Highlights
-
•
No commercial HEV RNA assay is validated for use in faecal samples.
-
•
Monitoring HEV faecal excretion is recommended for managing chronic HEV infection in solid-organ transplant recipients.
-
•
We evaluated the Altona assay by testing patients on ribavirin therapy.
Keywords: Hepatitis E virus, HEV RNA assay, Manual extraction, Automated extraction, Faeces
Abstract
Background
Detecting hepatitis E virus (HEV) RNA in faeces is useful for diagnosing and monitoring HEV infections, particularly in immunocompromised patients requiring ribavirin therapy.
Objectives
This study evaluated the performance of the Altona RealStar HEV RNA kit for detecting and quantifying HEV in faeces.
Study design
RNA was extracted from 94 stool samples by two methods: QIAamp Viral RNA Mini kit and MagNA Pure 96 automate. The Altona results were compared to a reference laboratory-developed accredited ISO15189 RT-PCR assay.
Results
The Altona and reference assays detect HEV RNA in 77/93 (82.8%) and 83/93 (89.2%) of the QIAamp extracted samples, respectively, after exclusion of invalid result; they detected HEV RNA in 67/92 (72.8%) and 66/92 (71.7%) of the MagNA Pure extracted samples, respectively, which emphasizes the importance of the RNA extraction method. The HEV RNA concentrations obtained with Altona RT-PCR and the reference RT-PCR were well correlated whatever the extraction method, and Bland Altman analyses indicated that the Altona values were higher than the reference assay values. The Altona values for QIAamp-extracted and MagNA Pure-extracted HEV RNA were very similar.
Conclusions
The Altona RealStar assay is suitable for quantifying HEV RNA in the faeces and monitoring HEV RNA shedding during ribavirin therapy. Extraction is critical for detecting faecal HEV with high performance RT-PCR assays.
1. Background
Over the last 10 years, it has become apparent that hepatitis E virus (HEV) is a pathogen of global significance. HEV infection is among the most frequent causes of acute hepatitis worldwide [1]. The HEV strains infecting humans are classified as one of 4 major genotypes. Peak viremia occurs during the incubation period and early phase of disease In patients with an acute HEV infection [2]. Viral RNA can be detected in the blood and faeces of these patients just before the onset of clinical symptoms. It does not persist in the blood, becoming undetectable about 3 weeks later, the onset of symptoms while remaining in faeces for longer [1].
Chronic HEV infections have been reported in solid-organ transplant recipients [[3], [4], [5]], hematology patients that given chemotherapy [[6], [7], [8]], and patients infected by human immunodeficiency virus (HIV) that have low CD4 counts [9,10]. Chronic HEV infection is defined by persistent HEV replication for more than three months [11,12]. These chronic infections are caused by genotype 3 and genotype 4 and may rapidly evolve to cirrhosis and loss of a liver graft [13,14]. Ribavirin has become the drug of choice for treating chronic HEV infections in immunocompromised [15,16]. While a decrease in HEV RNA in blood of 0.5 log copies/ml or greater by 7 days after initiation of ribavirin therapy is predictive of a response [17], detecting HEV RNA in the faeces despite a negative HEV RNA in blood is a strong predictor of a relapse in immunocompromised subjects [18]. Therefore, monitoring HEV faecal excretion could help determine the optimal duration of ribavirin therapy and has been recommended for managing chronic HEV infections in solid-organ transplant recipients [19]. However, the presence of PCR inhibitors in this particular matrix can make detecting HEV RNA difficult [[20], [21], [22]]. No commercial HEV RNA assay is presently validated for faecal samples.
2. Objectives
We evaluated the performance of two RNA extraction methods and that of a commercial assay, the Altona RealStar® HEV RT-PCR Kit, compared to a laboratory-developed test (LDT), for detecting and quantifying HEV RNA in the faeces. We determined the quantitative analytical performance of the Altona assay in two clinical setting: HEV shedding during acute hepatitis E and monitoring ribavirin therapy.
3. Study design
3.1. Patients and samples
We tested in our laboratory 94 faecal samples from patients with an HEV infection assessed by detecting HEV RNA in their blood with our ISO 15189-accredited assay LDT. All patients were followed in Toulouse University Hospital. Ten samples were collected at the acute phase of infection (<one month after the increase in ALT) from 10 untreated patients and 84 were from 13 solid-organ-transplant recipients with chronic HEV genotype 3 between 2011 and 2016. These 13 transplant recipients were treated with ribavirin monotherapy for 3 months and several faecal samples were collected during (n = 71) and after (n = 13) treatment. A sustained virological response was defined as a negative blood HEV RNA 6 months after treatment and relapse was defined as a positive blood HEV RNA after treatment withdrawal.
Faecal samples (50–100 mg) were weighted and suspended in 5 mL of Minimum Essential Eagle’s Medium (MEM, Sigma-Aldricht, France), vortexed, frozen at −20 °C for 24 h and then centrifuged at 3000 rpm for 10 min. The resulting supernatants were passed through a 45 μm filter and the filtrate was used for HEV RNA extraction and quantification by real time RT-PCR [18].
This non-interventional study included no additional procedures. Biological material and clinical data were obtained only for standard viral diagnosis following physicians’ orders (no specific sampling, no modified sampling protocol, no questions in addition to the national standardized questionnaire). Data were analysed using an anonymized database. The French Public Health law (CSP Art L 1121-1.1) does not require written informed consent for such a protocol.
3.2. Nucleic acid extraction
HEV RNA was extracted from the filtered faecal samples by 2 methods: the QIAamp Viral RNA Mini kit (Qiagen, Hilden, Germany), in which 140 μl of filtered-faeces were eluted in 50 μl according to the manufacturer’s instructions and the DNA and viral nucleic acid Small Volume 2.0 kit (Roche Diagnostics, Mannheim, Germany) implemented on the MagNA Pure 96 automate (Roche Diagnostics, Rotkreuz, Switzerland) with the Viral NA Universal Small Volume 3.1 protocol (200 μl of filtered- faeces eluted in 100 μl).
3.3. Detection and quantification
The HEV RNA in each eluate was quantified using an ISO 15189-accredited LDT reference RT-PCR assay [23]. This LDT has no internal control. It was also quantified using the Altona RealStar® HEV RT-PCR version 1 kit commercial assay. The internal control in the Altona RT-PCR kit was added during the RT-PCR mix preparation. The HEV RNA quantified by both assays was calculated with a standard curve generated from serial 10-fold dilutions of a transcribed RNA standard [23]. The limit of quantification for both assays was 100 copies/g.
3.4. Statistical analyses
HEV RNA values were log transformed and then analysed with GraphPad Prism 7 software. The Spearman test was used to test for correlation between the two assays. P values of less than 0.05 were taken to indicate statistical significance. A Bland-Altman analysis (a scatter plot of the differences between the paired measurements plotted against their means) was used on samples that were positive by both methods to assess the magnitude of disagreement between them and estimate the overall bias.
4. Results
4.1. Performance of the two RT-PCR assays using the QIAamp extraction method
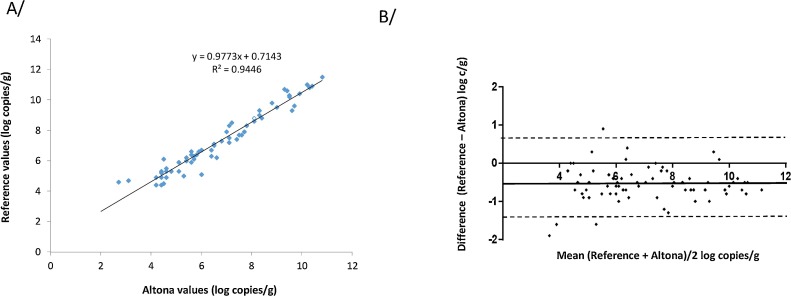
The 94 samples were tested with Altona assay and with the reference RT-PCR. The Altona internal control was negative in one sample, indicating inhibition of the RT-PCR; this sample tested positive with the reference assay (HEV RNA: 10.5 log copies/g). This sample was excluded from the analysis. The Altona RT-PCR detected HEV RNA in 77/93 (82.8%) samples and the reference RT-PCR detected HEV RNA in 83/93 (89.2%, p = 0.19) (Table 1 ). The 6 samples that tested negative with Altona assay tested positive with the reference assay (HEV RNA: 3.2 to 10.5 log copies/g). Thus, the assays were concordant for 87/93 (93.5%) faecal samples: 77 samples tested positive and 10 were negative with both assays. (Table 1). The Altona RT-PCR and reference RT-PCR HEV RNA concentrations were correlated (ρ = 0.90, p < 0.001) (Fig. 1 A). The Altona assay values were higher than the reference assay values. Bland-Altman analysis (Fig. 1B) produced a mean deviation between the assay results of -0.5 log10 c/g and a difference of > 0.5 log copies/g for 37 samples.
Table 1.
Detection of HEV RNA in faeces using the QIAamp extraction.
| Reference RT-PCR |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Altona RT-PCR | Positive | 77 | 0 | 77 |
| Negative | 6 | 10 | 16 | |
| Total | 83 | 10 | 93 | |
Fig. 1.
A/ Correlation between the Altona RT-PCR and reference RT-PCR assays after QIAamp extraction. B/ Bland Altman plot for bias analysis between the Altona RT-PCR and reference RT-PCR assays using QIAamp extraction. The solid line indicates the mean difference and dashed lines indicated upper and lower 95% limits of agreement (mean + 1.96 SD, mean − 1.96 SD; SD: standard deviation of the mean difference).
4.2. Performance of the two RT-PCR assays using the MagNA pure 96 extraction method
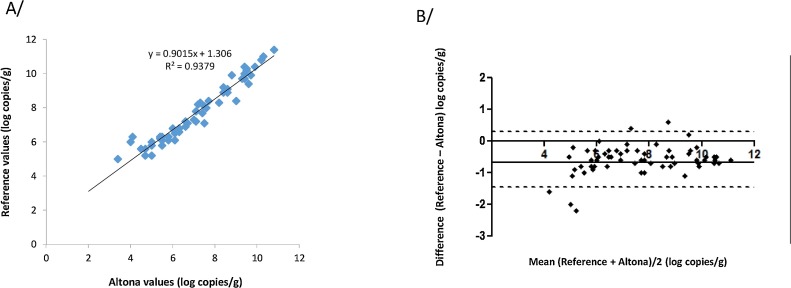
The Altona assay found 2 samples with negative internal controls; the reference test found one of these negative and the other positive (HEV RNA: 3.6 log copies/g). These samples were excluded from the analysis. The Altona RT-PCR detected HEV RNA in 67/92 (72.8%) samples and the reference assay detected HEV RNA in 66/92 (71.7%). Thus, the RT-PCR assays were concordant for 89/92 (96.7%) samples: 65 tested positive and 24 were negative with both assays. (Table 2 ). The 2 samples that were positive with the Altona assay (HEV RNA: 5 and 6 log copies/g) were negative with the reference assay. The Altona assay negative sample was positive with the reference assay (HEV RNA: 3.6 copies/g). The HEV RNA concentrations obtained with Altona RT-PCR and the reference RT-PCR were correlated (ρ = 0.98, p < 0.001) (Fig. 2 A) but the Altona assay values were higher than those from the reference assay. Bland-Altman analysis (Fig. 2B) gave a mean deviation between the reference RT-PCR and Altona results of -0.7 log10 c/g and a bias of > 0.5 log copies/g for 23 samples.
Table 2.
Detection of HEV RNA in faeces using the MagNa Pure extraction.
| Reference RT-PCR |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Altona RT-PCR | Positive | 65 | 2 | 67 |
| Negative | 1 | 24 | 25 | |
| Total | 66 | 26 | 92 | |
Fig. 2.
A/ Correlation between the Altona RT-PCR and reference RT-PCR assays after MagNA Pure extraction. B/ Bland Altman plot for bias analysis between the two assays using MagNA Pure extraction. The solid line indicates the mean difference and dashed lines indicated upper and lower 95% limits of agreement (mean + 1.96 SD, mean − 1.96 SD; SD: standard deviation of the mean difference).
4.3. The influence of extraction on Altona assay HEV RNA quantification
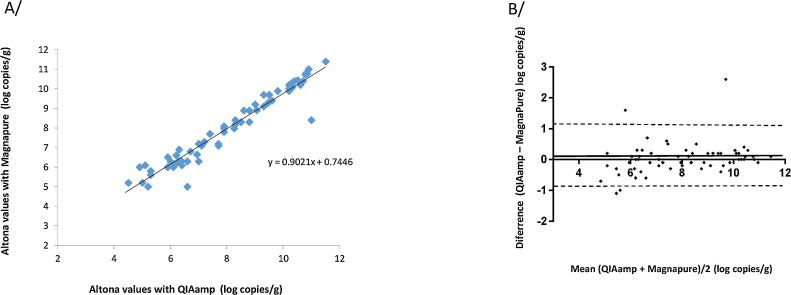
HEV RNA was detected more frequently (82.7%) after QIAamp extraction than after MagNA Pure extraction (72.8%) but the difference was not significant (p = 0.11). The HEV RNA concentrations obtained with the Altona RT-PCR after QIAamp extraction and MagNA Pure extraction were correlated (ρ = 0.96, p < 0.001) (Fig. 3 A). The Altona/QIAamp values and the Altona/MagNA Pure values were also similar. The mean deviation between them was 0.03 log10 c/g (Fig. 3B) and the bias was > 0.5 log copies/g for 9 samples.
Fig. 3.
Correlation between the Altona RT-PCR results obtained using QIAamp extraction and the MagNA Pure extraction. B/ Bland Altman plot for bias analysis between the Altona results obtained after QIAamp or MagNA Pure extraction. The solid line indicates the mean difference and dashed lines indicated upper and lower 95% limits of agreement (mean + 1.96 SD, mean − 1.96 SD; SD: standard deviation of the mean difference).
4.4. HEV RNA shedding in clinical setting
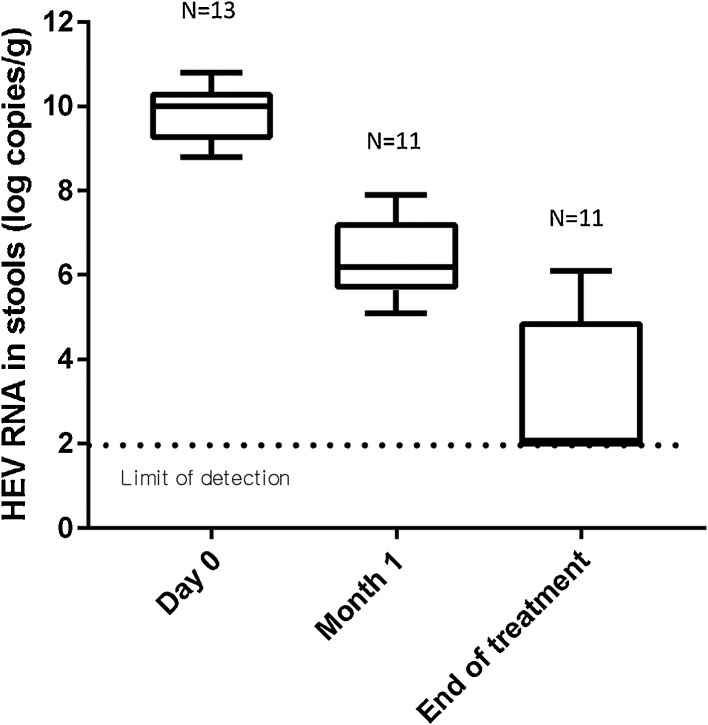
The Altona RT-PCR assay found the median HEV RNA concentration in 10 faeces samples from untreated patients to be 8.4 log10 copies/g faeces. The plasma HEV RNA concentration in the 13 patients treated with ribavirin decreased and that viremia became undetectable in 6 patients after one month of ribavirin therapy and in 7 patients after 2 months of ribavirin therapy. Their viremia remained undetectable until the end of treatment. The Altona assay found that the faecal shedding of HEV RNA decreased during the 3-months course of ribavirin (Fig. 4 ). Patients testing positive for HEV RNA at the end of treatment relapsed.
Fig. 4.
Box plot of HEV RNA shedding during ribavirin therapy using Altona assay.
5. Discussion
Our evaluation of the Altona Real Star RT-PCR kit for detecting and quantifying HEV RNA in faeces after each of two RNA extraction methods indicated that the Altona assay performed well and that its findings were correlated with those of the reference RT-PCR test.
We have previously investigated the performance of the Altona RT-PCR assay for detecting HEV RNA in the plasma [24]. Its analytical sensitivity was good and it was highly reproducible [24]. The present study evaluates the assay for detecting HEV RNA in faeces. While the values obtained with the Altona and reference assays were highly correlated, the reference assay gave lower HEV RNA values regardless of the extraction method. The Altona Kit may amplify the targeted region of the genome more efficiently than the reference assay.
Both RT-PCR assays found more positive samples after QIAamp extraction than after MagNA Pure treatment, although the difference was not significant. Clearly, the RNA extraction method is most important. Many PCR inhibitors are presumed to affect directly the RNA or the enzymes of the reaction [20,25]. Several inhibitors have been found in faeces: polysaccharides or chlorophyll originating from vegetables, bile salts, urea, glycolipids and heparin [[26], [27], [28]]. The QIAamp kit has been validated for extracting viruses from food [29,30] and this study indicates that it is suitable for extracting HEV RNA from faeces. The efficiency with which the RNA extraction method removes inhibitors or debris and produces nucleic acid extracts of high purity significantly affects the detection of the target by RT-PCR [20,29,31]. A study that evaluated six commercial extraction platforms for detecting SARS Coronavirus frm faeces indicated that the MagNA Pure method performed less well than other automated platform that used magnetic beads (miniMag by bioMérieux or the Magazorb by Cortex Biochem) [31]. Inhibitors can also be removed from faecal samples by sample dilution or using less faeces. This was successful when the Altona retested 2 negative samples. The Altona assay has an internal control that is introduced into the RT-PCR mix and thus acts as an amplification control (controlling only the performance of the PCR itself). A limitation of the reference LDT assay is the absence of an internal control. Nevertheless, a negative internal control was not always seen in results that were HEV RNA-positive with the reference RT-PCR and HEV RNA-negative with Altona RT-PCR. Moreover, the Altona assay package insert indicates that the internal control can also be placed in the lysis buffer during extraction. This may evaluate inhibition better in challenging matrice such as faeces.
We also measured virus shedding into the faeces in both treated and untreated patients. The median HEV RNA concentration for untreated patients, was 8.4 log10 copies/g faeces. The HEV RNA concentration in faeces is similar to the shedding of other enteric viruses. Sabria et al. reported virus concentrations of 7.5–6.5 log10 copies/g in symptomatic and asymptomatic patients with norovirus infections [32]. This high faecal HEV RNA load is in keeping with a recent observation on HEV-infected human-liver chimeric mice where the HEV RNA concentration is about 10-100-fold higher than in corresponding plasma samples [33].
We find that ribavirin therapy gradually reduced the HEV RNA concentration in the faeces, but that HEV was shed into the faeces for longer than the HEV viremia persists in these patients [18]. Our data indicate that the Altona RT-PCR assay can be used to follow the dynamics of HEV shedding into faeces. We have extended the treatment of those patients who still had HEV in their faeces at the end of the 3-months ribavirin therapy. Monitoring HEV faecal excretion is recommended for determining the optimal duration of ribavirin therapy [11,19]. While a qualitative assay could be sufficient, a validated quantitative assay can assess the decline and residual faecal virus shedding during ribavirin therapy.
We conclude that the Altona RT-PCR assay is suitable for quantifying HEV RNA in the faeces, particularly for monitoring HEV RNA shedding during ribavirin therapy. As extraction may influence HEV RNA detection in faeces sample, each laboratory should validate their extraction protocol for this matrice.
Author’s contribution
Florence Abravanel, Sébastien Lhomme and Jacques Izopet: designed the study, wrote the manuscript.
Jean-Marie Peron, Laurent Alric, Nassim Kamar: designed the study, followed the patients, reviewed the manuscript.
Audrey Lacipière, Martine Dubois, Luce Minier: performed virological analyses.
Funding
The RealStar® HEV RT-PCR kits were kindly provided by Altona. The National Reference Center for Hepatitis E is supported by a grant from the French Public Health authorities.
Conflict of interest
None to declare.
References
- 1.Kamar N., Dalton H.R., Abravanel F., Izopet J. Hepatitis E virus infection. Clin. Microbiol. Rev. 2014;27(1):116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamar N., Bendall R., Legrand-Abravanel F., Xia N.S., Ijaz S., Izopet J. Hepatitis E. Lancet. 2012;379(9835):2477–2488. doi: 10.1016/S0140-6736(11)61849-7. [DOI] [PubMed] [Google Scholar]
- 3.Gerolami R., Moal V., Colson P. Chronic hepatitis E with cirrhosis in a kidney-transplant recipient. N. Engl. J. Med. 2008;358(8):859–860. doi: 10.1056/NEJMc0708687. [DOI] [PubMed] [Google Scholar]
- 4.Halac U., Beland K., Lapierre P., Patey N., Ward P., Brassard J. Chronic hepatitis E infection in children with liver transplantation. Gut. 2012;61(4):597–603. doi: 10.1136/gutjnl-2011-300708. [DOI] [PubMed] [Google Scholar]
- 5.Kamar N., Selves J., Mansuy J.M., Ouezzani L., Peron J.M., Guitard J. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N. Engl. J. Med. 2008;358(8):811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 6.Halac U., Beland K., Lapierre P., Patey N., Ward P., Brassard J. Cirrhosis due to chronic hepatitis E infection in a child post-bone marrow transplant. J. Pediatr. 2012;160(5):871–874. doi: 10.1016/j.jpeds.2012.01.028. e1. [DOI] [PubMed] [Google Scholar]
- 7.Ollier L., Tieulie N., Sanderson F., Heudier P., Giordanengo V., Fuzibet J.G. Chronic hepatitis after hepatitis E virus infection in a patient with non-hodgkin lymphoma taking rituximab. Ann. Intern. Med. 2009;150(6):430–431. doi: 10.7326/0003-4819-150-6-200903170-00026. [DOI] [PubMed] [Google Scholar]
- 8.Tavitian S., Peron J.M., Huguet F., Kamar N., Abravanel F., Beyne-Rauzy O. Ribavirin for chronic hepatitis prevention among patients with hematologic malignancies. Emerg. Infect. Dis. 2015;21(8):1466–1469. doi: 10.3201/eid2108.150199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalton H.R., Bendall R.P., Keane F.E., Tedder R.S., Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N. Engl. J. Med. 2009;361(10):1025–1027. doi: 10.1056/NEJMc0903778. [DOI] [PubMed] [Google Scholar]
- 10.Kenfak-Foguena A., Schoni-Affolter F., Burgisser P., Witteck A., Darling K.E., Kovari H. Hepatitis E virus seroprevalence and chronic infections in patients with HIV. Switzerland Emerg. Infect. Dis. 2011;17(6):1074–1078. doi: 10.3201/eid1706.101067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Association for the Study of the Liver Electronic address eee, European association for the study of the L. EASL clinical practice guidelines on hepatitis E virus infection. J. Hepatol. 2018;68(6):1256–1271. doi: 10.1016/j.jhep.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Kamar N., Rostaing L., Legrand-Abravanel F., Izopet J. How should hepatitis E virus infection be defined in organ-transplant recipients? Am. J. Transplant. 2013;13(7):1935–1936. doi: 10.1111/ajt.12253. [DOI] [PubMed] [Google Scholar]
- 13.Kamar N., Mansuy J.M., Cointault O., Selves J., Abravanel F., Danjoux M. Hepatitis E virus-related cirrhosis in kidney- and kidney-pancreas-transplant recipients. Am. J. Transplant. 2008;8(8):1744–1748. doi: 10.1111/j.1600-6143.2008.02286.x. [DOI] [PubMed] [Google Scholar]
- 14.Schlosser B., Stein A., Neuhaus R., Pahl S., Ramez B., Kruger D.H. Liver transplant from a donor with occult HEV infection induced chronic hepatitis and cirrhosis in the recipient. J. Hepatol. 2011;56(2):500–502. doi: 10.1016/j.jhep.2011.06.021. [DOI] [PubMed] [Google Scholar]
- 15.Kamar N., Izopet J., Tripon S., Bismuth M., Hillaire S., Dumortier J. Ribavirin for chronic hepatitis E virus infection in transplant recipients. N. Engl. J. Med. 2014;370(12):1111–1120. doi: 10.1056/NEJMoa1215246. [DOI] [PubMed] [Google Scholar]
- 16.Kamar N., Rostaing L., Abravanel F., Garrouste C., Lhomme S., Esposito L. Ribavirin therapy inhibits viral replication on patients with chronic hepatitis e virus infection. Gastroenterology. 2010;139(5):1612–1618. doi: 10.1053/j.gastro.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Kamar N., Lhomme S., Abravanel F., Cointault O., Esposito L., Cardeau-Desangles I. An early viral response predicts the virological response to ribavirin in hepatitis E virus organ transplant patients. Transplantation. 2015;99(10):2124–2131. doi: 10.1097/TP.0000000000000850. [DOI] [PubMed] [Google Scholar]
- 18.Abravanel F., Lhomme S., Rostaing L., Kamar N., Izopet J. Protracted fecal shedding of HEV during ribavirin therapy predicts treatment relapse. Clin. Infect. Dis. 2015;60(1):96–99. doi: 10.1093/cid/ciu742. [DOI] [PubMed] [Google Scholar]
- 19.McPherson S., Elsharkawy A.M., Ankcorn M., Ijaz S., Powell J., Rowe I. Summary of the British Transplantation Society UK guidelines for hepatitis E and solid organ transplantation. Transplantation. 2018;102(1):15–20. doi: 10.1097/TP.0000000000001908. [DOI] [PubMed] [Google Scholar]
- 20.Schrader C., Schielke A., Ellerbroek L., Johne R. PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol. 2012;113(5):1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 21.Esona M.D., McDonald S., Kamili S., Kerin T., Gautam R., Bowen M.D. Comparative evaluation of commercially available manual and automated nucleic acid extraction methods for rotavirus RNA detection in stools. J. Virol. Methods. 2013;194(1-2):242–249. doi: 10.1016/j.jviromet.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verhaegen B., De Reu K., De Zutter L., Verstraete K., Heyndrickx M., Van Coillie E. Comparison of droplet digital PCR and qPCR for the quantification of shiga toxin-producing Escherichia coli in bovine feces. Toxins (Basel) 2016;8(5) doi: 10.3390/toxins8050157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abravanel F., Sandres-Saune K., Lhomme S., Dubois M., Mansuy J.M., Izopet J. Genotype 3 diversity and quantification of hepatitis e virus RNA. J. Clin. Microbiol. 2012;50(3):897–902. doi: 10.1128/JCM.05942-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abravanel F., Chapuy-Regaud S., Lhomme S., Dubois M., Peron J.M., Alric L. Performance of two commercial assays for detecting hepatitis E virus RNA in acute or chronic infections. J. Clin. Microbiol. 2013;51(6):1913–1916. doi: 10.1128/JCM.00661-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konet D.S., Mezencio J.M., Babcock G., Brown F. Inhibitors of RT-PCR in serum. J. Virol. Methods. 2000;84(1):95–98. doi: 10.1016/s0166-0934(99)00131-7. [DOI] [PubMed] [Google Scholar]
- 26.Oikarinen S., Tauriainen S., Viskari H., Simell O., Knip M., Virtanen S. PCR inhibition in stool samples in relation to age of infants. J. Clin. Virol. 2009;44(3):211–214. doi: 10.1016/j.jcv.2008.12.017. [DOI] [PubMed] [Google Scholar]
- 27.Abu Al-Soud W., Radstrom P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 2000;38(12):4463–4470. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pontiroli A., Travis E.R., Sweeney F.P., Porter D., Gaze W.H., Mason S. Pathogen quantitation in complex matrices: a multi-operator comparison of DNA extraction methods with a novel assessment of PCR inhibition. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu R., Shieh Y.C., Stewart D.S. Comparison of RNA extraction kits for the purification and detection of an enteric virus surrogate on green onions via RT-PCR. J. Virol. Methods. 2016;239:61–68. doi: 10.1016/j.jviromet.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Kim H.Y., Kwak I.S., Hwang I.G., Ko G. Optimization of methods for detecting norovirus on various fruit. J. Virol. Methods. 2008;153(2):104–110. doi: 10.1016/j.jviromet.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Petrich A., Mahony J., Chong S., Broukhanski G., Gharabaghi F., Johnson G. Multicenter comparison of nucleic acid extraction methods for detection of severe acute respiratory syndrome coronavirus RNA in stool specimens. J. Clin. Microbiol. 2006;44(8):2681–2688. doi: 10.1128/JCM.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabria A., Pinto R.M., Bosch A., Bartolome R., Cornejo T., Torner N. Norovirus shedding among food and healthcare workers exposed to the virus in outbreak settings. J. Clin. Virol. 2016;82:119–125. doi: 10.1016/j.jcv.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Sayed I.M., Verhoye L., Cocquerel L., Abravanel F., Foquet L., Montpellier C. Study of hepatitis E virus infection of genotype 1 and 3 in mice with humanised liver. Gut. 2017;66(5):920–929. doi: 10.1136/gutjnl-2015-311109. [DOI] [PubMed] [Google Scholar]