Abstract
The evolution of mankind has constantly been influenced by the pathogens encountered. The various populations of modern humans that ventured out of Africa adapted to different environments and faced a large variety of infectious agents, resulting in local adaptations of the immune system for these populations. The functional variation of immune genes as a result of evolution is relevant in the responses against infection, as well as in the emergence of autoimmune and inflammatory diseases observed in modern populations. Understanding how host–pathogen interactions have influenced the human immune system from an evolutionary perspective might contribute to unveiling the causes behind different immune-mediated disorders and promote the development of new strategies to detect and control such diseases.
Keywords: evolution, immunity, migrations, pathogens, selective pressure, autoimmune diseases
Pathogen Influence on the Evolution of Immunity
Infectious diseases are arguably the main source of evolutionary pressure that humanity has ever confronted. The dispersion of different human communities around the globe has exposed each population to different infectious agents, exerting a selective pressure (see Glossary) on them; thus, adaptation to the new environment has favored the selection of the most beneficial genetic variants for the host. As a result, infectious agents have caused the expansion of alleles behind the induction of either protection or tolerance to these diseases; heritable variations, that increased the survival to deadly infectious agents, may have been naturally selected before the hosts had the opportunity to reproduce [1]. Natural selection driven by pathogens is probably more remarkable for those infectious agents that have been among us for a longer time, namely the causative agents of well-known diseases such as leprosy, smallpox, malaria, or tuberculosis. The genetic imprint of pathogen-driven selection depends on the length and the virulence of the infections and also the geographical distribution.
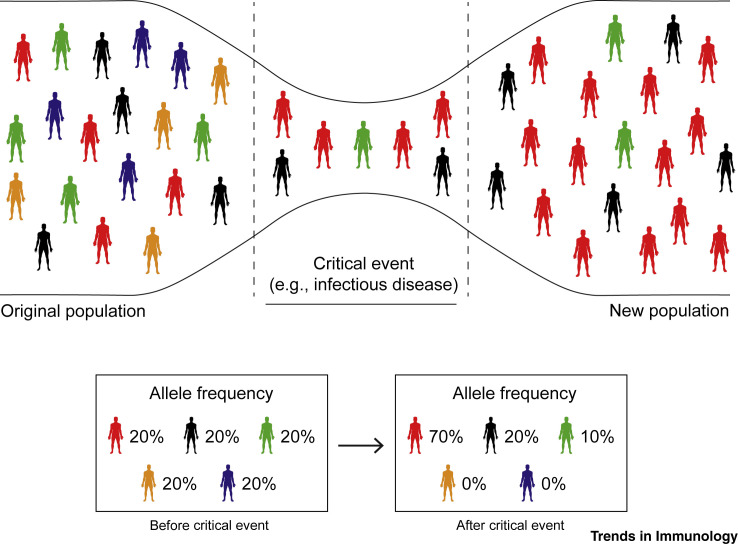
The human genome presents more than 5000 genetic loci with traces of selective pressure [2]. This group includes more than 300 immune-related genes with functional variations between populations, which are probably behind the variability of responses to immune-related diseases reported nowadays [3,4]. Besides natural selection, other evolutionary mechanisms, such as genetic drift, greatly influence the frequencies of the genetic variants found within diverse populations throughout the world [5] (Box 1 and Figure 1 ).
Box 1. Transmission of Genetic Variations.
The gene variations that pass from one generation to the next are often transmitted as a random process known as genetic drift, while selection of advantageous variants tends to be preferentially transmitted. Mutations, genetic drift, migration, and environmental selective pressure are among the fundamental processes behind the evolution of humans. The influence of these mechanisms in the diverse communities that were mobilized and then became isolated, as well as severe external factors such as epidemics, caused successive genetic bottlenecks in populations (see Figure 1 in main text) [90]. Human evolutionary studies are currently considered under ‘modern synthesis’, which merges Darwin's ideas of natural selection with theoretical population genetics and Mendelian principles, stating that evolution occurs via small genetic changes that are regulated by natural selection [91]. These beneficial adaptations subsequently expand within the members of a population and become evident in the ancestral specificity and the geographical distribution of the advantageous alleles in the genomes of contemporary humans.
Figure 1.
Genetic Bottlenecks.
Genetic bottlenecks occur when the number of individuals in a population is reduced drastically due to a catastrophic event such as an earthquake, a flood, a famine, or the outbreak of an infectious disease. These events limit the genetic variation of a population and can lead to genetic drift. As a result, a smaller population, with a correspondingly reduced genetic diversity, remains to transmit genes to future generations through sexual reproduction. Even if this reduction in the genetic diversity is temporary, it can lead to long-lasting effects on the genetic variation of the offspring populations.
With the burst of next-generation sequencing and the development of cutting-edge technologies such as transcriptomics, proteomics, and systems biology, we are starting to witness the great impact of evolutionary processes on human immunity and how the interactions between microorganisms and humans that took place millennia ago might play a fundamental role not only in the response against modern pathogenic threats, but also in the emergence of autoimmune and inflammatory diseases observed in modern populations worldwide. In this review we offer a novel perspective on the role of infectious diseases as agents of natural selection and as forces behind the evolutionary pressure encountered by human ancestors and modern humans in their migrations around the globe. Specific genetic variants selected throughout different periods of human history may have influenced immune responses of present-day populations against pathogenic microorganisms and may have played a role in the development of certain inflammatory and autoimmune diseases.
Examples of Evolutionary Pressures Providing Protection in African Populations
The majority of experts agree that Africa is where our species originated. Genetic studies conducted in diverse contemporary populations suggest connections with ancestors that lived on the continent up to 350 000 years ago [6].
Malaria
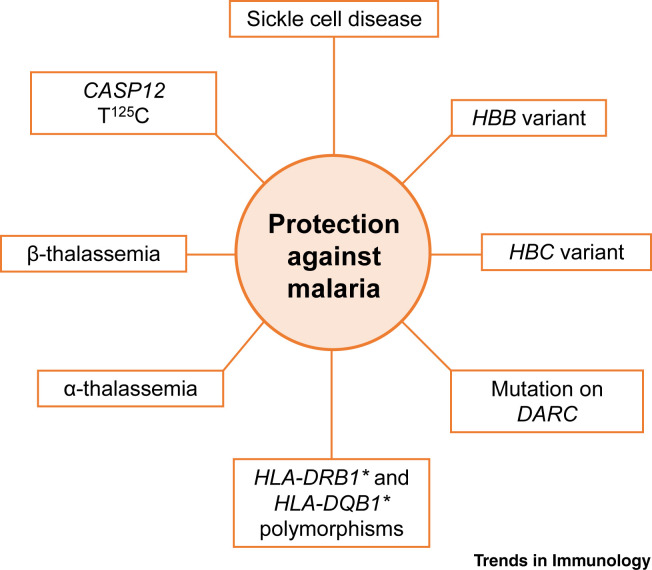
Pathogens have played a central role as agents of natural selection from those very early days. Among various infectious diseases, malaria has exerted the highest evolutionary pressure on the communities across the African continent (Figure 2 ) [7]. Populations remaining in sub-Saharan Africa have been exposed to malaria for such long periods of time that their genetic structures have been shaped by the severity of malaria (Plasmodium sp.) infections. In 1954, Allison described that sickle cell disease distribution was confined to Africa and was associated with the geographical presence of malaria [8]. This finding led to the more recent description of the existence of mutations in the Hemoglobin-B (HBB) gene as a result of natural selection driven by evolutionary pressure for protection against malaria [9] (Table 1 ). Similarly to HBB, some areas of West Africa with a high incidence of malaria present a high frequency of Hemoglobin-C (HBC) in their populations, which is associated with a 30–93% decrease in the possibility of developing the disease [10]. This is also the case for the Duffy antigen receptor gene (DARC) in erythrocytes and single nucleotide polymorphisms (SNPs) in human leukocyte antigen (HLA), which have been associated with protection against Plasmodium vivax malaria in certain areas of Africa where this disease is endemic [11,12]. Another example of natural selection driven by evolutionary pressure for protection against malaria are thalassemia (α and β) pathologies, a group of hemoglobin disorders that presents an incidence of up to 30% among communities of West Africa [13]. The human CASP12 T125C SNP, expression of which is restricted to the African subcontinent, South America, and certain areas of Asia, can modulate immune and inflammatory responses to malaria by antagonizing interleukin (IL)-1β and NF-κB signaling in innate immune cells; moreover, caspase 12-deficient mice (Casp12 –/–) exhibit decreased interferon (IFN)-γ production and clearance of the parasite, relative to wild type (WT) infected mice [14]. However, others have questioned these findings, given that caspase 12-deficient mice also lack caspase 11 expression, so the effects observed might not be specific to caspase 12 [15].
Figure 2.
Main Genetic Adaptations to Malaria.
Malaria is one of the greatest causes of morbidity and mortality in the history of humanity. Most human populations with a long history of endemic malaria have evolved genetic adaptations to malaria parasites due to the strong selective pressure that this infection has exerted. Since the parasite infects erythrocytes, the evolutionary pressure has selected genetic variants that affect red blood cells and, therefore, the survival of the parasite as well. Genetic variants conferring resistance to the disease have spread through human populations over time, including several abnormal hemoglobins that protect against malaria but usually cause erythrocyte-associated diseases in the populations where these adaptations are prevalent. These factors include the T125C polymorphism in the caspase 12 gene (CASP12); the Hemoglobin B (HBB) and Hemoglobin C (HBC) variants; mutations in the Duffy antigen receptor gene (DARC); thalassemias (α and β); sickle cell disease; and polymorphisms in the human leukocyte antigen (HLA) loci.
Table 1.
Examples of Pathogens and Their Relationships to Human Potential Resistance/Susceptibility Alleles
| Pathogen or disease | Gene or gene variants | Effect association | Refs |
|---|---|---|---|
| Plasmodium falciparum |
HBB, HBC, HBA1, HBA2 FCGR2B |
Protection (associated with hemoglobinopathies) Protection (associated with SLEa) |
[9,13,102] [69] |
| Plasmodium vivax | DARC, HLA-DRB1* and HLA-DQB1* | Protection | [11,12,103] |
| Bacterial sepsis | CASP12 (T125C) | Protection | [14] |
| Mycobacterium tuberculosis |
VDR, SLC11A1, TIRAP, HLA, CCL2, IL12A IFNG (874T/A) |
Protection Detrimental |
[17,18,104] [40] |
| Lassa virus | IL21 and LARGE | Protection | [20] |
| Trypanosoma brucei | APOL1 | Protection (associated with SLE) | [68] |
| Viral infections (e.g., HSV type 2, influenza, papillomavirus) | HLA-DQ2 and HLA-DQ8 | Protection (associated with CD) | [105,106] |
| Bacterial products (Escherichia coli LPS and muramyl dipeptide) | SH2B3 rs3184504*A | Protection (detrimental for CD) | [72] |
| Gram-negative bacterial infections and parasitic infections | NOD2 and TLR4/CD14 | Protection (detrimental for IBD) | [74,107, 108, 109] |
| HIV-1 | CCR5Δ32 | Protection | [98] |
Abbreviations: CD, Crohn’s disease; HSV, herpes simplex virus; IBD, inflammatory bowel disease; LPS, lipopolysaccharide; SLE, systemic lupus erythematosus.
Tuberculosis
Mycobacterium tuberculosis (MTB) has caused infections in our species and ancestors for at least 500 000 years [16]. This long-standing relationship between humans and MTB probably underlies the large variety of immune-related factors that modulate susceptibility to MTB infection, including vitamin D receptor (VDR), natural resistance-associated macrophage protein 1 (SLC11A1), TIR Domain Containing Adaptor Protein (TIRAP), HLA, monocyte chemoattractant protein 1 (MCP-1), and cytokines such as IL-12 and IFN-γ [17] (Table 1). Patients with African ancestry present a higher frequency of MTB-related genetic variants than individuals from other populations, including variants in the gene encoding for Toll-like receptor 6 (TLR6), mediating cellular responses to bacterial lipoproteins [18,19]. Selective pressure has also shaped the mechanisms that modulate the expression of genes implicated in immune responses against Lassa virus, such as IL-21 (IL21) and the glycosyltransferase-like protein LARGE1 (LARGE), suggesting that the natural selection exerted by the virus drove the expansion of genetic variants that enhance immunity against Lassa fever [20]. These examples indicate that infectious diseases have contributed to shaping the genetic landscape of African populations and their descendants, and highlight the great impact of pathogens as an evolutionary force in humans.
Neanderthals and Denisovans Contributed to Shaping the Modern Human Immune System
Our Homo sapiens ancestors were not the only species to venture out of Africa, with other Homo species performing a similar migration much earlier, such as Homo ergaster, Homo erectus, or Homo heidelbergensis [21]. From these early migrations, local populations such as the Denisovans and Neanderthals evolved [22]. These lineages were not geographically isolated, but lived side by side with modern humans and interbred with them, leaving a genetic footprint in their common progeny. Accordingly, 1–4% of the genome of European and Asian populations is thought to derive from these now-extinct hominid lineages [23].
Neanderthals spent close to 600 000 years adapting to their environment and their immune systems were shaped by the infections they faced. By interbreeding with archaic humans, modern humans incorporated these advantageous adaptations in the genome of their descendants. This was highlighted by different studies that showed that the introgression of diverse genes related to immune functions, such as the OAS cluster, TLR1, or the histocompatibility complex from Denisovans and Neanderthals shaped the genetic landscape of present-day Eurasian, but not African, communities. Genomic sequences and expression data from lymphoblastoid cell lines from 421 individuals of European and African ancestry confirmed that the TLR1–TLR6–TLR10 genetic loci, presenting signs of local positive selection and repeated introgression from both Neanderthal and Denisovan genomes [24, 25, 26], showed a significantly higher expression in individuals carrying archaic-like alleles than in individuals carrying the nonintrogressed modern human alleles [2,26,27]. The expression of these genes has shaped human immune responses against different types of pathogens. For example, the gp41 protein of the HIV-1 virus has been recently recognized as a TLR10 ligand [28]. In this regard, increased TLR10 expression has been correlated with higher IL-8 production by the macrophage cell line THP-1 and higher titers of HIV-1 in the breast milk of HIV-1-infected Nigerian women relative to controls [28]. TLR1 and TLR6 form dimers with TLR2, triggering immune responses against different types of bacteria, fungi, virus, and parasites [29]. Variation in TLR1–TLR6–TLR10 is the major genetic determinant of human interindividual differences in TLR1/2-mediated responses, including cytokine production to a number of clinically relevant pathogens such as Staphylococcus aureus and Listeria monocytogenes [30] (Table 1). This inheritance from archaic humans may have also left some human individuals more prone than others to developing asthma, hay fever, and other allergies (of 58 SNPs associated with susceptibility to allergic disease, 12 had a Neanderthal or Denisovan origin) [26], although these associations remain to be fully demonstrated [31].
These reports demonstrate that by interbreeding with archaic humans, modern humans incorporated a group of advantageous adaptations to the genome of their descendants and contributed to shaping immune responses in modern human populations.
Different Immune Responses in African and Eurasian Populations
The migration of our human ancestors out of Africa implied the exposure to different types of infectious diseases (Box 2 and Figure 3 ). One study tested the responsiveness of human macrophages to pathogenic bacteria in vitro, finding that almost 10% of the genes present in human macrophages infected with the bacteria Salmonella typhimurium or L. monocytogenes present different regulatory responses directly linked to the lineage of the donors and, also, that macrophages obtained from individuals of African origin display enhanced bactericidal activity compared with those from individuals of European lineage [32]. The trend towards lower inflammatory responses in European populations is strengthened by the fixation of a TLR1 gene variant that results in lower proinflammatory gene expression in populations with a European ancestry compared with those with an African one [33]. The largest population differences in gene expression between Africans and Europeans have been found in the macrophage receptor with collagenous structure (MARCO), a protein implicated in responses against viral infections and TLR-induced dendritic cell activation [34], the chemokine receptor CX3CR1, which mediates effector lymphocyte functions, and also several IFN-stimulated genes [35]. West Eurasian populations present a high frequency of TIRAP Ser180Leu SNPs [36]. TIRAP is an adaptor protein in TLR2 and TLR4 signaling pathways, involved in inflammatory responses and cytokine production. The heterozygous expression of the Ser180Leu SNP is protective against invasive pneumococcal disease, bacteremia, malaria, and tuberculosis, as shown in a case–control study of 6106 individuals from the UK, Vietnam, and several African countries, and it is associated with lower TLR2 signaling in humans [36]. This variant is considered to be a consequence of the natural selection that may have taken place in an early period following the migration of modern humans out of Africa [37].
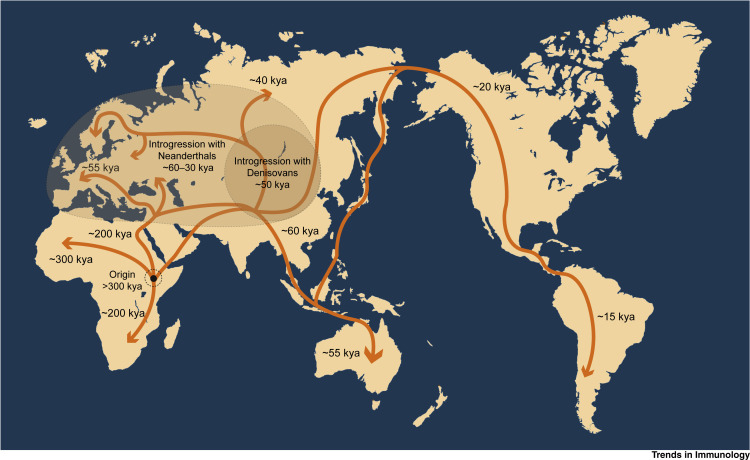
Box 2. Human Migration.
One of the most interesting aspects of humans is their ability to adapt to almost every ecosystem of the planet. The history of mankind is also the history of millions of individuals wandering around the world, looking for a better place to live. A glimpse to the migratory legacy of humanity around the globe reveals the great impact that the massive population movements defined the world as we know it today (see Figure 3 in main text). The distances ancient humans travelled are impressive, from the first hominids colonizing Africa to the conquest of the Americas in a time when the Bering Strait was not yet under water. The historical exodus of mankind started almost 2 million years ago with the migration of Homo erectus from Africa through Eurasia. From this event on, relatively isolated human populations evolved separately on different continents, leading to the emergence of different human species, such as Neanderthals in Europe, the Denisovans in Asia, and, later, modern Homo sapiens in Africa [92,93]. H. sapiens first colonized large areas of the continent around 300 000 years ago [6], spread towards the Middle East at some point between 150 000 and 80 000 years ago, and migrated through Eurasia, reaching Australia within 20 000 years [94]. Asian human ancestors went through the frozen waters of the Bering Strait in two distinct waves to colonize the American continent approximately 20 000 years before the present time [95].
Figure 3.
Map of Human Migrations.
Multiple migrations out of Africa took place between 150 000 and 100 000 years ago. The Sinai Peninsula to the north and the Bab-el-Mandeb strait to the east, worked as corridors for the past few million years, appearing to be the most likely routes of dispersal for our ancestors. While some of these early explorations certainly failed and became evolutionary dead ends, others survived, spreading across the different continents and interbreeding with Denisovans and Neanderthals [2,101]. Abbreviations: kya, thousand calendar years ago.
European populations present a selective adaptation of the IFN gene that allows a high production of IFN-γ in infectious scenarios due to the positive selection of IFNG variants +5173G and +874T; this suggests the existence of strong environmental pressures linked to higher IFN-γ concentrations in plasma during MTB infection in European individuals relative to other populations [38,39]. In line with this, a database meta-analysis showed that individuals expressing the +874T/A variant of IFNG presented higher susceptibility to tuberculosis MTB infection than individuals without it, which might be considered a putative prognostic factor for the development of tuberculosis [40], although this remains to be robustly demonstrated.
When humans ventured out of Africa, they faced different types of pathogens than the communities that stayed in the African continent. With time, the series of events faced by diverse populations has generated differences in the immune responses to pathogens in the populations with an African or Eurasian origin and which have spread throughout the world.
Colonization of New World and Immunity in the American Continent
The indigenous populations of South America are descendants of migrating populations of North-East Asians that crossed the Bering Strait around 20 000 years ago [41]. Five centuries ago, European settlers disembarked on the American continent, bringing a large collection of pathogens such as those causing measles, pneumonic plague, and influenza infections, which the indigenous populations had never faced before. These diseases rapidly spread and caused mortality rates above 90% [42].
The consequences of these pandemics are still visible in current populations; one report studied DNA from the bones of 25 ancient humans from the Tsimshian community, living in the British Columbia region in Canada until the 15th century, identifying marks of positive selection in a number of immune-related variants [43]. Specifically, the HLA-DQA1 variant was present in almost the totality of Tsimshian individuals, but only in one third of present-day humans studied; this suggested that ancient American genomes were evolutionarily selected to respond to local diseases but not to fight against pathogens brought by the Europeans [43]. Another study compiled information on infectious diseases that have killed more than 10 000 individuals among 59 indigenous communities of the Amazonia in the past two centuries, showing that the mortality rates and the incidence of infectious diseases rapidly decayed within the time following the first encounter with the pathogen, compatible with genetic adaptation [44]. European colonizers underwent purifying selection in situations of intense pressure. Such scenarios were documented when Dutch colonists migrated to Surinam and encountered epidemics of yellow and typhoid fever that caused a 60% mortality rate among settlers [45]. Variations in the frequencies of C3, GLO, and HLA-B genes among the descendants were not likely caused by genetic drift, but rather, it has been proposed that these populations were probably selected through genetic control of survival to the epidemics [45] (Box 3 ).
Box 3. The Case of Resistance Variants against HIV-1 Infection.
The origins of the HIV virus are still a matter of scientific discussion. The most accepted scenario argues that HIV originated in simians and was transmitted to humans in West Africa in the 1920s, likely due to local ingestion of ape meat infected with the simian immunodeficiency virus. Around 1960, the virus reached wide parts of the continent and finally spread overseas thanks to a group of Haitian professors coming back from Africa. In the following decades, the virus spread worldwide and generated the pandemic we now know. Today, there are approximately 37 million people worldwide living with HIV-1/AIDS [110]. CCR5 is a receptor of chemokines that plays a fundamental role in HIV-1 pathogenesis and it is also one of the most promising targets to restrict the infection, since mutations in this receptor turn individuals resistant [96]. The CCR5-Δ32 mutation results in a deletion that eliminates the HIV-1 co-receptor on lymphocytes, providing robust protection against HIV-1 and, therefore, AIDS [97]. CCR5-Δ32 allele frequencies reach 14% in northern Europe populations, whereas it is not present in populations with different ancestry, such as East Asian, native American, or African groups [96]. This regional distribution of CCR5-Δ32 variants is most likely related to a naturally selective episode that struck European populations around 700 years ago and involved a strong infectious agent that also employed CCR5 [98]. A mathematical model studying the changes in the European populations in the Middle Ages suggested Yersinia pestis (bubonic plague) as the most probable infectious agent behind the pressure that selected this particular genetic variant [99]. This is in agreement with the finding that European Rroma populations, but not Northwestern Indian populations that inhabit the area where the Rroma originally lived, present signatures of positive selection in TLR1–TLR6–TLR10, which influence cytokine responses in Y. pestis infections [100].
Africans and Americans with an African ancestry present a much higher number of genetic variants related to robust inflammatory reactions, increased cytokine secretion, and bactericidal activities compared with the other populations [32], including more than 250 genes with traces of recent selection, such as IL1A and IL1B gene variants [46]. The degree of African ancestry, analyzed by fine-mapping analysis refined to the Duffy-null allele of rs2814778, was correlated with an increased amount of the proinflammatory chemokines CCL2 and CCL11 in plasma relative to controls [12]. A study involving 12 000 African American and Hispanic American women found that the higher values of C-reactive protein (CRP) in blood found in these populations compared with European Americans were related to a CRP-associated variant of triggering receptors expressed by myeloid cells 2 (TREM2) [47]. Moreover, comparison of health record data from individuals with connective tissue diseases, including rheumatoid arthritis and systemic lupus erythematosus (SLE), as well as atherosclerotic cardiovascular disease from almost 300 000 African American and European American adults was conducted; the study reported for the latter, a prevalence of atherosclerotic cardiovascular disease in 29.7% African Americans (particularly high in young individuals), relative to 14.7% in European Americans [48]. These studies highlight certain genetic links to inflammatory predisposition/manifestation. However, increased proinflammatory activity is a double-edged sword. In the absence of regular pathogen challenges that require maintained modulation of the balance between inflammation and suppression of the immune response, the organism can overreact to inflammatory stimuli and trigger exacerbated responses. For instance, descendants of African populations generally present higher susceptibility to a variety of autoimmune syndromes such as inflammation-associated carcinomas, lupus, asthma, and multiple sclerosis (MS), the overall prevalence of which is up to three times higher in individuals with African ancestry relative to individuals with European ancestry [31,49,50].
There are extensive differences in immune cell gene expression between Americans with African and European ancestry. The increased proinflammatory responses observed in American individuals relative to other populations might be beneficial to combatting infections, but might also increase the chances of developing inflammatory and autoimmune disorders, which warrants further investigation.
Nonpathogenic Microbes: Mutualistic Bacteria and Viruses
Our gastrointestinal tract provides residence to both beneficial and potentially pathogenic microorganisms, harboring ten million different microbial genes in the human fecal microbiome [51]. The microbiome has its own evolutionary scenario across different populations with divergent lifestyles, nutrition, and exposure to environmental agents, generating extraordinary heterogeneity. The ongoing process of ‘lifestyle Westernization’ of different societies has an important impact on the mutualistic relationships between humans and commensal organisms worldwide. African tribes are adopting Western subsistence patterns, leading to remarkable changes in the composition of their microbiota [52]. The comparison of the intestinal flora of the BaAka hunter-gatherers and the Bantu agriculturalists (both from the Central Africa Republic), with a group of US-born African Americans showed a great example of the evolution of the human microbiome [52]. Specifically, the Bantu, still engaged in hunting, have a greater bacterial gut diversity than their BaAka neighbors, who left the jungle for agriculture, and even more than urbanized westerners (US African-Americans) [52]. This reduced microbiota diversity in Western societies has been associated with a higher incidence of the so-called ‘diseases of civilization’ such as cardiovascular diseases, diabetes, obesity, and autoimmune disorders, which are very unusual in hunter-gatherer societies compared with communities living a Western-type lifestyle [52,53].
Although viruses are mainly seen as pathogenic agents, they also play a fundamental role in the evolution and maturation of the human immune system [54,55]. Approximately 8% of the human genome is composed of endogenous retroviruses (ERVs), sequences derived from past retroviral infections and permanently inserted into different regions of the human genome [56]. One study showed that ERVs played a central role in the induction of IFN-dependent immune responses and that the removal of one or more of these viral DNA elements in the HeLa human cell line severely impaired the recruitment of transcription factors necessary to trigger the expression of IFNG against vaccinia virus infection relative to controls [57]. Viruses can also influence the severity of infections caused by other viruses. For example, cytomegalovirus infection in HIV-1 seropositive humans can potentiate the effects of HIV-1 infection by expanding the pool of circulating regulatory T cells (immunosuppressive); these were shown to inhibit the proliferation of autologous peripheral blood mononuclear cell (PBMC) in response to cytomegalovirus infection in vitro [58]. In one study, patients with chronic hepatitis C virus (HCV) infection and hepatitis A virus (HAV) superinfection presented lower titers of HCV RNA than patients harboring only HCV, suggesting that HCV replication might be potentially suppressed during HAV infection [59], although this will still require further investigation.
The relationships between humans and pathogenic or nonpathogenic organisms are extraordinarily complex and include tripartite evolutionary interactions between humans and microbes competing with each other. This is the case of parasites that infect other parasites, such as bacteriophage viruses, that can influence the outcome of bacterial infections. For example, in a cohort of individuals with chronic wounds, a report showed that the phage Pf, which coexists with Pseudomonas aeruginosa in infected wounds, triggered the production of type I IFN, the inhibition of tumor necrosis factor (TNF) production, and the suppression of phagocytosis in human primary monocytes and mouse bone marrow-derived macrophages, dampening the antibacterial response and promoting the bacterial infection [60]. However, bacteriophages can also provide protection to the human host by directly attacking pathogenic bacteria and by upregulating in human PBMCs the expression of proinflammatory genes such as IL1A, IL1B, IL6, TNFA, CXCL1, and CXCL5, as shown for several S. aureus and P. aeruginosa phages, including PNM, LUZ19, 14-1, and GE-vB_Pae-Kakheti25 [61].
Cooperative relationships between organisms are evolutionary processes themselves. The way microorganisms and their hosts associate can lead to interactions of mutualism, in which the interplay may be so intimate as to provide benefit for each party and influence immune responses against different types of pathogens.
A Role for Ancestry in the Susceptibility to Autoimmune Diseases
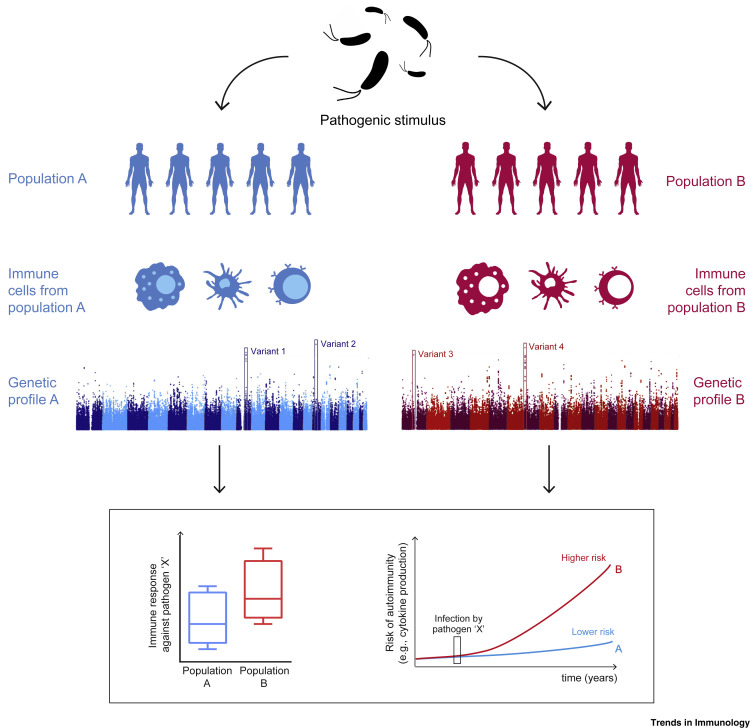
A great number of humans live far away from the original settlements of their ancestors and are subject to radically different environmental conditions. Between two and three million people with European genealogy suffer from autoimmune diseases, the prevalence of which is also increasing in other populations across the globe [62]. There is rising evidence that the emergence of autoimmune diseases is associated with the presence of a number of immune-related alleles that have been selected via evolutionary processes; and, furthermore, that the contrasting differences in the prevalence of autoimmune diseases between populations may be a result of different selective pressures [63]. Alleles associated with inflammatory diseases that present marks of modern positive selection include the risk allele FUT2 at rs601338 for Crohn’s disease (CD) or the risk variant SH2B3 at rs3184504 for celiac disease [64]; such variants have been linked to the development of several human autoimmune diseases, such as type 1 diabetes, MS, and celiac disease [64,65]. The analysis of the integrated haplotype score [66] of loci associated with SLE that might provide protection against infections, such as TNIP1, ITGAM, PTPN22, TNFSF4, UHRF1BP1, TET3-DGUOK, and BLK, has suggested that these loci exhibit robust signs of positive selection [67] (Figure 4 , Key Figure and Table 1). African and Asian human populations exposed to Trypanosoma brucei or Plasmodium sp. have presented positive selection of SNPs in the APOL1 and FCGR2B genes; indeed, by enhancing human macrophage-mediated phagocytosis of infected erythrocytes, despite their association with an SLE predisposition, these SNPs have been associated with protective roles against sleeping sickness and malaria, respectively [68,69].
Figure 4.
Key Figure. Impact of Ancestry on Immunity
The heterogeneity in the immune response to infectious diseases across different populations is under genetic control and is the result of evolutionary processes. Populations with different ancestry present different functional alleles in genes involved in the immune response. These different genetic variants allow the establishment of a strong inflammatory response against certain types of infections but can also increase the chances of developing certain autoimmune diseases and inflammatory disorders.
An analysis of human loci associated with inflammatory bowel disease (IBD) concluded that the majority of the loci associated with CD are also linked with a higher risk of developing ulcerative colitis [70] (Table 1). Many of these loci were also associated with the development of other autoimmune diseases, namely psoriasis and ankylosing spondylitis [62]. PBMCs from individuals carrying the SH2B3 variant rs3184504*A (associated with a risk for developing CD) [71] presented higher production of IL-6 and IL-1β after stimulation with lipopolysaccharide or muramyl dipeptide due to enhanced activity of the NOD2 inflammasome pathway relative to controls [72]; this has suggested an immune-related role for SH2B3 in the context of bacterial infections, which might help explain the positive selection of SH2B3 approximately 1500 years ago [72]. Others found an association between genetic variants in NOD2, CD14, and increased susceptibility to CD [73], in line with previous results showing that mutations in NOD2 and TLR4/CD14 are related to an increased risk of developing IBD [74].
From another angle, changes in hygiene patterns seen in the past two centuries brought vast improvements in sanitation, drinking water, and garbage collection, which greatly reduced the exposure to many infectious diseases. However, these conditions may have reduced the exposure to viral and microbial agents that help the immune system to develop tolerance during childhood. The hygiene hypothesis proposes that the lack of exposure to microbial agents in the early stages of life is related to a higher risk of developing hypersensitivity reactions, based on the fact that children that are exposed to higher amounts of microbial stimuli (e.g., by growing on farms) are less prone to develop allergies and asthma [75,76]. Moreover, reduced exposure to infectious agents can have a much wider effect than initially believed. Lack of exposure to microbes in childhood can cause aberrant responses to infection and potentiate the effects of ETV6–RUNX1 mutations in the pathogenesis of acute lymphoblastic leukemia [77]. By contrast, a meta-analysis of six observational studies, including 1902 participants, showed a correlation between low Helicobacter pylori infection and MS, suggesting that low H. pylori prevalence might be a putative protective factor in MS, although this remains to be experimentally validated [78]. One study also reported that antibodies against Toxoplasma gondii were detected less often in patients with MS compared with healthy controls [79]. However, these findings warrant further and robust investigation.
Overall, it is clear that evolutionary processes can drive the fixation of genetic variations that increase (or decrease) our defense against infections upon sensing microbial ligands, but can also lead to a greater risk of developing certain autoimmune diseases in which endogenous ligands can cause tissue damage and inflammation.
The Potential Role of Epigenetic Inheritance
A growing number of reports suggest that inheritance is not always governed by classical Darwinian evolutionary processes. Exposure to certain environmental stimuli can cause effects in the progeny of an exposed individual, even though the stimuli are no longer present. This type of transgenerational inheritance might be explained through the effects of epigenetic processes, which are hypothesized to be transmitted through the germline and passed on to the offspring [80]. For example, the worm Caenorhabditis elegans can transmit improved resistance to infections to pathogenic bacteria to their offspring through alterations of the histone landscape [81]. Indian meal moths exposed to low doses of virus are subsequently less susceptible to viral challenge, a protection offered to their offspring as well [82]. Transgenerational inheritance of diverse traits has also been observed in mice, in which the variation of the color of the coat is passed on the next generation [83,84]. Offspring of male rats subjected to a high-fat diet present glucose intolerance and reduced insulin secretion, linked to reduced methylation at the Il13ra2 gene relative to controls [85]; and mice fed scorpions are more resistant to a challenge with scorpion venom than mice on a normal diet [86]. Since infections are one of the strongest factors impacting survival, it is conceivable that transgenerational transmission of traits in mammals, including humans, evolved to improve host defense. The number of studies of the potential role of epigenetic inheritance in shaping the human immune system is still scarce. However, different experiences undergone by certain communities indicate that these mechanisms might be important. For example, the babies of pregnant women who suffered during the early stages of pregnancy (the effects of the Dutch Hunger Winter in 1944), 60 years later showed reduced DNA methylation marks in several genes that control metabolism and cell differentiation during development, such as IGF2, PIM3, TXNIP, ABCG1, PFKFB3, and METTL8, compared with their siblings [87,88]. This was related with higher rates of obesity, heart disease, cancer, and depression in individuals whose pregnant mothers suffered the effects of the famine [87,88]. Some of these effects seemed to be present in the progeny of this group, that is, in the grandchildren of those who had passed the famine during pregnancy [89]. The rapid growth in the number of reports covering the impact of epigenetic mechanisms in different human processes warrants further and robust studies on the impact of epigenetic inheritance in shaping the evolution of the human immune system.
Concluding Remarks
Human immune responses have been shaped by the evolutionary pressure exerted by microorganisms and viruses throughout history. Generating a broad range of genetic variations and immune functions in different populations favors the adaptation to new environments and increases the chance of survival of the human species against potential pandemics.
Much remains to be learned in this exciting field over the coming years in order to identify the main regulatory forces and the time window necessary for the fixation of an advantageous genetic trait in a population (see Outstanding Questions). The combination of the selective pressure caused by infectious diseases with other evolutionarily relevant processes, such as genetic drift, migratory events, bottlenecks, and introgressive hybridization, contribute to driving the expansion, fixation, or elimination of characteristic immune response-related traits in different populations around the world. These specific genetic variants are able to boost the host response against pathogens by improving the sensing of microbial ligands but can also lead to the development of autoimmune diseases, in which the immune system responds to endogenous ligands and induces abnormal responses targeted against the host’s own tissues. Of note, it is very difficult to assign certain variants, a specific role in the protection or induction of autoimmunity. To assign changes in the genetic landscape of human populations to certain diseases is an extraordinary challenge. Moreover, our species is in constant evolutionary interaction with various microorganisms and viruses. Populations of bacteria and their viruses (phages) undergo, under natural conditions, reciprocal evolution in terms of resistance and infection; this, in turn, also affects the evolutionary traits of our immune system. Thus, an extraordinarily complex scenario exists in which organisms of different phyla interact, compete, and coevolve, to ensure their own survival.
Outstanding Questions.
Which are the strongest evolutionary forces that drive the evolution of the human innate immune system?
How long does the genome of a given population take to adapt to a new infectious threat?
Are the mechanisms of resistance to infection transmitted only via genetic modifications, or can epigenetic adaptations to resistance also be transmitted to the progeny, and under what circumstances?
The development of single-cell sequencing technologies has opened new fields of study. How does genetic variation of the expression of a specific variant vary between different cell subsets and how does it affect the overall phenotype of an individual within a population?
Are there specific immune processes that are preferentially impacted by evolutionary pressures?
In Western societies we enjoy a life expectancy vastly superior to that of our predecessors, but at the same time, we suffer diseases that they did not suffer. Can some of the reasons for these changes lie among some of those bacteria that we have somehow lost in our microbiome? Identifying these bacteria and understanding their effects on the human body might be the first step to developing putative therapies based on bacterial restoration.
Since many of the variants causing autoimmune diseases are linked to an enhanced responsiveness to pathogens that is no longer needed in developed countries, could these genes and their related pathways be employed as targets for new putative therapeutic approaches against inflammatory/autoimmune syndromes?
Are other newly described genetic regulatory pathways, such as interfering RNA or long noncoding RNAs, also influenced by pathogen-driven evolutionary processes in different populations globally?
As novel tools, the development and refinement of methods that study large sections of the human genome, epigenome, and microbiota, will help to obtain genome-wide data in diverse human populations, allowing us to follow the evolutionary trails left from the encounters with different organisms, further unveiling the roots of human immunity. High-throughput biotechnology and an expanding computational capacity can enable the study of global population genomics and might contribute to decoding the origin and consequences of functional changes in adaptive alleles down to the single cell level. However, these methods also have limitations associated with the difficulty in linking gene variations to clinical phenotypes and disease, the generation of false positives, or the high number of samples necessary to reach reliable conclusions. Expanding the heterogeneity of populations studied for immune gene association studies relevant to disease will be key, as generally, a large focus is placed on certain European or American communities, thus generating results that are difficult to extrapolate to other populations.
The knowledge of the evolutionary and genetic basis of human immune traits and their impact on diverse pathologies (e.g., autoimmunity, infections, inflammatory diseases, cancer) increasingly suggests that the genetic basis of disease may be derived from a large number of rare variants of modest effect. The mechanisms described here acquire special importance in the current scenario of world globalization, in which the migration fluxes and the admixture of different populations are reaching unprecedented levels, allowing faster expansion of advantageous alleles. However, these processes may also accelerate the spread of new epidemics, as seen in the cases of HIV infection, or more recently, SARS-CoV, Ebola, and chikungunya viruses; as well as the emergence of multiresistant bacteria and fungi, such as methicillin-resistant S. aureus or Candida auris. This is just the starting point to unveil the evolutionary history of the relationships between pathogens, the immune system, and humans. Further investigation of the functional adaptations of human populations is warranted to provide a broad picture of the functional consequences of evolution in human immunity.
Acknowledgments
M.G.N. is supported by a Spinoza Grant of The Netherlands Organization for Scientific Research and an ERC Advanced Grant (#833247). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We thank Laszlo A. Groh for proofreading and correction of the manuscript. We additionally thank Srinivas Agra for providing the figure icon ‘Bacteria’ as deposited on https://thenounproject.com.
Glossary
- Bottlenecks
drastic decline in the number of members of a population due to a catastrophic event that decimates the population, such as a widespread infectious disease.
- Crohn’s disease (CD)
chronic inflammatory condition of the gastrointestinal tract with an autoimmune origin. It is a type of inflammatory bowel disease (IBD).
- Duffy antigen receptor gene
glycoprotein present in the surface of erythrocytes.
- European Rroma
community or ethnicity originating from the Indian subcontinent, with common cultural and genetic traits. Most of them (10–12 million people) currently live on the European continent.
- Genetic drift
changes in the allele frequencies of a population over generations due to chance.
- Genetic locus
fixed position on a chromosome (e.g., the position of a gene or a genetic marker).
- Histocompatibility complex
region of approximately 4000 kb, located on human chromosome 6, that contains a large number of genes whose products are expressed as proteins on immune cells. Of these genes, the best known are HLA genes.
- Introgressive hybridization
incorporation of genes from one species into the genetic reserves of another by interspecific hybridization and backcrossing with the parent species.
- Lassa fever
often fatal infection caused by Lassa virus, an arenavirus; it occurs most frequently in West Africa and can compromise multiple systems. In severe cases it can lead to bleeding, shock, and multiorgan failure.
- Modern synthesis
formulation of evolutionary theory that reconciled classical Darwinian selection theory with a newer population-oriented view of Mendelian genetics that attempted to explain the origin of biological diversity.
- OAS cluster
group of genes on human chromosome 12 that encode essential proteins involved in innate immune responses to viral infections.
- Single nucleotide polymorphisms (SNPs)
most common type of genetic variation. Each SNP represents a difference in a single DNA nucleotide. For example, an SNP may replace cytosine (C) with thymine (T) in a certain segment of DNA.
- Selective pressure
phenomenon that alters the behavior and fitness of living organisms within a given environment. It is the driving force of evolution and natural selection.
- Thalassemias (α and β)
inherited hemoglobinopathies characterized by a failure in the synthesis of the globin alpha or beta chains.
- Toll-like receptors
family of transmembrane pattern recognition receptors expressed by immune and nonimmune cells that recognize conserved pathogen-associated molecular patterns. They play a pivotal role in innate immunity.
- Transgenerational inheritance
transmission of traits from generation to generation.
References
- 1.Karlsson E.K., et al. Natural selection and infectious disease in human populations. Nat. Rev. Genet. 2014;15:379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deschamps M., et al. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 2016;98:5–21. doi: 10.1016/j.ajhg.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong X., et al. Genome-wide identification of regulatory sequences undergoing accelerated evolution in the human genome. Mol. Biol. Evol. 2016;33:2565–2575. doi: 10.1093/molbev/msw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian R., et al. Divergent selection of pattern recognition receptors in mammals with different ecological characteristics. J. Mol. Evol. 2018;86:138–149. doi: 10.1007/s00239-018-9832-1. [DOI] [PubMed] [Google Scholar]
- 5.Simons Y.B., et al. A population genetic interpretation of GWAS findings for human quantitative traits. PLoS Biol. 2018;16:e2002985. doi: 10.1371/journal.pbio.2002985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hublin J.-J., et al. New fossils from Jebel Irhoud, Morocco and the pan-African origin of Homo sapiens. Nature. 2017;546:289–292. doi: 10.1038/nature22336. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization . WHO; 2018. World Malaria Report 2018. [Google Scholar]
- 8.Allison A.C. Protection afforded by sickle-cell trait against subtertian malareal infection. Br. Med. J. 1954;1:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohashi J., et al. Extended linkage disequilibrium surrounding the hemoglobin E variant due to malarial selection. Am. J. Hum. Genet. 2004;74:1198–1208. doi: 10.1086/421330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modiano D., et al. Haemoglobin C protects against clinical Plasmodium falciparum malaria. Nature. 2001;414:305–308. doi: 10.1038/35104556. [DOI] [PubMed] [Google Scholar]
- 11.Sabeti P.C., et al. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 12.Yao S., et al. Genetic ancestry and population differences in levels of inflammatory cytokines in women: role for evolutionary selection and environmental factors. PLoS Genet. 2018;14:e1007368. doi: 10.1371/journal.pgen.1007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams T.N., Weatherall D.J. World distribution, population genetics, and health burden of the hemoglobinopathies. Cold Spring Harb. Perspect. Med. 2012;2:a011692. doi: 10.1101/cshperspect.a011692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labbe K., et al. Caspase-12 dampens the immune response to malaria independently of the inflammasome by targeting NF-B signaling. J. Immunol. 2010;185:5495–5502. doi: 10.4049/jimmunol.1002517. [DOI] [PubMed] [Google Scholar]
- 15.Vande Walle L., et al. Does caspase-12 suppress inflammasome activation? Nature. 2016;534:E1–E4. doi: 10.1038/nature17649. [DOI] [PubMed] [Google Scholar]
- 16.Kappelman J., et al. First Homo erectus from Turkey and implications for migrations into temperate Eurasia. Am. J. Phys. Anthropol. 2008;135:110–116. doi: 10.1002/ajpa.20739. [DOI] [PubMed] [Google Scholar]
- 17.Mboowa G. Genetics of sub-Saharan African human population: implications for HIV/AIDS, tuberculosis, and malaria. Int. J. Evol. Biol. 2014;2014:108291. doi: 10.1155/2014/108291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daya M., et al. The role of ancestry in TB susceptibility of an admixed South African population. Tuberculosis. 2014;94:413–420. doi: 10.1016/j.tube.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Zhang M., et al. Association of the TLR1 variant rs5743557 with susceptibility to tuberculosis. J. Thorac. Dis. 2019;11:583–594. doi: 10.21037/jtd.2019.01.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen K.G., et al. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos. Trans. R. Soc. B Biol. Sci. 2012;367:868–877. doi: 10.1098/rstb.2011.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacruz R.S., et al. The evolutionary history of the human face. Nat. Ecol. Evol. 2019;3:726–736. doi: 10.1038/s41559-019-0865-7. [DOI] [PubMed] [Google Scholar]
- 22.Clarkson C., et al. Human occupation of northern Australia by 65,000 years ago. Nature. 2017;547:306–310. doi: 10.1038/nature22968. [DOI] [PubMed] [Google Scholar]
- 23.Prüfer K., et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taskent R.O., et al. Variation and functional impact of Neanderthal ancestry in Western Asia. Genome Biol. Evol. 2017;9:3516–3524. doi: 10.1093/gbe/evx216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagoda E., et al. Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol. Biol. Evol. 2018;35:623–630. doi: 10.1093/molbev/msx314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dannemann M., et al. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human Toll-like receptors. Am. J. Hum. Genet. 2016;98:22–33. doi: 10.1016/j.ajhg.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abi-Rached L., et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henrick B.M., et al. TLR10 senses HIV-1 proteins and significantly enhances HIV-1 infection. Front. Immunol. 2019;10:482. doi: 10.3389/fimmu.2019.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oliveira-Nascimento L., et al. The role of TLR2 in infection and immunity. Front. Immunol. 2012;3:79. doi: 10.3389/fimmu.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikacenic C., et al. Variation in the TLR10/TLR1/TLR6 locus is the major genetic determinant of interindividual difference in TLR1/2-mediated responses. Genes Immun. 2013;14:52–57. doi: 10.1038/gene.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nyenhuis S.M., et al. Race is associated with differences in airway inflammation in patients with asthma. J. Allergy Clin. Immunol. 2017;140:257–265. doi: 10.1016/j.jaci.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nédélec Y., et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167:657–669. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 33.Quach H., et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maler M.D., et al. Key role of the scavenger receptor MARCO in mediating adenovirus infection and subsequent innate responses of macrophages. MBio. 2017;8 doi: 10.1128/mBio.00670-17. e00670–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lionakis M.S., et al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J. Clin. Invest. 2013;123:5035–5051. doi: 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khor C.C., et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat. Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferwerda B., et al. Functional and genetic evidence that the Mal/TIRAP allele variant 180L has been selected by providing protection against septic shock. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10272–10277. doi: 10.1073/pnas.0811273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manry J., et al. Evolutionary genetics evidence of an essential, nonredundant role of the IFN-γ pathway in protective immunity. Hum. Mutat. 2011;32:633–642. doi: 10.1002/humu.21484. [DOI] [PubMed] [Google Scholar]
- 39.Vallinoto A.C.R., et al. IFNG +874T/A polymorphism and cytokine plasma levels are associated with susceptibility to Mycobacterium tuberculosis infection and clinical manifestation of tuberculosis. Hum. Immunol. 2010;71:692–696. doi: 10.1016/j.humimm.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Wei Z., et al. A single nucleotide polymorphism in the interferon-γ gene (IFNG +874 T/A) is associated with susceptibility to tuberculosis. Oncotarget. 2017;8:50415–50429. doi: 10.18632/oncotarget.17304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoglund P., et al. Genetic evidence for two founding populations of the Americas. Nature. 2015;525:104. doi: 10.1038/nature14895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cook S.F. The significance of disease in the extinction of the New England Indians. Hum. Biol. 1973;45:485–508. [PubMed] [Google Scholar]
- 43.Lindo J., et al. A time transect of exomes from a Native American population before and after European contact. Nat. Commun. 2016;7:13175. doi: 10.1038/ncomms13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker R.S., et al. Mortality from contact-related epidemics among indigenous populations in Greater Amazonia. Sci. Rep. 2015;5:14032. doi: 10.1038/srep14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Vries R.R., et al. Genetic control of survival in epidemics. J. Immunogenet. 1979;6:271–287. doi: 10.1111/j.1744-313x.1979.tb00684.x. [DOI] [PubMed] [Google Scholar]
- 46.Ness R.B., et al. Differential distribution of allelic variants in cytokine genes among African Americans and White Americans. Am. J. Epidemiol. 2004;160:1033–1038. doi: 10.1093/aje/kwh325. [DOI] [PubMed] [Google Scholar]
- 47.Reiner A.P., et al. Genome-wide association and population genetic analysis of C-reactive protein in African American and Hispanic American women. Am. J. Hum. Genet. 2012;91:502–512. doi: 10.1016/j.ajhg.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alenghat F.J. The prevalence of atherosclerosis in those with inflammatory connective tissue disease by race, age and traditional risk factors. Sci. Rep. 2016;6:20303. doi: 10.1038/srep20303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pennington R., et al. Group differences in proneness to inflammation. Infect. Genet. Evol. 2009;9:1371–1380. doi: 10.1016/j.meegid.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 50.Brinkworth J.F., Barreiro L.B. The contribution of natural selection to present-day susceptibility to chronic inflammatory and autoimmune disease. Curr. Opin. Immunol. 2014;31:66–78. doi: 10.1016/j.coi.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 52.Gomez A., et al. Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns. Cell Rep. 2016;14:2142–2153. doi: 10.1016/j.celrep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 53.de la Cuesta-Zuluaga J., et al. Gut microbiota is associated with obesity and cardiometabolic disease in a population in the midst of Westernization. Sci. Rep. 2018;8:11356. doi: 10.1038/s41598-018-29687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roossinck M.J. The good viruses: viral mutualistic symbioses. Nat. Rev. Microbiol. 2011;9:99–108. doi: 10.1038/nrmicro2491. [DOI] [PubMed] [Google Scholar]
- 55.Ryan F. Collins; 2009. Virolution: The Most Important Evolutionary Book Since Dawkins’ Selfish Gene. [Google Scholar]
- 56.Wallace A.D., et al. To ERV is human: a phenotype-wide scan linking polymorphic human endogenous retrovirus-K insertions to complex phenotypes. Front. Genet. 2018;9:298. doi: 10.3389/fgene.2018.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chuong E.B., et al. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tovar-Salazar A., Weinberg A. Cytomegalovirus infection in HIV-infected and uninfected individuals is characterized by circulating regulatory T cells of unconstrained antigenic specificity. PLoS One. 2017;12:e0180691. doi: 10.1371/journal.pone.0180691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deterding K., et al. Hepatitis A virus infection suppresses hepatitis C virus replication and may lead to clearance of HCV. J. Hepatol. 2006;45:770–778. doi: 10.1016/j.jhep.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 60.Sweere J.M., et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science. 2019;363:eaat9691. doi: 10.1126/science.aat9691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Belleghem J.D., et al. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 2017;7:8004. doi: 10.1038/s41598-017-08336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jostins L., et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramos P.S., et al. Genetics of autoimmune diseases: insights from population genetics. J. Hum. Genet. 2015;60:657–664. doi: 10.1038/jhg.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raj T., et al. Common risk alleles for inflammatory diseases are targets of recent positive selection. Am. J. Hum. Genet. 2013;92:517–529. doi: 10.1016/j.ajhg.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fumagalli M., et al. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 2011;7:e1002355. doi: 10.1371/journal.pgen.1002355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voight B.F., et al. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramos P.S., et al. Genes associated with SLE are targets of recent positive selection. Autoimmune Dis. 2014;2014:203435. doi: 10.1155/2014/203435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Freedman B.I., et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol. 2014;66:390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clatworthy M.R., et al. Systemic lupus erythematosus-associated defects in the inhibitory receptor Fc RIIb reduce susceptibility to malaria. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7169–7174. doi: 10.1073/pnas.0608889104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cleynen I., et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet (London, England) 2016;387:156–167. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hunt K.A., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhernakova A., et al. Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am. J. Hum. Genet. 2010;86:970–977. doi: 10.1016/j.ajhg.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Azzam N., et al. CARD15/NOD2, CD14 and toll-like 4 receptor gene polymorphisms in Saudi patients with Crohn’s disease. Int. J. Mol. Sci. 2012;13:4268–4280. doi: 10.3390/ijms13044268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gazouli M., et al. Association between polymorphisms in the Toll-like receptor 4, CD14, and CARD15/NOD2 and inflammatory bowel disease in the Greek population. World J. Gastroenterol. 2005;11:681–685. doi: 10.3748/wjg.v11.i5.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Campbell B., et al. The effects of growing up on a farm on adult lung function and allergic phenotypes: an international population-based study. Thorax. 2017;72:236–244. doi: 10.1136/thoraxjnl-2015-208154. [DOI] [PubMed] [Google Scholar]
- 76.Stein M.M., et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N. Engl. J. Med. 2016;375:411–421. doi: 10.1056/NEJMoa1508749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greaves M. A causal mechanism for childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer. 2018;18:471–484. doi: 10.1038/s41568-018-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jaruvongvanich V., et al. Association between Helicobacter pylori infection and multiple sclerosis: a systematic review and meta-analysis. Mult. Scler. Relat. Disord. 2016;7:92–97. doi: 10.1016/j.msard.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 79.Koskderelioglu A., et al. Is Toxoplasma gondii infection protective against multiple sclerosis risk? Mult. Scler. Relat. Disord. 2017;15:7–10. doi: 10.1016/j.msard.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 80.Horsthemke B. A critical view on transgenerational epigenetic inheritance in humans. Nat. Commun. 2018;9:2973. doi: 10.1038/s41467-018-05445-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Belicard T., et al. The piRNA pathway responds to environmental signals to establish intergenerational adaptation to stress. BMC Biol. 2018;16:103. doi: 10.1186/s12915-018-0571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tidbury H.J., et al. Within and transgenerational immune priming in an insect to a DNA virus. Proc. R. Soc. B Biol. Sci. 2011;278:871–876. doi: 10.1098/rspb.2010.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morgan H.D., et al. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 84.Oey H., et al. Genetic and epigenetic variation among inbred mouse littermates: identification of inter-individual differentially methylated regions. Epigenetics Chromatin. 2015;8:54. doi: 10.1186/s13072-015-0047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ng S.-F., et al. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 86.Rowe A.H., Rowe M.P. Physiological resistance of grasshopper mice (Onychomys spp.) to Arizona bark scorpion (Centruroides exilicauda) venom. Toxicon. 2008;52:597–605. doi: 10.1016/j.toxicon.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 87.Heijmans B.T., et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tobi E.W., et al. DNA methylation as a mediator of the association between prenatal adversity and risk factors for metabolic disease in adulthood. Sci. Adv. 2018;4:eaao4364. doi: 10.1126/sciadv.aao4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Veenendaal M., et al. Transgenerational effects of prenatal exposure to the 1944-45 Dutch famine. BJOG. 2013;120:548–553. doi: 10.1111/1471-0528.12136. [DOI] [PubMed] [Google Scholar]
- 90.Abel S., et al. Analysis of bottlenecks in experimental models of infection. PLoS Pathog. 2015;11:e1004823. doi: 10.1371/journal.ppat.1004823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarkar S. Haldane’s The Causes of Evolution and the modern synthesis in evolutionary biology. J. Genet. 2017;96:753–763. doi: 10.1007/s12041-017-0840-5. [DOI] [PubMed] [Google Scholar]
- 92.Slon V., et al. The genome of the offspring of a Neanderthal mother and a Denisovan father. Nature. 2018;561:113–116. doi: 10.1038/s41586-018-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jacobs Z., et al. Timing of archaic hominin occupation of Denisova Cave in southern Siberia. Nature. 2019;565:594–599. doi: 10.1038/s41586-018-0843-2. [DOI] [PubMed] [Google Scholar]
- 94.Bae C.J., et al. On the origin of modern humans: Asian perspectives. Science. 2017;358:eaai9067. doi: 10.1126/science.aai9067. [DOI] [PubMed] [Google Scholar]
- 95.Moreno-Mayar J.V., et al. Early human dispersals within the Americas. Science. 2018;362:eaav2621. doi: 10.1126/science.aav2621. [DOI] [PubMed] [Google Scholar]
- 96.Stephens J.C., et al. Dating the origin of the CCR5-Δ32 AIDS-resistance allele by the coalescence of haplotypes. Am. J. Hum. Genet. 1998;62:1507–1515. doi: 10.1086/301867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Church J.A. Long-term control of HIV by CCR5 delta32/delta32 stem-cell transplantation. Pediatrics. 2009;124:S159. [Google Scholar]
- 98.Galvani A.P., Slatkin M. Evaluating plague and smallpox as historical selective pressures for the CCR5-delta 32 HIV-resistance allele. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15276–15279. doi: 10.1073/pnas.2435085100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duncan S.R., et al. Reappraisal of the historical selective pressures for the CCR5-D32 mutation. J. Med. Genet. 2005;42:205–208. doi: 10.1136/jmg.2004.025346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Laayouni H., et al. Convergent evolution in European and Rroma populations reveals pressure exerted by plague on Toll-like receptors. Proc. Natl. Acad. Sci. U S A. 2014;111:2668–2673. doi: 10.1073/pnas.1317723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vattathil S., Akey J.M. Small amounts of archaic admixture provide big insights into human history. Cell. 2015;163:281–284. doi: 10.1016/j.cell.2015.09.042. [DOI] [PubMed] [Google Scholar]
- 102.Himeidan Y.E., Kweka E.J. Malaria in East African highlands during the past 30 years: impact of environmental changes. Front. Physiol. 2012;3:315. doi: 10.3389/fphys.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hill A.V.S., et al. Common West African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 104.Ma X., et al. Full-exon resequencing reveals Toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One. 2007;2:e1318. doi: 10.1371/journal.pone.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dubois P.C.A., et al. Multiple common variants for celiac disease influencing immune gene expression. Nat. Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hung S.-C., et al. Epitope selection for HLA-DQ2 presentation: implications for celiac disease and viral defense. J. Immunol. 2019;202:2558–2569. doi: 10.4049/jimmunol.1801454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lesage S., et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am. J. Hum. Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hugot J.-P., et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 109.Keestra-Gounder A.M., Tsolis R.M. NOD1 and NOD2: beyond peptidoglycan sensing. Trends Immunol. 2017;38:758–767. doi: 10.1016/j.it.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Marsh K., et al. Global, regional and country-level 90-90-90 estimates for 2018: assessing progress towards the 2020 targe. AIDS. 2019 doi: 10.1097/QAD.0000000000002355. Published online September 3, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]