promoter region. Following reports that NEAT1 expression is increased after viral infection of mouse neural cells [40], Imamura et al. [23] examined the role of NEAT1 in the antiviral response within HeLa and A549 epithelial cells. Activation of TLR3 using poly(IC), or infection with influenza virus or herpes simplex virus, was shown to stimulate NEAT1 expression, resulting in NEAT1-dependent increases in paraspeckle formation [23]. Significantly, increased CXCL8 expression also occurred as a result of the translocation of SFPQ from the CXCL8 promoter sites to the paraspeckles (Figure 1D). Of relevance, knockdown and overexpression identified 85 additional genes that were regulated by NEAT1, including several that are involved in the antiviral response [23].
Lnc-DC regulates differentiation of human monocytes into dendritic cells
Profiling of lncRNA expression during differentiation of monocytes into dendritic cells identified LOC645638, renamed lnc-DC, as being uniquely upregulated [26]. Lnc-DC knockdown impacted upon the expression of 664 protein-coding genes, resulting in impaired antigen uptake, reduced allogenic CD4+ T cell production and attenuated cytokine release. Interestingly, lnc-DC is located within the cytoplasm and its action is mediated through activation of STAT3, a transcription factor that is important for dendritic cell differentiation. Detailed analysis showed an interaction between the 3′-end of lnc-DC (residues 265–417) and the C-terminus of STAT3 (residues 583–770). This prevented de-phosphorylation of the Tyr705 (Y705) by Src homology region 2 domain-containing phosphatase-1 (SHP-1) and maintained STAT3 in its active phosphorylated form (Figure 1E).
PACER mediates COX-2 expression in human monocytes
PACER is located directly upstream of the Cox2 transcriptional start-site and expressed in the antisense direction [16]. Increased PACER expression, following PMA-induced differentiation of the monocytic U937 cell into macrophages and subsequent LPS-stimulation, was required for PMA/LPS-induced COX2 expression. This action was mediated through an interaction between PACER and the inhibitory p50 homodimer (of NF-κB). PACER decreased p50–p50 occupancy at the Cox2 promoter and permitted the binding of the active p50–p65 form of NF-κB and assembly of the RNA polymerase II pre-initiation complex [16] (Figure 1F). This event was associated with the recruitment of p300 histone acetyltransferase and increases in histone acetylation. Of relevance, these researchers demonstrate that baseline PACER expression and access to the Cox2 promoter were established and maintained by the chromatin boundary/insulator factor CTCF, which opened the chromatin structure in this region [16].
In summary, these reports have described widespread changes in lncRNA expression following activation of the innate immune response and have shown that these can regulate gene expression, the production of inflammatory mediators, and the differentiation of monocytes into macrophage and dendritic cells. In addition to the innate immune response, publications have also begun to reveal a role for lncRNAs in T cell biology and the adaptive immune response.
LncRNAs regulate T cell activation, development, and differentiation
Although the existence of individual lncRNAs in T cells, including the noncoding transcript in CD4+ T cells (NTT) [41], growth-arrest-specific transcript 5 (Gas5) 42, 43, and noncoding repressor of NFAT (NRON) [24] (Table 1), has been known for several years, the first widespread screen was undertaken in human and mouse CD8+ T cells by Pang et al. [44]. Using microarrays, these investigators uncovered 100s of lymphoid-specific lncRNAs that showed altered expression during CD8+ T cell activation and following differentiation into CD8+ memory and effector T cells [44]. More recently, a comprehensive analysis of lincRNA expression during development and differentiation of 42 mouse T cell subsets identified 1524 lincRNA genes [14]. Focusing on the polarization of CD4+ T cells into TH1 and TH2 subsets, they showed that the expression of TH1-specific lincRNAs was preferentially induced by the TH1-related transcription factors, STAT4 and T-box transcription factor (T-bet). Similarly, the TH2-specific transcription factor STAT6 and GATA binding protein 3 (GATA3) preferentially regulated TH2 lincRNA expression. Once again, there is now merging evidence linking individual lncRNAs with T cell function.
NRON represses nuclear translocation of NFAT in resting T cells
Nuclear factor of activated T cells (NFAT) is a Ca2+-activated transcription factor that is an important mediator of T cell activation including the induction of IL-2 expression. The noncoding repressor of NFAT (NRON), an intronic lncRNA, was first identified in 2005 during a short hairpin RNA (shRNA) library screen against 512 lncRNAs that had been characterised in the mouse genome and shown to have significant homology to humans [24]. Mechanistic studies showed that interaction between NRON and karyopherin importin-β1 (KPNB1) blocked the nucleocytoplasmic transport and therefore the transcriptional activity of NFAT [24] (Figure 2A). Subsequent studies in resting cells indicated that the heavily phosphorylated NFAT is located within a large cytoplasmic RNA–protein complex that contains NRON, a scaffold protein [IQ motif containing GTPase activating protein (IQGAP)], and three NFAT kinases [casein kinase 1 (CK), glycogen synthase 3 (GSK), and dual specificity tyrosine phosphorylation regulated kinase (DYRK)] [45]. Knockdown studies showed a functional synergy between NRON and IQGAP1 in blocking NFAT de-phosphorylation, a process that is required for nuclear translocation/activation and induction of IL-2 [45] (Figure 2A). NRON is therefore a constitutively expressed, intronic lncRNA that forms a complex with other proteins to bind the inactive NFAT and localise this transcription factor in the cytoplasm.
Figure 2.
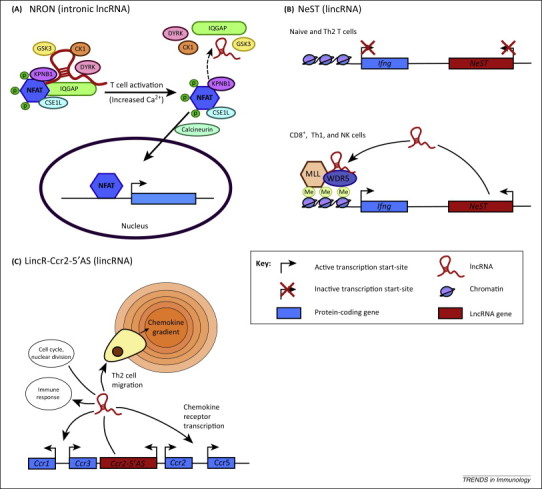
LncRNAs in the adaptive immune response. (A) The NFAT transcription factor is held inactive in the cytoplasm as part of a complex including the lncRNA NRON. Upon T cell activation, several of the proteins and NRON disassociate from the complex, and increased intracellular levels of Ca2+ activate calcineurin to facilitate the dephosphorylation of NFAT, thereby allowing it translocate to the nucleus 24, 45. (B)NeST is a lincRNA located downstream of Ifng which promotes the transcription of Ifng in TH1 CD4+ T cells, CD8+ T cells, and natural killer cells in mice. NeST binds to the methyltransferase WDR5 leading to methylation of the Ifng promoter 30, 47. (C)LincR-Ccr2-5′AS positively regulates the expression of genes involved in immunity and defence but negatively regulates genes involved in the cell cycle and nuclear division. Specifically, lincR-Ccr2-5′AS regulates the transcription of several chemokine receptor genes (located in the same loci as the lincRNA-Ccr2-5′AS gene) in mouse CD4+ TH2 cells that are required for cell migration [14].
NeST/Tmevpg1 induces interferon γ (IFN-γ) expression in T cells
Nettoie Salmonella pas Theilers's (NeST), which translates to ‘clean-up Salmonella but not Theiler‘s’, is a lincRNA that is located ∼45 kb downstream and transcribed in a convergent manner to the Ifng gene in mice [46]. NeST, which was formally known as Tmevpg1, is expressed in TH1 CD4+ T cells, CD8+ T cells, and natural killer cells [46]. Earlier work showed that expression of NeST was increased in TH1 but not TH2 polarised cells, correlated with Ifng expression, and was dependent upon the TH1-specific transcription factors STAT4 and T-bet [47]. By using mouse strains overexpressing NeST, Gomez et al. [30] showed increased clearance of Salmonella infection but reduced resistance to Theiler's virus, a mouse picornavirus. Mechanistic analysis indicated that nucleus-located NeST induced the expression of Ifng in trans in activated CD8+ T cells through an interaction with WDR5, a core subunit of the MLL H3K4 methyltransferase complex, leading to histone methylation at the Ifng locus [30] (Figure 2B). Overall, this milestone publication is the first to have shown that lncRNAs play a central role in regulating the adaptive immune response using an in vivo infection model.
lincR-Ccr2-5′AS regulates mouse TH2 migration into the lung
The TH2-specific lincR-Ccr2-5′AS, which is located at the 5′ end of Ccr2 and transcribed in the antisense direction, was identified during lincRNA expression profiling of 42 T cell subsets in mice [14]. shRNA-mediated lincR-Ccr2-5′AS knockdown (delivered via lentivirus) in mouse TH2 cells resulted in the upregulation of 709 mRNAs that were shown to be preferentially expressed in TH2 cells and enriched for genes involved in the immune response. There was also downregulation of 656 genes associated with the cell cycle and nuclear division [14]. Its mechanism of action is currently unknown, although knockdown had no effect upon H3K4me3 levels, DNase hypersensitivity, and RNA polymerase II binding, thus indicating that lincR-Ccr2-5′AS does not function by modifying epigenetic marks and chromatin accessibility (Figure 2C). Given that lincR-Ccr2-5′AS expression was highly correlated with seven of the 23 genes implicated in chemokine-mediated signaling pathways, including six that were located in the same genomic region, these investigators proceeded to examine its role in cell migration. Interestingly, they found that lincR-Ccr2-5′AS knockdown in TH2 cells impaired migration of TH2 cells to the lung.
Once again, early publications indicate that lncRNAs have an important role in mediating the differentiation, activation, and migration of T cells. Crucially, the report by Gomez et al. [30] is the first demonstration that lncRNAs can regulate the immune response in an animal model of infection. By comparison, little is known about B cell function, although Bolland et al. [48] have described a role for lncRNAs in the chromatin remodelling associated with the variable, diversity, and joining (V(D)J) recombination required to produce antigen receptors (Ig or TCR). A subsequent publication has shown that transcription of these antisense and sense lncRNAs is linked to looping of VH regions into close proximity with the DJH region during recombination in pro-B cells [49]. The process occurs within transcription factories but the mechanism has yet to be defined [49].
LncRNAs in host–pathogen interactions
Many pathogens produce lncRNAs which are believed to be important both in the pathogen life cycle and/or in the interaction between intracellular pathogens and their host cells. An example of the former is NEAT1, which controls HIV-1 replication by regulating the nuclear-to-cytoplasmic export of Rev-dependent instability element (INS) containing HIV mRNA [50]. Another well-characterised example is polyadenylated nuclear (PAN) RNA produced by Kaposi's sarcoma-associated herpesvirus (KSHV) that is able to modulate viral gene expression as well as subvert the host immune response. Thus, induction of PAN RNA is important in the switch from latent to lytic infection 51, 52, a process mediated by an interaction between PAN and the demethylases UTX and JMJD3 to remove the suppressive H3K23me3 mark within the KSHV viral genome (Figure 3A) [31]. Of note, PAN also physically interacts with LANA (latency associated nuclear antigen), a protein that maintains latency by binding to the KSHV genome, and has a role in regulating the dissociation of the LANA protein upon viral activation [53]. In addition to regulating the viral life cycle, PAN RNA also suppressed the expression of host genes involved in the inflammatory and antiviral response [52], a process mediated through activation of the polycomb repression complex 2 (PRC2) (Figure 3A) [29]. Overall, these studies imply that the viral non-coding PAN RNA is a regulator of both viral and host gene expression.
Figure 3.
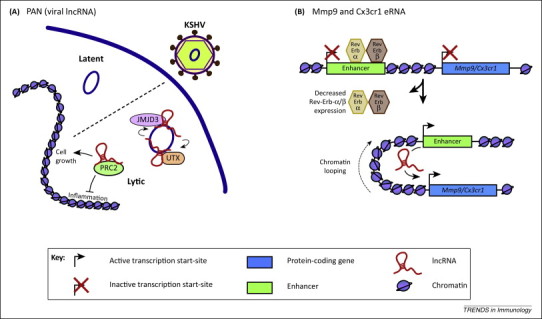
Viral lncRNAs and enhancer RNAs. (A) The viral lncRNA PAN recruits histone-modifying complexes to the KSHV genome to promote the switch from latent to lytic infection. PAN also regulates host gene expression through PRC2 to repress the inflammatory response and promote cell growth and survival 53, 82(B) The Mmp9- and Cx3cr1-eRNAs promote the transcription of Mmp9 and Cx3cr1 in mouse bone marrow-derived macrophages in cis. The nuclear receptors Rev-Erb-α and β repress the expression of Mmp9 and Cx3cr1 by binding to the enhancers and inhibiting eRNA transcription [58].
Enhancer RNAs are required for gene expression in macrophages and monocytes
Enhancer regions regulate the expression of protein-coding genes, often over large distances and in an orientation-independent manner. These regions are characterised by increased sensitivity to nuclease, p300/CBP acetyltransferase, and RNA polymerase II (RNAP) binding, as well as by deposition of chromatin marks including histone H3K4 monomethylation (H3K4me1) and H3K27 acetylation (H3K27ac) [54]. Recent studies have also demonstrated that these sites are associated with active transcription to produce enhancer RNAs (eRNAs) ∼0.5–5 kb in length 1, 22, 55, 56. Of relevance, the recent release of the FANTOM 5 consortium has identified >43 000 eRNAs across 808 cells/tissues [57]. However, until recently it was unclear whether eRNAs represented transcriptional noise or were instrumental in mediating the action of the enhancer regions. To address this question, Lam et al. [58] investigated the role of eRNA transcription during Rev-Erb-α (nuclear receptor subfamily 1, group D, member 1; Nr1d1) and Rev-Erb-β (nuclear receptor subfamily 1, group D, member 2; Nr1d2)-mediated gene repression in mouse macrophages, a process occurring through the recruitment of the nuclear receptor corepressor (NcoR)-HDAC3 to promoter and enhancer sites. Chromatin immunoprecipitation of Rev-Erb-α and β and subsequent sequencing showed that the majority (90%) of binding sites were located >1 kb from transcription start sites and within putative enhancer regions. Subsequent global run-on sequencing revealed that 76% of these enhancers were transcribed. To provide direct evidence that transcription of these eRNAs was linked to mRNA expression, these investigators focused on two eRNAs and their nearest neighbouring genes; Cx3cr1 and Mmp9. Knockdown of these eRNAs reduced expression of Cx3cr1 and Mmp9 but not that of other local genes (Figure 3B). Furthermore, cloning of the enhancer region revealed that although the core regions containing the transcription factor binding sites were able to enhance gene expression in cis, the magnitude of cis regulation was notably increased when the eRNA region was also cloned. Similarly, we recently described widespread expression of eRNAs following exposure of human monocytes to LPS and that LPS-induced expression of IL1B is dependent on a downstream eRNA that also appeared to regulate CXCL8 expression [13].
In addition to regulating gene expression, the transcription of eRNAs may also be important in the establishment of active enhancer regions. This was demonstrated in a series of elegant experiments by Kaikkonen et al. [59] that examined the sequential changes that occurred during the remodelling of the enhancer landscape following activation of mouse bone BMDM. Specifically, Kaikkonen et al. [59] examined the changes associated with the selection and activation of ∼3000 new enhancer regions following exposure to the TLR4 agonist, Kdo2-lipid A (KLA). This showed an initial interaction at the enhancer site between the TLR4-induced p65 component of NF-κB and the macrophage-specific transcription factors, PU.1 and C/EBP. However, the subsequent establishment of the enhancer region, through the acetylation of histone H4K5/8, was coupled to eRNA transcription. Thus transcription of eRNAs appears to play a role both in the establishment of enhancer regions [59] and in enhancer-mediated gene expression in cis [58]. This conclusion is supported by studies in other cell types 60, 61, 62, 63, as well as by the FANTOM 5 data which showed that eRNA expression was predominantly bidirectional and strongly correlated with enhancer activity [57].
Concluding remarks and future directions
In concluding, we need to return to the question of whether lncRNAs are novel regulators of the immune response? As a starting point, investigators often address this question by using RNA sequencing to detect the differential expression of known and novel lncRNAs. However, as with all profiling studies, one of the great challenges is the identification of functionally relevant lncRNAs from the large lists that are commonly produced. This problem is compounded by the fact that lncRNAs generally demonstrate poor evolutionary sequence conservation, thereby preventing the use of this traditional approach for the identification of functionality. Interestingly, although this indicates that most lncRNAs are likely to be non-functional, even those lncRNAs that are known to be conserved across multiple species (such as Malat1, Cyrano, and Xist) are not reported using existing alignment programmes [6]. This has led to speculation that these algorithms, which are based upon whole-genome alignments, may be an inappropriate approach for detecting conservation in rapidly evolving lncRNAs. This underlines the need for better bioinformatics tools that can be employed to identify ‘conserved’ lncRNAs and their homologues [6]. Despite these limitations, cell-based studies have identified several lncRNAs that regulate the innate immune response. Importantly, these studies have also characterised their mechanism of action and, as this number increases, it is hoped that this information can be employed to refine the bioinformatics tools required for the identification of functional sequences and/or structural motifs.
Given the complexity of the immune response, arguably the strongest evidence to support a role of lncRNAs would be provided by animal models of infection and/or disease. Indeed, animal models provide the only available approach for investigating many aspects of the acquired immune response. This therefore underlines the importance of the report by Gomez et al. [30] that demonstrated a role for NeST in a mouse model of viral and bacterial infection. However, the lack of conservation across species will once again represent a major hurdle to the standard approach of extrapolating from mouse to human.
Before finishing it also worth mentioning two recent potential areas of interest related to lncRNAs. First, given the importance of miRNAs in the immune response [64], it will be important to determine whether lncRNAs and miRNAs interact to regulate mRNA expression. This possibility is supported by reports showing that lncRNAs can reduce miRNA levels by acting either as sponges [19] or through base-pairing with primary miRNAs to block their processing into mature miRNAs [65]. Second, it has also been suggested that lncRNAs might actually code for peptides and small proteins 66, 67, an intriguing prospect given the importance of peptides in the immune response 68, 69.
Overall, as with miRNAs [64], there is emerging evidence that lncRNAs are important regulators of the immune response. It is likely that there are many additional immune-related lncRNAs to be discovered and that these will act via multiple different mechanisms. Future studies will need to examine whether aberrant lncRNA expression is also linked to the development of autoimmune and allergic disease, as well as the inflammation associated with many chronic diseases. Indeed, several studies have reported differential expression of lncRNAs in various inflammatory conditions 15, 70, 71, 72, 73, 74, 75 although further work is necessary to determine whether these have an active role in pathogenesis.
Acknowledgements
Mark A. Lindsay and James A. Heward are supported by the BBSRC (BB/K006223/1).
References
- 1.Djebali S. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doolittle W.F. Is junk DNA bunk? A critique of ENCODE. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5294–5300. doi: 10.1073/pnas.1221376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graur D. On the immortality of television sets: ‘function’ in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 2013;5:578–590. doi: 10.1093/gbe/evt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derrien T. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutter C. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabili M.N. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Necsulea A. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 10.Khalil A.M. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maass P.G. Long non-coding RNA in health and disease. J. Mol. Med. 2014;92:337–346. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 12.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.IIott N.E. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014;5:3979. doi: 10.1038/ncomms4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu G. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 2013;14:1190–1198. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. U.S.A. 2013;111:1002–1007. doi: 10.1073/pnas.1313768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krawczyk M., Emerson B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife. 2014;3:e01776. doi: 10.7554/eLife.01776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui H. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 2014;44:2085–2095. doi: 10.1002/eji.201344126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem. Sci. 2014;39:35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tay Y. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ørom U.A., Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamura K. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 24.Willingham A.T. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 25.Rapicavoli N.A. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang P. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 27.Kino T. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carpenter S. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341:789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossetto C.C. Regulation of viral and cellular gene expression by Kaposi's sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013;87:5540–5553. doi: 10.1128/JVI.03111-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez J.A. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossetto C.C., Pari G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012;8:e1002680. doi: 10.1371/journal.ppat.1002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng X. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio. 2010;1:e00206–e210. doi: 10.1128/mBio.00206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dave R.K. Regulated expression of PTPRJ/CD148 and an antisense long noncoding RNA in macrophages by proinflammatory stimuli. PLoS ONE. 2013;8:e68306. doi: 10.1371/journal.pone.0068306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garmire L.X. A global clustering algorithm to identify long intergenic non-coding RNA – with applications in mouse macrophages. PLoS ONE. 2011;6:e24051. doi: 10.1371/journal.pone.0024051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dreyfuss G. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 37.Sauvageau M. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hutchinson J.N. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clemson C.M. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saha S. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J. Gen. Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 41.Liu A.Y. The human NTT gene: identification of a novel 17 kb noncoding nuclear RNA expressed in activated CD4+ T cells. Genomics. 1997;39:171–184. doi: 10.1006/geno.1996.4463. [DOI] [PubMed] [Google Scholar]
- 42.Mourtada-Maarabouni M. Growth arrest in human T cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J. Cell Sci. 2008;121:939–946. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 43.Mourtada-Maarabouni M. Inhibition of human T cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5) Mol. Pharmacol. 2010;78:19–28. doi: 10.1124/mol.110.064055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang K.C. Genome-wide identification of long noncoding RNAs in CD8+ T cells. J. Immunol. 2009;182:7738–7748. doi: 10.4049/jimmunol.0900603. [DOI] [PubMed] [Google Scholar]
- 45.Sharma S. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA–protein scaffold complex. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11381–11386. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vigneau S. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J. Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collier S.P. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J. Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolland D.J. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 2004;5:630–637. doi: 10.1038/ni1068. [DOI] [PubMed] [Google Scholar]
- 49.Verma-Gaur J. Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 2012;109:17004–17009. doi: 10.1073/pnas.1208398109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Q. NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio. 2013;4:e00596. doi: 10.1128/mBio.00596-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borah S. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog. 2011;7:e1002300. doi: 10.1371/journal.ppat.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rossetto C.C., Pari G.S. Kaposi's sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 2011;85:13290–13297. doi: 10.1128/JVI.05886-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell M. A lytic viral long non-coding RNA modulates the function of a latent protein. J. Virol. 2013;88:1843–1848. doi: 10.1128/JVI.03251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bulger M., Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ørom U.A. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.De Santa F. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersson R. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lam M.T.Y. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaikkonen M.U. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Melo C.A. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 61.Mousavi K. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsieh C.L. Enhancer RNAs participate in androgen-driven looping that selectively enhances gene activation. Proc. Natl. Acad. Sci. USA. 2014;111:7319–7924. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O’Connell R.M. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 65.Liz J. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol. Cell. 2014;55:138–147. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 66.Slavoff S.A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ingolia N.T. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kopf M. Averting inflammation by targeting the cytokine environment. Nat. Rev. Drug Discov. 2010;9:703–718. doi: 10.1038/nrd2805. [DOI] [PubMed] [Google Scholar]
- 69.Lai Y., Gallo R.L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsitsiou E. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J. Allergy Clin. Immunol. 2012;129:95–103. doi: 10.1016/j.jaci.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 71.Zhang H. Profiling of human CD4+ T cell subsets identifies the TH2-specific noncoding RNA GATA3-AS1. J. Allergy Clin. Immunol. 2013;132:1005–1008. doi: 10.1016/j.jaci.2013.05.033. [DOI] [PubMed] [Google Scholar]
- 72.Stuhlmüller B. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am. J. Pathol. 2003;163:901–911. doi: 10.1016/S0002-9440(10)63450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B. Transcriptome analysis of psoriasis in a large case–control sample: RNA-seq provides insights into disease mechanisms. J. Invest. Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Q. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis Rheumatol. 2014;66:969–978. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 75.Müller N. Interleukin-6 and tumour necrosis factor-α differentially regulate lincRNA transcripts in cells of the innate immune system in vivo in human subjects with rheumatoid arthritis. Cytokine. 2014;68:65–68. doi: 10.1016/j.cyto.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 76.Wight M., Werner A. The functions of natural antisense transcripts. Essays Biochem. 2013;54:91–101. doi: 10.1042/bse0540091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cabili M.N. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;15:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pink R.C. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17:792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heintzman N.D. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 80.Natoli G., Andrau J-C. Noncoding transcription at enhancers: general principles and functional models. Annu. Rev. Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 81.Shi L. Noncoding RNAs and LRRFIP1 regulate TNF expression. J. Immunol. 2014;192:3057–3067. doi: 10.4049/jimmunol.1302063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rossetto C.C. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14+ monocytes and CD34+ cells. PLoS Pathog. 2013;9:e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]


