Summary
Background
A new betacoronavirus—Middle East respiratory syndrome coronavirus (MERS-CoV)—has been identified in patients with severe acute respiratory infection. Although related viruses infect bats, molecular clock analyses have been unable to identify direct ancestors of MERS-CoV. Anecdotal exposure histories suggest that patients had been in contact with dromedary camels or goats. We investigated possible animal reservoirs of MERS-CoV by assessing specific serum antibodies in livestock.
Methods
We took sera from animals in the Middle East (Oman) and from elsewhere (Spain, Netherlands, Chile). Cattle (n=80), sheep (n=40), goats (n=40), dromedary camels (n=155), and various other camelid species (n=34) were tested for specific serum IgG by protein microarray using the receptor-binding S1 subunits of spike proteins of MERS-CoV, severe acute respiratory syndrome coronavirus, and human coronavirus OC43. Results were confirmed by virus neutralisation tests for MERS-CoV and bovine coronavirus.
Findings
50 of 50 (100%) sera from Omani camels and 15 of 105 (14%) from Spanish camels had protein-specific antibodies against MERS-CoV spike. Sera from European sheep, goats, cattle, and other camelids had no such antibodies. MERS-CoV neutralising antibody titres varied between 1/320 and 1/2560 for the Omani camel sera and between 1/20 and 1/320 for the Spanish camel sera. There was no evidence for cross-neutralisation by bovine coronavirus antibodies.
Interpretation
MERS-CoV or a related virus has infected camel populations. Both titres and seroprevalences in sera from different locations in Oman suggest widespread infection.
Funding
European Union, European Centre For Disease Prevention and Control, Deutsche Forschungsgemeinschaft.
Introduction
In 2012, a new betacoronavirus—Middle East respiratory syndrome coronavirus (MERS-CoV)—was identified in patients with severe respiratory disease in the Middle East. As of Aug 2, 2013, 94 laboratory-confirmed cases, including 46 deaths, have been reported to WHO.1 Illness associated with MERS-CoV infection is characterised primarily by mild-to-severe respiratory complaints, most requiring hospital admission for acute respiratory distress syndrome. Comorbidities and immunosuppression seem to predispose for infection and severe disease,2, 3, 4, 5, 6 and unpublished serological studies suggest that asymptomatic infections occur.7
All cases reported so far have been linked to Jordan, Qatar, Saudi Arabia, and United Arab Emirates. Human-to-human transmission has been reported, particularly in health-care settings, but on the basis of available evidence the basic reproduction number (R0) is thought to be low, suggesting that the virus is not transmitted readily.6, 8 Therefore, the primary reservoir of MERS-CoV is probably animals. Different coronaviruses have various hosts including wildlife, livestock, poultry, pets, and human beings. Coronaviruses can adapt to new host species, as shown by the zoonotic origin of several human coronaviruses.9 Human coronavirus OC43 has recent common ancestry with bovine coronaviruses.10 Rhinolophid bats were identified as a likely reservoir for severe acute respiratory syndrome coronavirus (SARS-CoV), which emerged in people in 2002–03, through intermediate carnivorous hosts.11 Molecular clock analysis12 showed that bat and civet strains of viruses closely related to SARS-CoV only diverged a few years before the outbreak. Human coronavirus 229E has a common ancestor with coronaviruses found in Ghanaian Hipposideros spp bats.13
MERS-CoV is able to replicate in various bat cell lines14 and phylogenetic analyses show that it is closely related to betacoronavirus lineage C viruses from Pipistrellus spp bats in Europe and Asia.15, 16, 17, 18 Molecular clock dating of epidemiologically unlinked isolates of human MERS-CoV estimated their divergence from a common ancestor in mid-2011,4, 19 with a cluster of isolates from the eastern Arabian peninsula diverging in late 2012.4 This finding could suggest that the diversity of MERS-CoV in people is the result of multiple independent, geographically structured, zoonotic events in the Middle East.4, 19
Possible animal reservoirs need to be identified to determine how circulation of MERS-CoV is maintained and to break the chain of transmission.20 MERS-CoV can infect cells of several species, including human beings and bats.14 The functional receptor is conserved between species, suggesting that receptor use is not an important barrier to cross-species transmission.21 Data for exposure history of patients are scarce, but suggest contact with livestock, including dromedary camels and goats.2, 4, 5 Food and Agriculture Organization data from 2011 show that cows, goats, sheep, and dromedary camels are the main sources of meat and milk in Jordan, Saudi Arabia, and United Arab Emirates.22
Serological studies are best suited to screen animal populations, but have not yet been reported for MERS-CoV in animals, although several methods have been described for testing antibodies of people.23, 24 For specificity, WHO recommends use of a combination of screening assays with recombinant spike protein, and confirmatory testing by neutralisation assays. Here, we describe antibody profiling of serum samples from major livestock species that might be relevant to the epidemiology of MERS-CoV in the Middle East, using samples collected from herds inside and outside the region.
Methods
Serum sample collection
We sampled a cohort of 105 dromedary camels (Camelus dromedarius) from two herds on the Canary Islands. 50 were male, 55 were female, 88 were adults, nine were age 3–4 years, seven were age 2 years, and one was age 3 months. Both herds had the same owner, with frequent exchange of animals between the herds. One herd is from a coastal dune habitat with no other livestock, while the other herd is in an inland valley close to a tropical fruit farm, in particular mango and papaya—which could attract fruit bats—and nearby (roughly 500 m) to horse and goat farms with 25 and 300 animals, respectively. The camels were born in the Canary Islands except for three adults, which were imported from Morocco.25 The camels are used in the tourist industry. 80 sera were taken April–June, 2012, nine in May, 2013, and 16 paired sera were taken in these months in 2012, and 2013, all for routine veterinary purposes. Samples were obtained by jugular puncture.
50 female dromedary camels from Oman were sampled in March, 2013. The camels were aged 8–12 years and belonged to different owners from separate locations. The camels are retired racing camels now used for breeding, and blood was taken by jugular puncture for routine screening for brucellosis.
Sera were collected for veterinary purposes from two llamas (Lama glama), six alpacas (Vicugna pacos), and two Bactrian camels (Camelus bactrianus) in the Netherlands. Sera were collected for veterinary purposes from two Bactrian camels, 18 alpacas, five llamas, and two guanaco (Lama guanicoe) in Buin Zoo in Chile. Sera from cattle (n=40), domestic goats (n=40), and sheep (n=40) were from routine submission to the Dutch Animal Health Service. Sera from Spanish domestic goats (n=40) were provided by the Instituto de Investigación en Recursos Cinegéticos (Ciudad Real, Spain) from submissions for tuberculosis control in 2011. All sera were obtained in agreement with local regulations and Dutch import regulations with regard to animal disease legislation. Positive human control sera for the three antigens used on the microarray were taken as described previously.24 All samples were stored at −20°C until testing.
Laboratory procedures
We tested the sera for the presence of IgG antibodies reactive with MERS-CoV, SARS-CoV, and human coronavirus OC43 S1 antigens in a protein microarray. The receptor-binding domains, which contain the S1 subunit of spike proteins of MERS-CoV (residues 1–747), SARS-CoV (residues 1–676), and human coronavirus OC43 (residues 1–760) were expressed, purified, and spotted on glass slides. Slides were incubated with serum and species-specific conjugates, as previously described.24 Goat sera were incubated with Alexa Fluor 647-conjugated rabbit anti-goat IgG Fc fragment (Jackson Immuno Research, West Grove, PA, USA); cow sera with Alexa Fluor 647-conjugated goat anti-bovine IgG (H+L; Jackson Immuno Research); sheep sera with Alexa Fluor 647-conjugated donkey anti-sheep IgG (H+L; Millipore, Temecula, CA, USA); and camelid sera with Dylight 650-conjugated goat anti-llama IgG (H+L; Agrisera, Vännas, Sweden). Fluorescence signals were quantified as described previously.24 We tested the sera for IgG reactivity at a dilution of 1/20 and set an arbitrary cutoff at an average signal intensity of 20 000 relative fluorescence units (RFU). This high cutoff was chosen to reduce cross-reactive false positives.24 We present results as RFU for each set of quadruplicate spots per antigen. Negative fluorescent intensities (caused by background correction) were assigned to 0. Analyses were done with GraphPad Prism (version 6.02).
Sera were heat-inactivated before virus neutralisation by incubation for 30 min at 56°C. Two-fold serial dilutions of sera were prepared using 96-well plates, starting dilution 1/10. MERS-CoV was diluted in Iscove's modified Dulbecco's medium (IMDM) supplemented with clemizole penicillin (penicillin G), streptomycin, and 1% fetal bovine serum, to a dilution of 2000 tissue culture infective dose50 per mL. 50 μL virus suspension was added to the plates and the plates were incubated at 37°C for 1 h. The mixtures of virus and serum were then incubated on 96-well plates containing Vero cells for 1 h followed by washing with phosphate buffered saline and incubation with IMDM and 1% fetal bovine serum for 3–4 days, after which endpoint titres were measured. All tests were repeated twice independently.
We tested neutralisation activity of sera against MERS-CoV (Erasmus MC isolate) and bovine coronavirus (Nebraska strain) by plaque reduction neutralisation test (90% plaque reduction) with African green monkey kidney cells (cell line Vero B4; DSMZ ACC 33) or bovine kidney cells (cell line PT; CCLV-RIE11) in a 24-well plate format. Virus (30–60 plaque-forming units) and heat-inactivated sera (diluted from 1/40 to 1/640) were pre-incubated in 200 μL of serum-free OPTIpro medium (Life Technologies, Karlsruhe, Germany) at 37°C for 1 h. Virus adsorption was done at 37°C for 1 h. Supernatants were removed and overlaid with Avicel resin (FCM BioPolymer, Brussels, Belgium).5 Assays were stopped after 3 days by fixation with 8% paraformaldehyde for 30 min. All samples were tested in duplicate and titres were expressed as the serum dilution resulting in a plaque reduction of at least 90%.
Role of the funding source
The sponsors had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
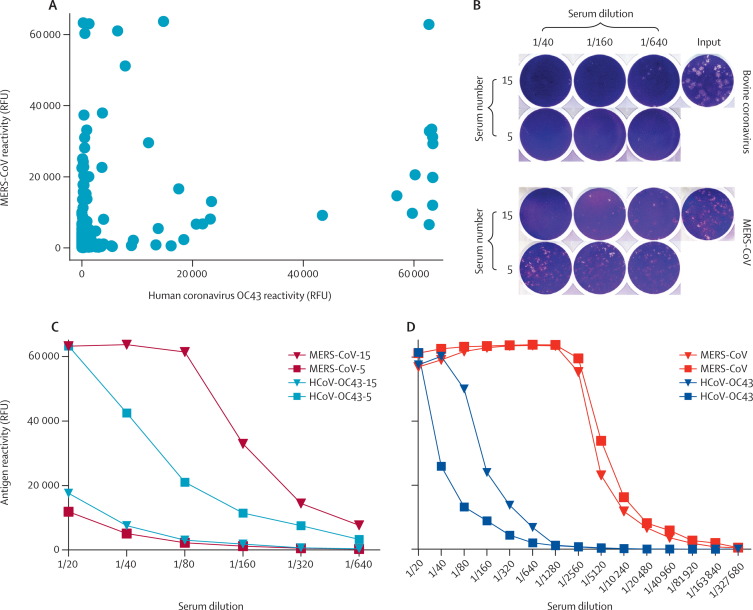
Sera were tested for IgG antibodies reactive with MERS-CoV, SARS-CoV, and human coronavirus OC43 S1 antigens in a protein microarray (figure 1 ). Human coronavirus OC43 is serologically closely related to bovine coronavirus,26 diverging at the end of the 19th century.10 Bovine coronavirus circulates in cows, sheep, goats, and Old and New World camelids.27, 28, 29, 30 Because bovine coronavirus S1 was not available, human coronavirus OC43 S1 antigen was used as a proxy. Sera from three llamas, four alpacas, one guanaco, and two Bactrian camels reacted with human coronavirus OC43 antigen. One cow and one goat serum reacted with human coronavirus OC43 antigen as did sera from 16 of 105 (15%) Spanish dromedary camels. All sera from cattle, sheep, and goats tested negative for MERS-CoV antigen, but sera from 15 Spanish camels (14%) did react with MERS-CoV antigen (figure 1). The reactivity was highly specific—the same sera did not bind to SARS-CoV antigen but a positive control specimen did. No correlation existed between the reactivity of sera with MERS-CoV antigen and human coronavirus OC43 antigen (figure 2 ). All but one serum sample that reacted with MERS-CoV antigen were from adult animals. One reactive serum was from a 2-year-old animal.
Figure 1.
Reactivity of livestock sera with three coronavirus S1 antigens
Fluorescent intensities per antigen at a serum dilution of 1/20. Black lines indicate median. Dashed line is cutoff of the assay. RFU=relative fluorescence units. SARS-CoV=severe acute respiratory syndrome coronavirus. HCoV=human coronavirus. MERS-CoV=Middle East respiratory syndrome coronavirus.
Figure 2.
MERS-CoV and human coronavirus OC43 or bovine coronavirus cross-reactivity
Combinations of the mean fluorescent intensities of reactions of sera with MERS-CoV and human coronavirus OC43 antigens from 105 Spanish dromedary camels (A). plaque reduction neutralisation tests for bovine coronavirus and MERS-CoV (B): two representative sera are shown (numbers 15 and 5, corresponding to camel ID numbers in table 2) in dilutions of 1/40, 1/160, and 1/640 as well as the virus input control. All samples were tested in duplicates (only one well shown) and titres were expressed as the serum dilution resulting in a plaque reduction of at least 90%. IgG reactivity of both camel sera to MERS-CoV antigen and human coronavirus OC43 antigen in a two-step dilution series in the microarray (C). IgG reactivity of two two-step serially diluted Omani dromedary camel sera with human coronavirus EMC antigen and human coronavirus OC43 antigen in the microarray (D). RFU=relative fluorescence units. MERS-CoV=Middle East respiratory coronavirus.
To confirm the presence of MERS-CoV specific IgG in the Spanish camel sera, we used a MERS-CoV neutralisation assay to test a subset of 49 camel sera with different degrees of reactivity with MERS-CoV and human coronavirus OC43 antigen according to microarray. Nine Spanish camels had MERS-CoV neutralising antibodies with titres varying between 1/20 and 1/320 (table 1 ). Three of the 12 sera reacted with MERS-CoV spike antigen but did not neutralise MERS-CoV, most likely because of recognition of non-neutralising epitopes. All MERS-CoV neutralising sera had (almost) saturating reactivity with MERS-CoV antigen on the microarray, whereas reactivity with human coronavirus OC43 antigen varied from negative to 50% of saturating reactivity (table 1). The variable human coronavirus OC43 signals suggest that MERS-CoV did not generally cross-react with human coronavirus OC43 or bovine coronavirus antigens. All nine camels with MERS-CoV neutralising antibodies were born and raised on the Canary Islands; seven were female, two were male. Eight camels were adults, one was 2 years old.
Table 1.
Results of neutralising assay for MERS-CoV from Spanish and Omani camel serum samples
| Number of serum samples | Positive MERS-CoV neutralisation titre (n; %) | Titre range | ||
|---|---|---|---|---|
| Spanish samples (no geographic link) | ||||
| MERS-CoV antigen array signal (RFU) | ||||
| <10 000 | 31 | 0 (0%) | .. | |
| 10 000–20 000 | 3 | 0 (0%) | .. | |
| 20 000–30 000 | 3 | 0 (0%) | .. | |
| 30 000–40 000 | 0 | 0 (0%) | .. | |
| >40 000 | 12 | 9 (75%) | 1/20 to 1/320 | |
| Human coronavirus OC43 antigen array signal (RFU) | ||||
| <10 000 | 26 | 1 (4%) | .. | |
| 10 000–20 000 | 8 | 3 (38%) | .. | |
| 20 000–30 000 | 5 | 2 (40%) | .. | |
| 30 000–40 000 | 6 | 3 (50%) | .. | |
| >40 000 | 4 | 0 (0%) | .. | |
| Omani camel samples (geographic link) | ||||
| MERS-CoV antigen array signal (RFU) | ||||
| 0–40 000 | 0 | 0 (0%) | .. | |
| >40 000 | 50 | 50 (100%) | 1/320 to 1/2560 | |
| Human coronavirus OC43 antigen array signal (RFU) | ||||
| <10 000 | 3 | 3 (100%) | .. | |
| 10 000–20 000 | 4 | 4 (100%) | .. | |
| 20 000–30 000 | 4 | 4 (100%) | .. | |
| 30 000–40 000 | 5 | 5 (100%) | .. | |
| >40 000 | 34 | 34 (100%) | .. | |
RFU=relative fluorescence units. MERS-CoV=Middle East respiratory syndrome coronavirus.
To show that the reactivity of the camel sera with human coronavirus OC43 antigen according to the microarray was caused by the presence of bovine coronavirus IgG and to further exclude MERS-CoV neutralising activity caused by cross-neutralisation by the bovine coronavirus antibodies, we tested camels that had sufficient serum left (n=15) in a comparative MERS-CoV and bovine coronavirus plaque reduction neutralisation test (figure 2, table 2 ). All camel sera neutralised bovine coronavirus, but with varying titres, suggesting a lower cutoff than 20 000 RFU for OC43 in the microarray (figure 1). Five camels had high neutralising antibody titres against bovine coronavirus (and a mean signal intensity of greater than 50 000 RFU for human coronavirus OC43 antigen on microarray) but were negative for MERS-CoV neutralisation, suggesting that cross-neutralisation in this direction did not occur and that the MERS-CoV neutralising activity was not caused by the presence of bovine coronavirus neutralising antibodies. A serum sample from a patient who had MERS, neutralised MERS-CoV with a high titre (1/640) but neutralised bovine coronavirus less efficiently (titre 1/80). The latter finding was most probably caused by previous infection with human coronavirus OC43—this patient had a high titre (1/>5120) in a human coronavirus OC43 recombinant spike immunofluorescence assay and a saturating signal with human coronavirus OC43 antigen in the microarray. Two human serum samples positive for human coronavirus OC43 did not neutralise MERS-CoV, one of which neutralised bovine coronavirus at a titre of 1/80 (table 2).
Table 2.
Protein microarray and PRNT results from sera from 15 Spanish dromedary camels and three people
|
PRNT* |
PRNT† |
Microarray (RFU) |
||
|---|---|---|---|---|
| MERS-CoV | BCoV | MERS-CoV antigen | HCoV OC43 antigen | |
| Camel | ||||
| 1 | Negative | >1/640 | 7848 | 51 147 |
| 2 | Negative | 1/320 | 23 235 | 8164 |
| 3 | Negative | 1/160 | 1273 | 20 064 |
| 4 | Negative | >1/640 | 3725 | 37 972 |
| 5 | Negative | >1/640 | 6493 | 61 046 |
| 6 | Negative | 1/160 | 1321 | 63 015 |
| 7 | Negative | 1/640 | 62 748 | 62 837 |
| 8 | Negative | 1/40 | 18 421 | 2376 |
| 9 | 1/80 | 1/160 | 62 775 | 6554 |
| 10 | 1/40 | 1/80 | 59 729 | 9726 |
| 11 | 1/40 | 1/160 | 63 433 | 29 333 |
| 12 | 1/40 | 1/640 | 63 377 | 31 207 |
| 13 | Negative | 1/160 | 13 806 | 5483 |
| 14 | 1/160 | 1/>640 | 63 438 | 19 775 |
| 15 | 1/160 | 1/320 | 63 402 | 12 029 |
| Human | ||||
| MERS-CoV | 1/640 | 1/80 | 64 353 | 63 437 |
| HCoV-OC43 (13 DPI) | Negative | 1/80 | 2848 | >55 000 |
| HCoV-OC43 (10 DPI) | Negative | 1/<40 | 2826 | >55 000 |
PRNT=plaque reduction neutralisation test. MERS-CoV=Middle East respiratory syndrome coronavirus. BCoV=bovine coronavirus. HCoV=human coronavirus. DPI=days post-infection. RFU=relative fluorescence units.
Titration range 1/40 to 1/1280.
Titration range 1/40 to 1/640.
We tested 50 sera from dromedary camels in Oman at a dilution of 1/20 by microarray and MERS-CoV neutralisation test. All the sera showed saturating reactivity with MERS-CoV antigen on the microarray, no SARS-CoV antigen reactivity, and human coronavirus OC43 antigen reactivity varying between negative (below the cutoff of 20 000 RFU) and saturating signals (figure 1, table 1). Serial dilution of two sera with saturating reactivity for both antigens at the initial dilution of 1/20 showed that MERS-CoV antigen reactivity was still above the cutoff at 1/5120, whereas human coronavirus OC43 antigen reactivity fell below the cutoff at dilutions of 1/80 to 1/320 (figure 2D). Consistent with the microarray data, all sera had high MERS-CoV neutralising capacity, with titres varying between 1/320 (seven of 50 samples) and 1/2560 or more (16 or 50 samples).
Discussion
In this study we describe the presence of MERS-CoV neutralising antibodies in dromedary camels both in a MERS-CoV linked (Oman) and unlinked regions (Canary Islands). All the sera from dromedary camels from Oman and some from Spain had specific IgG reactivity with the MERS-CoV receptor binding domain S1. We confirmed our expectation that another betacoronavirus—bovine coronavirus—circulated in these camelids.29, 30 However, all Omani camels and nine Spanish camels (9%) had specific neutralising antibodies against MERS-CoV that were clearly not caused by cross-neutralisation by bovine coronavirus antibodies.
Our study is the first in which animals have been tested for the presence of antibodies specific to MERS-CoV (panel ). Animal screening is necessary to understand the epidemiology of MERS-CoV. At present, bats are thought to be the ultimate reservoirs for several established human coronaviruses as well as SARS-CoV. Accordingly, phylogenetic analysis has shown that MERS-CoV is related to betacoronavirus lineage C viruses found in Pipistrellus spp bats.15, 16 However, direct transmission of MERS-CoV to people from bats seems unlikely.4, 19 The identification of possible intermediate hosts that are probably in closer contact with people (eg, livestock) is urgently needed. Common livestock species in the Middle East include dromedary camels but also cattle, sheep, and goats. Based on the available data, we cannot rule out circulation of a MERS-related coronavirus in these species—sera were not available from epidemiologically linked regions.
Panel. Research in context.
Systematic review
We searched PubMed for “novel coronavirus EMC” or “MERS-CoV”, we identified 43 reports in English linked to the Middle East respiratory syndrome coronavirus (MERS-CoV) published before July 22, 2013. None of these reports described a serological study for MERS-CoV-specific antibodies in animals.
Interpretation
Our report describes the first MERS-CoV serological study of major livestock relevant to the Middle East. Our study shows that MERS-CoV or a related virus has infected dromedary camel populations. Both titre levels and seroprevalences in sera from different locations in Oman suggest widespread infection of camelids with MERS-CoV or a closely related virus. Targeted studies are needed to confirm our findings and their possible relevance to human cases of MERS-CoV. Comparative seroprevalence testing of historical and more recent samples from camels from different regions for which epidemiological background information is available, as well as virological assessment of samples from seroconverting animals are needed to identify and characterise this MERS-CoV-related virus. In the meantime, we recommend a detailed case history of confirmed MERS-CoV cases, with review of any animal exposures including animal products, and targeted, prospective serosurveys to establish whether camels or their products are a potential source of human infections.
The high prevalence of MERS-CoV neutralising antibodies in dromedary camels from Oman suggests circulation of MERS-CoV or a closely related virus in this population. However, attempts to identify viral sequences in Spanish camel sera and faecal samples using pancoronavirus and specific betacoronavirus 2C PCR methods15, 31, 32 were unsuccessful (unpublished data), as was untargeted amplification followed by deep sequencing of faecal samples (unpublished data). These results imply that the camels were not actively shedding the virus at the time of sampling.
Less than 10% of the animals in the Canary Islands had MERS-CoV neutralising sera with titres up to 1/320. This low seroprevalence means either that exposure of the animals to other putative reservoirs is rare33 or that the virus is absent in this closed-off population of roughly 2000 animals.25 We cannot rule out that the population might have once had an outbreak but that by the time of sampling, antibody titres had waned and no new introductions of the virus had occurred. The camels have contact with wild rodents, rabbits, pigeons, and doves and possibly also with bats. Seven insectivorous bat species, including three Pipistrellus spp, are native to the Canary Islands, while Egyptian fruit bats (Rousettus aegyptiacus) have been introduced.34
The 100% seroprevalence with high titres in Omani camels from different owners and locations suggests a different situation in the Middle East, with widespread circulation of MERS-CoV or a closely related virus. This difference of epidemiology might be because the virus circulating in the Middle East is different to that circulating in Spain, with increased animal transmissibility and human infections.35 In addition, the Omani camels were once racing camels now held for breeding and might be kept in circumstances that favour virus transmission. For cattle, a relation has been established between the incidence and effects of respiratory diseases, management practices, and animal transport.36, 37
To our knowledge, the camel populations in Oman and the Canary Islands are not connected. Camels on the Canary Islands were originally imported in the 15th century from the Horn of Africa for labour and transport. Nowadays, import of animals from Africa is banned because of the risk of foot-and-mouth disease. Only three camels in our study were originally imported from Morocco, more than 18 years ago. Because the closest relatives of MERS-CoV were identified very recently in Neoromicia zuluensis bats from Africa,38 the introduction of MERS-CoV or related viruses into some African camel populations could have occurred decades ago, giving a possible explanation for MERS-CoV antibodies in camels from the Canary Islands. In the Middle East, huge numbers of camels are imported from Africa to meet the demand for meat. The top five camel breeding countries are all African, and Saudi Arabia and United Arab Emirates are in the top five camel meat producing countries.22 This increased turnover of animals in the Middle East compared with the Canary Islands could also affect the epidemiology of a virus, through more frequent influx of immunologically naive animals.
Targeted studies are needed to confirm our findings and their possible relevance in relation to the human cases of MERS-CoV. Comparative seroprevalence testing of historical and more recent samples from camels for which epidemiological background information is available, as well as virological assessment of specimens from seroconverting animals are needed to identify and characterise this MERS-CoV-related virus. In the meantime we recommend a detailed case history of people with MERS-CoV, with review of any animal exposures including animal products, and targeted, prospective serosurveys to establish whether camels or their products are a potential source of human infections.
Acknowledgments
Acknowledgments
We thank Prof MC Horzinek for helpful suggestions. RDS was funded by the European Public Health Training Program (Euphem), ECDC, Stockholm, Sweden. Contributions to the study were funded through the European Union FP7 projects EMPERIE (contract number 223498; to BLH, SLS, AO, CD) and ANTIGONE (contract number 278976; to CG, CD, MPGK, AO). Work in Bonn was also funded by Deutsche Forschungsgemeinschaft (DFG grant DR772/3-1 to CD).
Contributors
CBEMR designed and coordinated the study, wrote the report, searched the published work, analysed and interpreted data, and produced the figures. BLH and MAM wrote the report, produced the figures, and analysed and interpreted data. CG collected samples and wrote the report. G-JG, BM, and DM did laboratory testing and analysed, and interpreted data. VSR, LS-DV, VMC, and PR generated and analysed data. J-FD and B-JB did laboratory testing, analysed data, and wrote the report. SLS did laboratory testing, analysed data, wrote the report, and produced the figures. YEET, KvM, and EH-H collected samples. RDR searched the published work. JvB analysed data and produced the figures. NN collected samples and interpreted data. AO, CD, and MPGK interpreted data and wrote the report. CG-S collected samples, wrote the report, and coordinated the study. MPGK designed the study.
Conflicts of interest
BLH, AO, VSR, and B-JB have a patent pending for a MERS-CoV receptor. The other authors declare that they have no conflicts of interest.
References
- 1.WHO Global Alert and Response (GAR): novel coronavirus infection. http://www.who.int/csr/don/2013_08_01/en/index.html (accessed Aug 1, 2013).
- 2.Albarrak AM, Stephens GM, Hewson R, Memish ZA. Recovery from severe novel coronavirus infection. Saudi Med J. 2012;33:1265–1269. [PubMed] [Google Scholar]
- 3.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368:2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 4.Drosten C, Seilmaier M, Corman VM. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013 doi: 10.1016/S1473-3099(13)70154-3. published online June 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz U, Müller MA, Nitsche A. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October–November 2012. Euro Surveill. 2013;18:20406. [PubMed] [Google Scholar]
- 6.Assiri A, McGeer A, Perl TM. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013 doi: 10.1056/NEJMoa1306742. http://www.nejm.org/doi/full/10.1056/NEJMoa1306742 published online June 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Middle East respiratory syndrome coronavirus (MERS-CoV)–update. http://www.who.int/csr/don/2013_06_22/en/index.html [DOI] [PMC free article] [PubMed]
- 8.Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013 doi: 10.1016/S0140-6736(13)61492-0. published online July 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. 2009;234:1117–1127. doi: 10.3181/0903-MR-94. [DOI] [PubMed] [Google Scholar]
- 10.Vijgen L, Keyaerts E, Lemey P. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. 2006;80:7270–7274. doi: 10.1128/JVI.02675-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SK, Woo PC, Li KS. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau SK, Li KS, Huang Y. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. 2010;84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfefferle S, Oppong S, Drexler JF. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis. 2009;15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller MA, Raj VS, Muth D. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. mBio. 2012;3:e00515. doi: 10.1128/mBio.00515-12. e00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Annan A, Baldwin HJ, Corman VM. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau SK, Li KS, Tsang AK. Genetic characterization of betacoronavirus lineage C viruses in bats revealed marked sequence divergence in the spike protein of Pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications on the origin of the novel Middle East respiratory syndrome coronavirus. J Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reusken CB, Lina PH, Pielaat A. Circulation of group 2 coronaviruses in a bat species common to urban areas in Western Europe. Vector Borne Zoonotic Dis. 2010;10:785–791. doi: 10.1089/vbz.2009.0173. [DOI] [PubMed] [Google Scholar]
- 18.Woo PC, Wang M, Lau SK. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–1585. doi: 10.1128/JVI.02182-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotten M, Lam TT, Watson SJ. Full-genome deep sequencing and phylogenetic analysis of novel human betacoronavirus. Emerg Infect Dis. 2013;19:736–742. doi: 10.3201/eid1905.130057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cauchemez S, Van Kerkhove MD, Riley S, Donelly CA, Fraser C, Ferguson NM. Transmission scenarios for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and how to tell them apart. Euro Surveill. 2013;18:20503. [PMC free article] [PubMed] [Google Scholar]
- 21.Raj VS, Mou H, Smits SL. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.FAOSTAT 2013. http://faostat3.fao.org/home/index.html (accessed June 17, 2013).
- 23.Corman V, Muller M, Costabel U. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 24.Reusken C, Mou H, Godeke G. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveill. 2013;18:20441. doi: 10.2807/1560-7917.es2013.18.14.20441. [DOI] [PubMed] [Google Scholar]
- 25.Gutierrez C, Juste MC, Corbera JA, Magnus E, Verloo D, Montoya JA. Camel trypanosomosis in the Canary Islands: assessment of seroprevalence and infection rates using the card agglutination test (CATT/T. evansi) and parasite detection tests. Vet Parasitol. 2000;90:155–159. doi: 10.1016/s0304-4017(00)00225-9. [DOI] [PubMed] [Google Scholar]
- 26.Gerna G, Cereda PM, Revello MG, Cattaneo E, Battaglia M, Gerna MT. Antigenic and biological relationships between human coronavirus OC43 and neonatal calf diarrhoea coronavirus. J Gen Virol. 1981;54:91–102. doi: 10.1099/0022-1317-54-1-91. [DOI] [PubMed] [Google Scholar]
- 27.Traven M, Carlsson U, Lunden A, Larsson B. Serum antibodies to bovine coronavirus in Swedish sheep. Acta Veterinaria Scandinavica. 1999;40:69–74. doi: 10.1186/BF03547042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang DK, Hwang IJ, Kim BH. Serosurveillance of viral diseases in Korean native goats (Capra hircus) J Vet Med Sci. 2008;70:977–979. doi: 10.1292/jvms.70.977. [DOI] [PubMed] [Google Scholar]
- 29.Wunschmann A, Frank R, Pomeroy K, Kapil S. Enteric coronavirus infection in a juvenile dromedary (Camelus dromedarius) J Vet Diagn Invest. 2002;14:441–444. doi: 10.1177/104063870201400518. [DOI] [PubMed] [Google Scholar]
- 30.Jin L, Cebra CK, Baker RJ. Analysis of the genome sequence of an alpaca coronavirus. Virology. 2007;365:198–203. doi: 10.1016/j.virol.2007.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Silva Filho LV, Zerbinati RM, Tateno AF. The differential clinical impact of human coronavirus species in children with cystic fibrosis. J Infect Dis. 2012;206:384–388. doi: 10.1093/infdis/jis274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Souza Luna LK, Heiser V, Regamey N. Generic detection of coronaviruses and differentiation at the prototype strain level by reverse transcription-PCR and nonfluorescent low-density microarray. J Clin Microbiol. 2007;45:1049–1052. doi: 10.1128/JCM.02426-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci USA. 2004;101:15124–15129. doi: 10.1073/pnas.0308344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trujillo D, Barone R., III La fauna de quiropteros del archipelgo canario. 2006. http://www.magrama.gob.es/es/biodiversidad/temas/conservacion-de-especies-amenazadas/090471228015efe4_tcm7-21160.pdf (accessed July 14, 2013) [in Spanish].
- 35.Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohlson A, Heuer C, Lockhart C, Traven M, Emanuelson U, Alenius S. Risk factors for seropositivity to bovine coronavirus and bovine respiratory syncytial virus in dairy herds. Vet Rec. 2010;167:201–206. doi: 10.1136/vr.c4119. [DOI] [PubMed] [Google Scholar]
- 37.Cernicchiaro N, White BJ, Renter DG, Babcock AH, Kelly L, Slattery R. Associations between the distance traveled from sale barns to commercial feedlots in the United States and overall performance, risk of respiratory disease, and cumulative mortality in feeder cattle during 1997 to 2009. J Anim Sci. 2012;90:1929–1939. doi: 10.2527/jas.2011-4599. [DOI] [PubMed] [Google Scholar]
- 38.Ithete NL, Stoffberg S, Corman VM. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013 doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]