Highlights
-
•
Gargle samples were collected from children and tested respiratory viruses.
-
•
In 45(45/200; 22.5%) episodes, some respiratory viruses detected without symptoms.
-
•
Under asymptomatic conditions, detected viruses were mainly RVs and EV/RV untyped.
-
•
PIVs, RSV and hCoV OC43 were detected only when clinical symptom was seen.
-
•
Asymptomatic infections may play an important role in the viral circulation.
Abbreviations: PCR, polymerase chain reaction; PIV, parainfluenza virus; RSV, respiratory syncytial virus; hMPV, human metapneumovirus; EV, enterovirus; RV, rhinovirus; RVA, rhinovirus genogroup A; RVB, rhinovirus genogroup B; RVC, rhinovirus genogroup C; hBoV, human bocavirus; hPeV, human parechovirus; AdV, adenovirus; hCoV, human coronavirus; FluV, influenza virus; RT, reverse transcription
Keywords: Respiratory viruses, Coinfection, Real-time PCR, Healthy children, Gargle specimen
Abstract
Background
Respiratory tract viral infection is one of the most common and important diseases in children. Polymerase chain reaction (PCR) tests are often used to detect viruses in samples, it is difficult to interpret the clinical significance of PCR positivity, which may reflect a past, imminent or active asymptomatic infection due to their high sensitivity. Although single respiratory viruses have been detected in samples from children with symptoms, other respiratory viruses can also be detected simultaneously. However, the clinical importance of these findings for the symptoms is not known.
Objectives
To investigate the prevalence of respiratory viruses among children without any symptoms such as acute respiratory illness and/or fever.
Study design
From week twenty-five 2013 to week twenty-six 2014, gargle samples were collected from children once a week and these samples were subjected to real-time PCR to detect respiratory viruses. On each sampling day, we asked the parents about their children’s health condition.
Results
Among the 286 samples collected, 200 were from asymptomatic children. In the asymptomatic condition, human parechovirus, adenovirus, enterovirus, rhinovirus, coronavirus 229E and HKU1 were observed in 45 episodes. In samples from symptomatic children, parainfluenza viruses, respiratory syncytial virus and coronavirus OC43 were detected in addition to those mentioned above.
Conclusions
Various viruses of different species were detected in the specimens from the children regardless of their health status. It might be speculated that host factors such as the function of the immune system influence the clinical outcome of the infection. However, this needs to be studied further.
1. Background
Respiratory tract viral infection is one of the most common and important disease conditions in children. Recently, PCR based assays have made it possible for novel viruses to be discovered, leading to appraisal of the clinical impacts of these viruses and several other well-known respiratory viruses [1], [2], [3], [4]. Some of these viruses are detected alone in specimens from patients with respiratory symptoms (sometimes in those of inpatients) but their pathogenicity is not clear because they are detected simultaneously with other viruses in many cases [5], [6], [7]. As a result, the clinical importance of these findings for the symptoms is not known.
2. Objectives
In this study, we investigated how often and what respiratory viruses were detected in specimens from asymptomatic children. Gargle specimens (obtained by rinsing the throat with distilled water) were collected from children once a week and the samples were subjected to two-step real-time PCR to detect respiratory viruses. Singleplex real-time PCR procedures were employed for detection of the following 15 respiratory viral pathogens: parainfluenza viruses (PIV) 1–4, respiratory syncytial virus (RSV), human metapneumovirus (hMPV), enterovirus (EV)/rhinovirus (RV), human bocavirus (hBoV), human parechovirus (hPeV), adenovirus (AdV), and human coronaviruses (hCoV) OC43, NL63, 229E, and HKU-1 (Table 1 ), and one-step real-time reverse transcription (RT)–PCR was used for detection of influenza viruses (FluV) A and B (Table 1).
Table 1.
Primers and probes used in this study.
| Virus | Target | Product size(bp) | Specific primers and probes | Detection limit (copy/uL) | Reference |
|---|---|---|---|---|---|
| PIV1 | HN | 135 | Antisense 5′ GTCCTTCCTGCTGGTGTGTTAAT 3′ | 6.55 × 102 | |
| Sense 5′ CCAACCTACAAGGCAACAACATC 3′ | [27] | ||||
| Probe 5′ (FAM)CAAACGATGGCTGAAAA(TAMRA) 3′ | |||||
| PIV3 | HN | 161 | Antisense 5′ TTGTTATAGTGTGTAATGCAGCTCGT 3′ | 5.30 × 102 | |
| Sense 5′ GGGAGCATTGTGTCATCTGTCA 3′ | [27] | ||||
| Probe 5′ (FAM)CCCAGTCATAACTTACTC(TAMRA) 3′ | |||||
| PIV2 | NP | 65 | Antisense 5′ TCYTCAGCTAATGCTTCRAARGC 3′ | 1.0 × 102 | |
| Sense 5′ ATTCCAGATGCTCGATCAACTATG 3′ | [28] | ||||
| Probe 5′ (FAM)AGCACYTCTCCTCTGG(TAMRA) 3′ | |||||
| PIV4 | NP | 123 | Antisense 5′ ATGTGGCCTGTAAGGAAAGCA 3′ | 1.0 × 101 | |
| Sense 5′ CAAAYGATCCACAGCAAAGATTC 3′ | [29] | ||||
| Probe 5′ (FAM)GTATCATCATCTGCCAAATCGGCAATTAAACA(TAMRA) 3′ | |||||
| RSV | F | 89 | Antisense 5′ CGATTTTTATTGGATGCTGTACATTT 3′ | 2.22 × 102 | |
| Sense 5′ AACAGATGTAAGCAGCTCCGTTATC 3′ | [30] | ||||
| Probe 5′(FAM)TGCCATAGCATGACACAATGGCTCCT(TAMRA) 3′ | |||||
| hPMV | M | 152 | Antisense 5′ CATCAGCCYYATCWGTGTTTCTTAAAA 3′ | 2.47 × 102 | |
| Sense 5′ GGCTCCATGCAAATATGAAGTG 3′ | [31] | ||||
| Probe 5′ (FAM)CTAACGAGTGTGCGCAAG(TAMRA) 3′ | |||||
| EV/RV | 5'NTRb | 203 | Antisense 5′ GAAACACGGACACCCAAAGTAGT 3v | Echo 9.76 × 10 | |
| Sense 5′ AGCCTGCGTGGCKGCC 3′ | RVC 2.98 × 102 | [32] | |||
| Probe 5′ (FAM) CTCCGGCCCCTGAATGYGGCTAA(TAMRA) 3′ | |||||
| hBoV | NP-1 | 75 | Antisense 5' TGGACTCCCTTTTCTTTTGTAGGA 3′ | 5.05 × 102 | |
| Sense 5' GCACAGCCACGTGACGAA 3′ | [33] | ||||
| Probe 5′ (FAM)TGAGCTCAGGGAATATGAAAGACAAGCATCG(TAMRA) 3′ | |||||
| hCoV229E | NC | 80 | Antisense 5′ TCTTTTCCACCGTGGCTTTT 3′ | 1.0 × 102 | |
| Sense 5′ CTGCCAAGAGTCTTGCTCGTT 3′ | [28] | ||||
| Probe 5′ (FAM)AGAACAAAAGCATGAAATG(TAMRA) 3′ | |||||
| hCoVNL63 | NC | 61 | Antisense 5′ CGAGGACCAAAGCACTGAATAA 3′ | 1.17 × 102 | |
| Sense 5′ AACCTCGTTGGAAGCGTGTT 3′ | [28] | ||||
| Probe 5′ (FAM)ATTTTCCTCTCTGGTAG(TAMRA) 3′ | |||||
| hCoVOC43 | NC | 67 | Antisense 5′ GCTGAGGTTTAGTGGCATCCTT 3′ | 2.19 × 102 | |
| Sense 5′ GACATGGCTGATCAAATTGCTAGT 3′ | [28] | ||||
| Probe 5′ (FAM)TCTGGCAAAACTTGG(TAMRA) 3′ | |||||
| hCoV HKU | ORF 1a/b | 61 | Antisense 5′ CATTCATTCGCAAGGCGATA 3′ | 1.11 × 102 | |
| Sense 5′ CCCGCAAACATGAATTTTGTT 3′ | [28] | ||||
| Probe 5′ (FAM)AATCTATCACCATGTGAA (TAMRA) 3′ | |||||
| hPeV | 5'NTR | 194 | Antisense 5′ GGCCCCWGRTCAGATCCAYAGT 3′ | 1.0 × 102 | |
| Sense 5′ GTAACASWWGCCTCTGGGSCCAAAAG 3′ | [34] | ||||
| Probe 5′(FAM)CCTRYGGGTACCTYCWGGGCATCCTTC(TAMRA) 3′ | |||||
| AdV(ACDF) | Hexon | 85 | Antisense 5′ AAACTTGTTATTCAGGCTGAAGTACGT3′ | 1.0 × 102 | |
| Sense 5′ CCAGGACGCCTCGGAGTA 3′ | [35] | ||||
| Probe 5′ (FAM)AGTTTGCCCGCGCCACCG(TAMRA) 3′ | |||||
| AdV(BE) | Hexon | 81 | Antisense 5′ CTTGTTCCCCAGACTGAAGTAGGT 3′ | 1.0 × 102 | |
| Sense 5′ GGACAGGACGCTTCGGAGTA 3′ | [35] | ||||
| Probe 5′ (FAM)CAGTTCGCCCGYGCMACAG(TAMRA) 3′ | |||||
| FluV typeA | MP | 149 | Antisense 5′ TGACAGRATYGGTCTTGTCTTTAGCCAYTCCA | 7.5a | |
| Sense 5′ CCMAGGTCGAAACGTAYGTTCTCTCTATC | [36] | ||||
| Probe 5′ (FAM)ATYTCGGCTTTGAGGGGGCCTG(MGB) 3′ | |||||
| FluV | HA | 187 | Antisense 5′ TGTTTCCACAATGTARGACCAT | 6.8a | |
| AH1pdm09 | Sense 5′ AGAAAAGAATGTAACAGTAACACACTCTGT | [36] | |||
| Probe 5′ (FAM)CAGCCAGCAATRTTRCATTTACC(MGB) 3′ | |||||
| FluV AH3 | HA | 178 | Antisense 5′GTCATTGGGRATGCTTCCATTTGG | 7.1a | |
| Sense 5′ CTATTGGACAATAGTAAAACCGGGRGA | [36] | ||||
| Probe 5′ (FAM)AAGTAACCCCKAGGAGCAATTAG(MGB) 3′ | |||||
| FluV B | NS | 105 | Antisense 5′GTKTAGGCGGTCTTGACCAG | 8.2a | |
| Sense 5′ GGAGCAACCAATGCCAC | [37] | ||||
| Probe 5′ (FAM)ATAAACTTTGAAGCAGGAAT(MGB) 3′ |
From reference data.
NTR: non translated region.
3. Study design
3.1. Subjects
Twelve children aged 3–10 years old were enrolled. From week twenty-five 2013 to week twenty-six 2014, throat gargle samples were obtained from the children once a week. Their parents noted the existence of respiratory symptoms (cough, sore throat or nasal mucus) and systemic symptoms (fever or rash) at the time of sampling. Written informed consent was obtained from the parents.
3.2. Molecular analysis
Nucleic acids were extracted from 200 μL specimens using the Magtration System with a MagDEA viral DNA/RNA 200 kit (Precision System Science Co., Ltd., Chiba, Japan) as 50 μL of elution volume. RT reactions were performed using a ReverTra Ace qPCR RT kit (TOYOBO Co., Ltd., Osaka, Japan) following the manufacturer’s instructions. The cDNA was then amplified using Realtime PCR Master Mix (TOYOBO) with a total volume of 25 μL. Each sample was amplified containing primers and probes specific for each of the targets as described in Table 1 [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37]. The sensitivity of each of the real-time PCR methods was evaluated by detecting serial dilutions of quantitated plasmids that contained each target DNA clone. For detection of FluV A and B, we used the one-step real-time RT–PCR method because of its increased sensitivity. Enteroviruses and rhinoviruses were genotyped by direct sequencing. Amplification of the VP4/VP2 region of the enterovirus or rhinovirus for typing was performed with semi-nested RT–PCR as previously described [8]. The purified PCR products were subjected to direct sequencing with a BigDye Terminator v1.1 kit as per the manufacturer’s instructions (Applied Biosystems, CA, USA). Sequence analysis was performed using the DNADynamo program (Blue Tractor Software, UK). Using MEGA5.2 (Tamura et al., 2011, Ver5.2.2), we employed the neighbor-joining method [14] to construct phylogenetic trees from the VP4/VP2 region (420nt) sequences retrieved from GenBank of prototype isolates of each rhinovirus type commonly used in epidemiologic studies of human rhinoviruses [9], [10], [11] and new types proposed previously [9], [12], [13]. Genotypes were assigned on the basis of their clustering with known prototype reference strains.
4. Results
Four children were excluded because of insufficient sampling frequency. For the asymptomatic condition, the criteria were the absence of respiratory symptoms (cough, sniffle or sore throat) and systemic symptoms (fever or rash) from one week before to two days after sampling. Of the 286 samples, 200 were from children who were asymptomatic (Fig. 1 ). When RNA was EV/RV positive by real-time PCR but the viral VP4/VP2 region could not be amplified by semi-nested RT–PCR, we defined it as EV/RV untyped. The threshold cycle (Ct.) of real-time PCR is a relative measure of the concentration of the target in the PCR reaction. If the Ct. value of the EV/RV real-time PCR test is high (over 36.0), the nucleic acids cannot be amplified by the semi-nested PCR used for genotyping (data not shown).
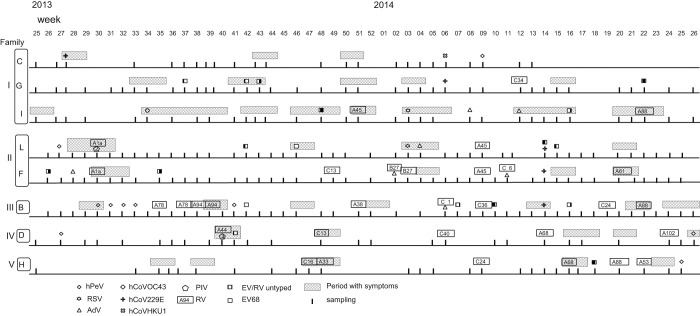
Fig. 1.
Relations between viruses detected in gargle specimens of 8 children and respiratory and/or systemic symptoms. Detected viruses are shown using the symbols noted in the explanatory notes. Vertical lines indicate sampling time. The letters and numbers in rectangles indicate RV genotypes. Children C, G and I are three of four siblings. F and L are a girl and her older brother.
Of the 200 samples, 45 (22.5%) were real-time PCR positive. Four of the 45 positive samples contained two viruses. The prevalence of respiratory viruses among asymptomatic children varied from 9.1% (1/11) to 42.9% (15/35) and that in the symptomatic period ranged from 18.2% (2/11) to 57.1% (4/7) (Table 2 ). The most frequently detected virus was RV genogroup A (RVA) (n = 10) (Table 3 ). EV/RV from 11 samples could not be genotyped. Two of the 4 samples with codetection contained RVC and adenovirus, one RVB and adenovirus, and one EV/RV untyped and hCoV 229E.
Table 2.
Prevalence of respiratory viruses in gargle specimens of children.
| Family | Child | Age (years) | Sex | Total no. of sample | Condition (n) | Prevalence% (positive sample) |
|---|---|---|---|---|---|---|
| I | C | 9 | M | 24 | Asymptomatic (19) | 10.5 (2) |
| Symptomatic (5) | 20.0 (1) | |||||
| G | 6 | F | 38 | Asymptomatic (27) | 14.8 (4) | |
| Symptomatic (11) | 18.2 (2) | |||||
| I | 3 | M | 33 | Asymptomatic (11) | 9.1 (1) | |
| Symptomatic (22) | 31.8 (7) | |||||
| II | L | 6 | M | 44 | Asymptomatic (35) | 14.3 (5) |
| Symptomatic (9) | 44.4 (4) | |||||
| F | 3 | F | 45 | Asymptomatic (36) | 25.0 (9) | |
| Symptomatic (9) | 22.2 (2) | |||||
| III | B | 4 | M | 48 | Asymptomatic (35) | 42.9 (15) |
| Symptomatic (13) | 30.8 (4) | |||||
| IV | D | 3 | M | 27 | Asymptomatic (20) | 20.0 (4) |
| Symptomatic (7) | 57.1 (4) | |||||
| V | H | 5 | F | 27 | Asymptomatic (19) | 26.3 (5) |
| Symptomatic (8) | 37.5 (3) |
Table 3.
Detection of respiratory viruses in gargle specimens of children.
| Virus | Condition | No. of detections(%) |
|---|---|---|
| Enterovirus 68 | Asymptomatic | 1 (0.5) |
| Symptomatic | 1 (1.2) | |
| Human rhinovirus A | Asymptomatic | 10 (5.0) |
| Symptomatic | 10 (11.6) | |
| Human rhinovirus B | Asymptomatic | 2 (1.0) |
| Symptomatic | 0 (0) | |
| Human rhinovirus C | Asymptomatic | 8 (4.0) |
| Symptomatic | 2 (2.3) | |
| Human parechovirus | Asymptomatic | 8 (4.0) |
| Symptomatic | 2 (2.3) | |
| Human coronavirus HKU-1 | Asymptomatic | 1 (0.5) |
| Symptomatic | 0 (0) | |
| Human coronavirus 229 E | Asymptomatic | 3 (1.5) |
| Symptomatic | 2 (2.3) | |
| Human coronavirus OC43 | Asymptomatic | 0 (0) |
| Symptomatic | 1 (1.2) | |
| Parainfluenza virus 2 | Asymptomatic | 0 (0) |
| Symptomatic | 1 (1.2) | |
| Parainfluenza virus 4 | Asymptomatic | 0 (0) |
| Symptomatic | 1 (1.2) | |
| RS virus | Asymptomatic | 0 (0) |
| Symptomatic | 2 (2.3) | |
| Adenovirus | Asymptomatic | 5 (2.5) |
| Symptomatic | 2 (2.3) | |
| EVRV untyped | Asymptomatic | 11 (5.5) |
| Symptomatic | 5 (5.8) |
Human PeV was detected in 8 samples. After hPeV was detected in a sample from a symptomatic child, it was subsequently detected for more than three weeks without any symptoms (Fig. 1, Child B).
In samples from symptomatic children, PIV, RSV and hCoV OC43 were detected in addition to the viruses detected in those from asymptomatic children (27/86; 31.4%). The most commonly detected virus was RVA (10/27; 37.0%). Among the 27 samples, 2 contained PIV and RVA. FluV, hBoV and hMPV were not detected.
5. Discussion
Gargle specimens from 8 children were collected once a week and the samples were subjected to real-time PCR to detect respiratory viruses. RVs and EV/RV untyped were the viruses most frequently detected in samples from asymptomatic children. Current diagnosis of respiratory infections is mainly done using PCR methods. Due to their high sensitivity, it is difficult to determine the exact explanation for positivity in individual participants (e.g., post-viral shedding, asymptomatic infection, or incubation before symptomatic infection). We were able to clarify the active asymptomatic infection by testing gargle specimens of the same children once a week for one year.
RVs are most commonly isolated from persons experiencing mild upper respiratory illness (common cold). Recent studies have reported that those viruses are responsible for severe infections of the lower respiratory tract in children. These viruses play a critical role in exacerbating asthma and chronic lung diseases [15], [16]. However, most studies were conducted with symptomatic patients. Few studies have investigated the existence of the viruses in children without any respiratory symptoms [17], [18]. One study reported that, after the onset of symptomatic respiratory infection, rhinovirus RNA may take a long time (5–6 weeks) to disappear from nasal mucus [19]. In this study, the children who could gargle might have been relatively older, but RVs were often detected in their throats at a time without symptoms. It seems that RV infection is in most cases asymptomatic or mild. As the sensitivity of the real-time PCR was 100 copies, it can be assumed that the virus might have replicated to some extend. The same RV genotype was detected in two consecutive samples of a child and another RV genotype was detected in the next sample. These findings suggest that RVs do not exist in the upper respiratory tract for a long time even if a child does not show symptoms which were probably the result of interferon response to a virus multiplication.
HPeV was also detected in samples from asymptomatic children. Recent studies have investigated the involvement of hPeVs in respiratory diseases, reporting a low frequency of detection and a lack of clear disease association. In addition to a low hPeV prevalence in respiratory samples, a high rate of coinfection with other respiratory viruses has been observed in hPeV-positive samples [1], [20]. With monthly sampling, hPeV was detected in the stools of 48% of healthy Finnish infants by the age of 22 months [21]. In this study, the duration of parechovirus shedding in gargle specimens was calculated to be 3 weeks after the disappearance of the respiratory symptoms.
On the other hand, for PIVs, RSV, and hCoV OC43, which were detected only when clinical symptoms were seen, it is thought that, if these viruses grow in the airway, certain host reactions such as respiratory symptoms or fever will be triggered [22], [23], [24], [25], [26].
FluV, hBoV, hMPV and hCoV NL63 were not detected during the study period, probably because the children in this study did not live in a viral epidemic area.
Since various viruses were detected in the children regardless of their health condition, it might be speculated that the clinical outcome of the respiratory viral infection is affected predominantly such as the function of immune system. Most respiratory viruses infect the upper or lower airway and replicate in airway epithelial cells. In patients with normal immunity, these viruses are cleared immediately and it is generally thought that prolonged infection is rare. Therefore these respiratory viruses must repeat human-to-human transmission to continue to be present in the human population. As PCR is a nucleic acid amplification method, it remains unknown whether the respiratory viruses detected in the specimens from asymptomatic children are infective or not. Respiratory viral infection without any symptoms may play an important role in the viral circulation in human populations.
Competing interest
None declared.
Ethical approval
This study was approved by the Osaka Prefectural Institute of Public Health ethical committee (No. 1302-05-01).
Acknowledgments
The authors would like to thank Maki Otsuka for helpful technical assistance in amplification and Kim Barrymore for editing the manuscript. This work was supported by Grant-in-Aid for Scientific Research (C) Grant number 24590839 from the Japan Society for the Promotion of Science.
Contributor Information
Saeko Morikawa, Email: morikawa@iph.pref.osaka.jp.
Satoshi Hiroi, Email: hiroi@iph.pref.osaka.jp.
Tetsuo Kase, Email: kasetetsuo@iph.pref.osaka.jp.
References
- 1.Debiaggi M., Canducci F., Ceresola E.R., Clementi M. The role of infections and co-infections with newly identified and emerging respiratory viruses in children. Virol. J. 2012;9:247. doi: 10.1186/1743-422X-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali S.A., Gern J.E., Hertert T.V., Edwards K.M., Griffin M.R., Miller E.K. Real-world comparison of two molecular methods for detection of respiratory viruses. Virol. J. 2011;8:332. doi: 10.1186/1743-422X-8-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Hoogen B.G., de jong J.C., Groen J., Kuiten T., de Groot R., Fouchier R.A., Osterhaus A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001;7:719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prill M.M., Iwane M.K., Edward K.M., Williams J.V., Weinberg G.A., Staat M.A. Human coronavirus in young children hospitalized for acute respiratory illness and asymptomatic controls. Pediatr. Infect. Dis. J. 2012;31(3):235–240. doi: 10.1097/INF.0b013e31823e07fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Linstow M.L., Hogh M., Hogh B. Clinical and epidemiologic characteristics of human bocavirus in Danish infants: results from a prospective birth cohort study. Pediatr. Infect. Dis. J. 2008;27:897–902. doi: 10.1097/INF.0b013e3181757b16. [DOI] [PubMed] [Google Scholar]
- 7.Jin Y., Yuan X.H., Xie Z.P., Gao H.C., Song J.R., Zhang R.F. Prevalence and clinical characterization of a newly identified human rhinovirus C species in children with acute respiratory tract infections. J. Clin. Microviol. 2009;47:2895–2900. doi: 10.1128/JCM.00745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishiko H., Shimada Y., Yonaha M., Hashimoto O., Hayashi A., Sakae K., Takeda N. Molecular diagnosis of human enteroviruses by phylogeny-based classification by use of the VP4 sequence. J. Infect. Dis. 2002;185(6):744–754. doi: 10.1086/339298. [DOI] [PubMed] [Google Scholar]
- 9.Mclntyre C.L., Knowles N.J., Simmonds P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J. Gen. Virol. 2013;94:1791–1806. doi: 10.1099/vir.0.053686-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henquell C., Mirand A., Deusebis A.L., Regagnon C., Archimbaud C., Chambon M. Prospective genotyping of human rhinoviruses in children and adults during the winter of 2009–2010. J. Clin. Virol. 2012;53:280–284. doi: 10.1016/j.jcv.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Kaida A., Kubo H., Takakura K., Togawa M., Shiomi M., Koudera U., Iritani N. Molecular epidemiology of human rhinovirus C in patients with acute respiratory tract infections in Osaka City, Japan. Jpn. J. Infect. Dis. 2011;64:488–492. [PubMed] [Google Scholar]
- 12.Simmonds P., Mclntyre C., Savolainen-kopra C., Tapparel C., Mackay I.M., Hovi T. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J. Gen. Virol. 2010;91:2409–2419. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- 13.Picornaviridae Study Group. http://www.picornaviridae.com (accessed 02.09.14.).
- 14.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Imakita M., Shiraki K., Yutani C., Ishibashi-Umeda H. Pneumonia caused by rhinovirus. Clin. Infect. Dis. 2000;30:611–612. doi: 10.1086/313723. [DOI] [PubMed] [Google Scholar]
- 16.Jackson D.J., Gangnon R.E., Evans M.D., Roberg K.A., Anderson E.L., Pappas T.E. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care. Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen R.R., Wieringa J., Koekkoek S.M., Visser C.E., Pajkrt D., Molenkamp R. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J. Clin. Microviol. 2011;49(7):2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camargo C.N., Carraro E., Granato C.F., Bellei N. Human rhinovirus infections in symptomatic and asymptomatic subjects. Braz. J. Microbiol. 2012;43(4):1641–1645. doi: 10.1590/S1517-838220120004000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jartti T., Lehtinen P., Vuorinen T., Koskenvuo Ruuskanen O. Persistence of rhinovirus and enterovirus RNA after acute respiratory illness in children. J. Med. Virol. 2004;72:695–699. doi: 10.1002/jmv.20027. [DOI] [PubMed] [Google Scholar]
- 20.Harvala H., Robertson I., McWilliam Leitch E.C., BenschopK. Wolthers K.C., Templeton K., Simmonds P. Epidemiology and clinical associations of human parechovirus respiratory infections. J. Clin. Microbiol. 2008;46(10):3446–3453. doi: 10.1128/JCM.01207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolehmainen P., Oikarinen S., Koskiniemi M., Simell O., Ilonen J., Knip M., Hyöty H., Tauriainen S. Human parechoviruses are frequently detected in stool of healthy Finnish children. J. Clin. Virol. 2012;54(2):156–161. doi: 10.1016/j.jcv.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Hara M., Takao S., Shimazu Y., Nishimura T. Three-year study of viral etiology and features of febrile respiratory tract infections in Japanese pediatric outpatients. Pediatr. Infect. Dis. J. 2014;33:687–692. doi: 10.1097/INF.0000000000000227. [DOI] [PubMed] [Google Scholar]
- 23.Brittain-Long R., Andersson L.M., Olofsson S., Lindh M., Westin J. Seasonal variations of 15 respiratory agents illustrated by the application of a multiplex polymerase chain reaction assay. Scand. J. Infect. Dis. 2012;44:9–17. doi: 10.3109/00365548.2011.598876. [DOI] [PubMed] [Google Scholar]
- 24.Kusel M.M., de Klerk N.H., Holt P.G., Kebadze T., Johnston S.L., Sly P.D. Role of respiratory viruses in acute upper and lower respiratory tract illness in the first year of life: a birth cohort study. Pediatr. Infect. Dis. J. 2006;25:680–686. doi: 10.1097/01.inf.0000226912.88900.a3. [DOI] [PubMed] [Google Scholar]
- 25.Fairchok M.P., Martin E.T., Chambers S., Kuypers J., Behrens M., Braun L.E., Englund J.A. Epidemiology of viral respiratory tract infections in a prospective cohort of infants and toddlers attending daycare. J. Clin. Virol. 2010;49:16–20. doi: 10.1016/j.jcv.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishaupt J.O., Russcher A., Smeets L.C., Versteegh F.G.A., Hartwig N.G. Clinical impact of RT–PCR for pediatric acute respiratory infections: a controlled clinical trial. Pediatrics. 2011;128:e1113–e1120. doi: 10.1542/peds.2010-2779. [DOI] [PubMed] [Google Scholar]
- 27.Templeton K.E., Scheltinga S.A., Beersma M.F.C., Kroes A.C.M., Claas E.C.J. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses: respiratory syncytial virus, and parainfluenza viruses 1,2,3, and 4. J. Clin. Microbiol. 2004;42(4):1564–1569. doi: 10.1128/JCM.42.4.1564-1569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chidlow G.R., Harnett G.B., Shellam G.R., Smith D.W. An economical tandem multiplex real-time PCR technique for the detection of a comprehensive range of respiratory pathogens. Viruses. 2009;1:42–56. doi: 10.3390/v1010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Pol A.C., van Loon A.M., Wolfs T.F.W., Jansen N.J.G., Nijhuis M., Breteler E.K., Schuurman R., Rossen J.W.A. Increased detection of respiratory syncytial virus: parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J. Clin. Microbiol. 2007;45(7):2260–2262. doi: 10.1128/JCM.00848-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mentel R., Wegner U., Bruns R., Gürtler L. Real-time to improve the diagnosis of respiratory syncytial virus infection. J. Med. Microbiol. 2003;52:893–896. doi: 10.1099/jmm.0.05290-0. [DOI] [PubMed] [Google Scholar]
- 31.Reymond F., Carbonneau J., Boucher N., Robitaille L., Boisvert S., Wu W. Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J. Clin. Microbiol. 2009;47(3):743–750. doi: 10.1128/JCM.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tapparel C., Cordey S., Belle S.V., Turin L., Lee W., Regamey N. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J. Clin. Microbiol. 2009;47(6):1742–1749. doi: 10.1128/JCM.02339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neske F., Blessing K., Tollmann F., Schubert J., Rethwilm A., Kreth H.W., Real-time Weissbrich B. for diagnosis of human bocavirus infections and phylogenetic analysis. J. Clin. Microbiol. 2007;45(7):2116–2122. doi: 10.1128/JCM.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nix W., Maher K., Johansson E.S., Niklasson B., Lindberg A.M., Pallansch M.A., Oberste M.S. Detection of all known parechoviruses by real-time PCR. J. Clin. Microbiol. 2008;46(8):2519–2524. doi: 10.1128/JCM.00277-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong S., Pabbaraju K., Pang X.L., Lee B.E., Fox J.D. Detection of a broad range of human adenoviruses in respiratory tract samples using a sensitive multiplex real-time PCR assay. J. Med. Virol. 2008;80(5):856–865. doi: 10.1002/jmv.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakauchi M., Yasui Y., Miyoshi T., Minagawa H., Tanaka T., Tashiro M., Kageyama T. One-step real-time reverse transcription-PCR assays for detecting and subtyping pandemic influenza A/H1N1 2009, seasonal influenza A/H1N1, and seasonal influenza A/H3N2 viruses. J. Virol. Methods. 2011;171:156–162. doi: 10.1016/j.jviromet.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakauchi M., Takayama I., Takahashi H., Oba K., Kubo H., Kaida A., Tashiro M., Kageyama T. Real-time RT–PCR assays for discriminating influenza B virus Yamagata and Victoria lineages. J. Virol. Methods. 2014;205:110–115. doi: 10.1016/j.jviromet.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]