Summary
Background
Middle East respiratory syndrome coronavirus (MERS-CoV) causes severe lower respiratory tract infection in people. Previous studies suggested dromedary camels were a reservoir for this virus. We tested for the presence of MERS-CoV in dromedary camels from a farm in Qatar linked to two human cases of the infection in October, 2013.
Methods
We took nose swabs, rectal swabs, and blood samples from all camels on the Qatari farm. We tested swabs with RT-PCR, with amplification targeting the E gene (upE), nucleocapsid (N) gene, and open reading frame (ORF) 1a. PCR positive samples were tested by different MERS-CoV specific PCRs and obtained sequences were used for phylogentic analysis together with sequences from the linked human cases and other human cases. We tested serum samples from the camels for IgG immunofluorescence assay, protein microarray, and virus neutralisation assay.
Findings
We obtained samples from 14 camels on Oct 17, 2013. We detected MERS-CoV in nose swabs from three camels by three independent RT-PCRs and sequencing. The nucleotide sequence of an ORF1a fragment (940 nucleotides) and a 4·2 kb concatenated fragment were very similar to the MERS-CoV from two human cases on the same farm and a MERS-CoV isolate from Hafr-Al-Batin. Eight additional camel nose swabs were positive on one or more RT-PCRs, but could not be confirmed by sequencing. All camels had MERS-CoV spike-binding antibodies that correlated well with the presence of neutralising antibodies to MERS-CoV.
Interpretation
Our study provides virological confirmation of MERS-CoV in camels and suggests a recent outbreak affecting both human beings and camels. We cannot conclude whether the people on the farm were infected by the camels or vice versa, or if a third source was responsible.
Funding
European Union projects EMPERIE (contract number 223498), ANTIGONE (contract number 278976), and the VIRGO consortium.
Introduction
An emerging betacoronavirus—Middle East respiratory syndrome coronavirus (MERS-CoV)—causes illness characterised predominantly by mild-to-severe respiratory complaints, with most patients requiring admission to hospital because of pneumonitis or acute respiratory distress syndrome. Old age and the presence of comorbidities or immunosuppression seem to increase the risk of infection and are associated with severe forms of the disease. However, some patients can remain asymptomatic or mildly symptomatic and atypical presentations such as gastroenteritis have occurred.1 As of Dec 2, 2013, 163 laboratory-confirmed cases, including 71 deaths, have been reported to WHO.2 All of these cases were directly or indirectly linked to the Middle East region, including Saudi Arabia, Qatar, United Arab Emirates, Kuwait, Oman, and Jordan.
Coronaviruses reside in many animal hosts and can adapt to different species, including people. MERS-CoV belongs to the lineage C betacoronaviruses, which are associated with bats3 and the closest relatives of MERS-CoV have been identified in vesper bats from Europe, Asia, and South Africa.4, 5, 6 Notably, this coronovirus is able to replicate in various bat cell lines.7
Molecular clock dating of epidemiologically unlinked human MERS-CoV isolates estimated their divergence from a common ancestor in mid-2011, with a cluster of isolates from the eastern parts of the Arabian peninsula diverging in late 2012. These findings suggest that the reported MERS-CoV diversity in human beings is the result of several independent, geographically structured, zoonotic events from an unknown reservoir in the Middle East.8, 9 Human-to-human transmission has been noted, especially in health-care settings and households, but is thought to be relatively inefficient.1
To determine how circulation of MERS-CoV in human beings is maintained and to mitigate the chain of transmission, identification of possible animal reservoirs and modes of transmission is needed.10 Limited available exposure history of patients suggest that contact with livestock, including dromedary camels, might have a role.1, 9, 11, 12 In addition, our recent investigations have provided evidence for the circulation of MERS-CoV or a related virus in dromedary camels.13 Both MERS-CoV spike protein binding antibodies and virus neutralising antibodies were reported in dromedary camels from different regions, including Oman and Egypt, but no virus shedding could be detected and, therefore, the significance of these observations remained an issue of debate.13, 14 In this study we aimed to assess presence of MERS-CoV in dromedary camels from a farm in Qatar that was linked to two human cases of MERS-CoV.
Methods
Outbreak investigation
In October, 2013, the Qatar Supreme Council of Health recorded two cases of laboratory confirmed MERS-CoV. Case 1 involved a 61-year-old man who was the owner of a farm that he visited regularly. The patient had substantial contact with animals including camels, sheep, pigeons, and hens, and no history of travel outside of Qatar in the 2 weeks before becoming ill. MERS-CoV infection was diagnosed by E gene (upE) PCR on a sputum sample collected on Oct 13, 2013, in the National Virology Laboratory of Qatar. Case 2 involved a 23-year-old male employee of the farm owned by case 1 and was identified in the close-contact investigation of that case. He was reported on Oct 17, 2013, and had no underlying medical conditions and no recent travel history, but had regular contact with the animals on the farm. MERS-CoV infection was diagnosed in a throat swab taken on Oct 17, 2013, by the National Virology Laboratory of Qatar. Diagnosis for both cases was confirmed (ORF1b and N gene PCR) by Public Health England.15, 16 Qatari authorities, with support from WHO, did an epidemiological investigation at the farm within 1 week of diagnosis of the first case, which included the collection of various clinical samples from the farm's dromedary camels. Full details of the outbreak investigation will be described elsewhere.
Sample collection
We collected serum, rectal swabs, and nasal swabs from all camels present on the premises where the two men had been in contact with the animals. After an experienced animal handler fixated the camel by nose pinching, samples were obtained through swabbing with flocked swabs (FLOQSwabs, Copan Improve Diagnostics, Brescia, Italy) that were inserted deep into one of the nostrils of the camel. After sampling, swabs were put into tubes containing viral transport medium-UTM (Copan Diagnostics, Brescia, Italy). In addition, we obtained five stool samples from three different cages. The sample collection was done jointly by the Communicable Disease Control Outbreaks Rapid Response Team of the Public Health Department and the Animal Health Team, which used full personal-protective equipment including N95 masks, goggles, disposable gowns, gloves, and head covers during the handling of the animals, and during the environmental and human sampling. Serum samples were stored at −20°C and all other samples were stored at −80°C and were shipped to the Netherlands on dry ice. We tested nose swabs with two independent RT-PCR targets specific for MERS-CoV (upE and nucleocapsid [N gene]) in one laboratory (Department of Viroscience, Erasmus Medical Center, Rotterdam, Netherlands), and did RT-PCR testing with a third target (ORF1a) in another laboratory (National Institute of Public Health and the Environment, Bilthoven, Netherlands).
Laboratory procedures
We inoculated Vero and Huh-7 cells for 1 h with cell-culture medium containing samples from camel nose or rectal swabs, or with MERS-CoV (EMC isolate) as a positive control. After washing, the cells were incubated with medium containing 1% fetal bovine serum at 37°C. We stained formaldehyde-fixed Huh-7 cells infected with MERS-CoV by use of heat-inactivated (at 56°C for 30 min) camel sera or a MERS-CoV patient's serum according to standard protocols with fluorescein isothiocyanate-conjugated antibodies as a second step.
We isolated RNA from 140 μL of swab medium or culture supernatant with the QIAamp viral RNA minikit (Qiagen, Hilden, Germany) and eluted it in 50 μL. Camel MERS-CoV RNA was quantified on the ABI prism 7700 with the TaqMan Fast Virus 1-Step Master Mix (Applied Biosystems, Bleiswijk, Netherlands), with 10 μL isolated RNA, one TaqMan mix, 0·5 U of uracil-N-glycosylase, primer, and probes targeting the N gene, upE, or ORF1a as described elsewhere.17, 18 We used RNA dilutions isolated from a MERS-CoV isolate EMC stock as a standard control.
10 μL of RNA was reverse transcribed with the Superscript III first strand synthesis system (Invitrogen, Bleiswijk, Netherlands) with random hexamers. cDNA was used to amplify six different camel MERS-CoV fragments, including a partial spike gene by use of pfu Ultra II fusion HS DNA polymerase (Agilent technologies, Santa Clara, CA, USA). We did the PCR as follows: one initial denaturation step of 95°C for 5 min, followed by 39 cycles of 95°C for 20 s, 50°C for 20 s, 72°C for 2 min, and a final extension of 72°C for 5 min. The amplified camel MERS-CoV fragments were sequenced directly on both strands with the BigDye Terminator version 3.1 cycle sequencing kit on an ABI PRISM 3100 genetic analyser (Applied Biosystems). We generated PhyML phylogenetic trees with Seaview 4 software with the approximate likelihood-ratio test based on a Shimodaira-Hasegawa-like procedure, which used a general time reversible as substitution model. We used nearest neighbour interchange and subtree pruning and regrafting-based tree search algorithms to estimate the tree topologies.19
RNA from human samples (sputum for Qatar 3 case and throat swab for Qatar 4 case) was extracted with the NucliSENS easyMag system (bioMérieux, Basingstoke, UK). Viral sequences from the two human cases were isolated (with the same techniques as for camel cases) in the Reference Department, Public Health England, London, UK. We used primers designed against sequences of MERS-CoV JX869059_EMC/2012 and bat coronaviruses HKU4 and HKU5 to amplify the entire genomes from human cases for sequencing on an Illumina MiSeq sequencer, subsequently indicated as Qatar_3_2013 (case 1) and Qatar_4_2013 (case 2).
The sequences obtained in this study were deposited in GenBank under the following accession numbers: Camel_MERS-CoV/Qatar_1_2013_ORF1a_1_partial, KF933380; Camel_MERS-CoV/Qatar_1_2013_ORF1a_3_partial, KF933381; Camel_MERS-CoV/Qatar_1_2013_ORF1b_partial, KF933382; Camel_MERS-CoV/Qatar_1_2013_Spike_partial, KF933383; Camel_MERS-CoV/Qatar_1_2013_ORF4b_full, KF933384; Camel_MERS-CoV/Qatar_1_2013_ORF1a_2_partial, KF933385; Qatar_3_2013, KF961221; and Qatar_4_2013, KF961222.
We tested all sera for the presence of IgG antibodies reactive with MERS-CoV (residues 1–747), severe acute respiratory syndrome coronavirus (SARS-CoV; residues 1–676), and human coronavirus OC43 (residues 1–760) S1 antigens as described previously.13, 20 We report results as the relative mean fluorescent intensity (RFU) for each set of quadruplicate spots per antigen. We did IgG antibody testing only because anti-camel IgM conjugates were not available.
For virus neutralisation, serum samples were heat-inactivated by incubation for 30 min at 56°C. We prepared twofold serial dilutions with 96 well plates, starting dilution at 1/10. MERS-CoV was diluted in Iscove's modified Dulbecco's medium (IMDM) supplemented with penicillin, streptomycin, and 1% fetal bovine serum, to a dilution of 2000 TCID50 per mL. We then added 50 μL of virus suspension to the plates and the plates were incubated at 37°C for 1 h. Next, we incubated virus-serum mixtures on 96 well plates containing Vero cells for 1 h, and then washed them with phosphate buffer saline and incubated them with IMDM and 1% fetal bovine serum for 5 days.
Role of the funding source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
Samples from 14 camels were obtained on Oct 17, 2013 (table , figure 1 ). Nose swab specimens from five camels were positive for all three assays (upE, N gene, and ORF1a), one sample reacted in two assays, and five camels tested positive in only one RT-PCR assay. Subsequently, all samples were subjected to a PCR assay targeting the spike gene of MERS-CoV (nucleotides 22449–22806) to obtain a fragment for sequencing as definitive confirmation. Such sequencing confirmed the presence of sequences specific to MERS-CoV in three camels.
Table.
Detection of genomes and antibodies specific for MERS-CoV in 14 dromedary camels
|
MERS-CoV detection |
MERS-CoV serology |
|||||||
|---|---|---|---|---|---|---|---|---|
| upE | ORF1a | Nucleocapsid | Spike* | Virus isolation | IFA | VNT† | Array‡ | |
| Camel 1 | Negative | 37·77 | 38·16 | Negative | Negative | Positive | 640 | 25 978 |
| Camel 2 | 38·73 | 37·01 | 37·11 | Negative | Negative | Positive | 5120 | 63 645 |
| Camel 3 | Negative | Negative | 36·53 | Negative | Negative | Positive | 640 | 14 684 |
| Camel 4 | Negative | Negative | Negative | Negative | Negative | Positive | 2560 | 53 746 |
| Camel 5 | 32·52 | 33·05 | 30·64 | Positive | Negative | Positive | 640 | 12 007 |
| Camel 6 | Negative | 37·16 | Negative | Negative | Negative | Positive | 2560 | 57 667 |
| Camel 7 | 33·97 | 34·43 | 32·72 | Positive | 37·71 | Positive | 640 | 10 740 |
| Camel 8 | Negative | Negative | 37·38 | Negative | Negative | Positive | 5120 | 63 775 |
| Camel 9 | 38·04 | 38·35 | 36·63 | Negative | Negative | Positive | 2560 | 63 465 |
| Camel 10 | Negative | Negative | 38·22 | Negative | Negative | Positive | 160 | 8002 |
| Camel 11 | 37·09 | 34·99 | 34·90 | Positive | Negative | Positive | 2560 | 37 612 |
| Camel 12 | Negative | Negative | 36·94 | Negative | Negative | Positive | 640 | 12 396 |
| Camel 13 | Negative | Negative | Negative | Negative | Negative | Positive | 2560 | 63 521 |
| Camel 14 | Negative | Negative | Negative | Negative | Negative | Positive | 1280 | 27 527 |
Presence of MERS-CoV E gene (upE), ORF1a, and nucleocapsid was assessed with a specific TaqMan assay. Virus isolation was done in Vero cells; values shown are cycle threshold values by upE test on day 4 after inoculation. IFA was done with fixed MERS-CoV infected Huh-7 cells tested at a 1/200 dilution. MERS-CoV=Middle East respiratory syndrome coronavirus. IFA=immunofluorescence assay. VNT=virus neutralisation titre.
PCR by MERS-CoV spike specific primers and subsequent sequence confirmation.
Lower detection limit of 20.
Relative fluorescence units are shown of 1/2560 dilution of sera when tested in a microarray format.
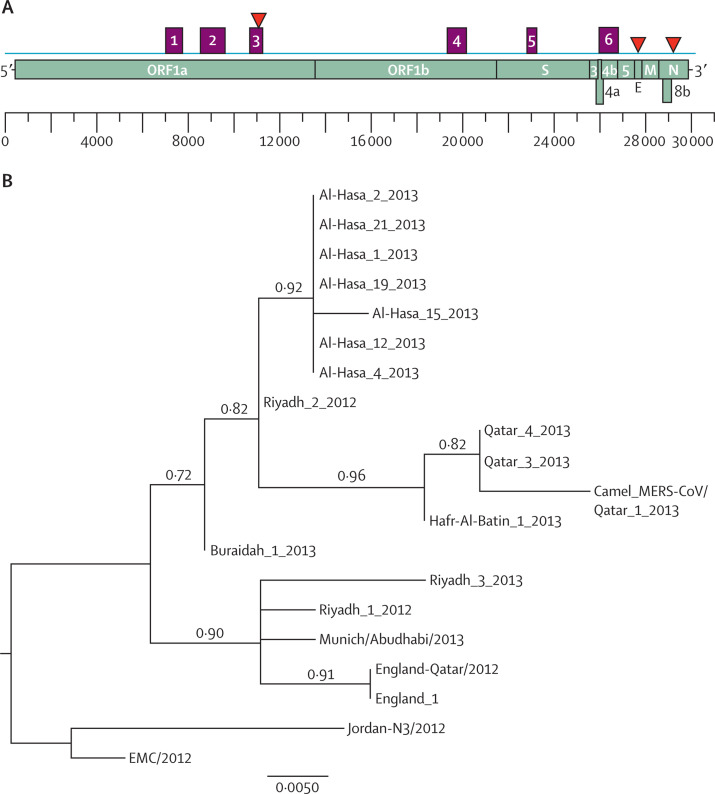
Figure 1.
Characterisation of dromedary camel Middle East respiratory syndrome coronavirus (MERS-CoV) genome sequences
(A) Nucleotide sequence fragments obtained from the nose swab of camel 5 that cover different parts of the MERS-CoV genome; purple boxes (1–6) show different fragments obtained with primers specific to MERS-CoV; red triangles show the position of the real-time TaqMan probes targeting different regions of the MERS-CoV genome. (B) Nucleotide sequences of representative MERS-CoVs' concatenated 4·2 kb sequences were analysed and a phylogenetic tree was constructed by the PhyML method; values at the branches show the result of the approximate likelihood-ratio, with values less than 0·70 not depicted.
Alignment of these partial spike gene sequences with known human MERS-CoV sequences, including those determined from the two related human cases and some other human MERS-CoV isolates such as Hafr-Al-Batin_1_2013 (KF600628) and Riyadh_3_2013 (KF600613), showed 100% identity, but the sequences differed from EMC MERS-CoV, which is used routinely in the Erasmus MC laboratory, by one nucleotide. The three camel MERS-CoV sequences obtained by this approach were identical. Virus culture attempts were not successful, although one sample tested for camel 7 yielded a positive RT-PCR of the supernatant (table) and staining of the fixed virus-infected cells (data not shown). Further passage of the virus on Vero cells, however, was not successful. In addition, serum from camel 9 showed low levels of MERS-CoV as determined by RT-PCR, and all other camel sera tested negative (data not shown). Rectal swabs and faecal materials were negative for MERS-CoV when tested for reactivity in the upE and N gene RT-PCRs.
Further sequencing of the virus from camel 5 with primers specific to MERS-CoV rendered six different fragments covering 4·2 kb across the MERS-CoV genome (figure 1). Fragments 1 (nucleotides 6708–7459), 2 (8095–9034), and 3 (10073–10658) were located in ORF1a, fragment 4 (18792–19584) in ORF1b, fragment 5 (22449–22806) in the spike gene, and fragment 6 (26074–26846) in ORF4b. The sequences obtained from the human cases from the same farm differed by one nucleotide difference in ORF1a and one in ORF4b. The MERS-CoV EMC isolate differs by eight nucleotides from the camel MERS-CoV in ORF1a. Alignment of the divergent 940 nucleotide ORF1a fragment (data not shown) and a 4·2 kb concatenated sequence fragment was used for PhyML phylogenetic analysis. The camel MERS-CoV clustered with viral sequences obtained from the two human cases related to the farm and with a sequence from Hafr-Al-Batin as the next closest relative (figure 1). Less stringent methods of analysis do show limited bootstrap support (data not shown), in line with relatively limited sequence variation within this fragment.
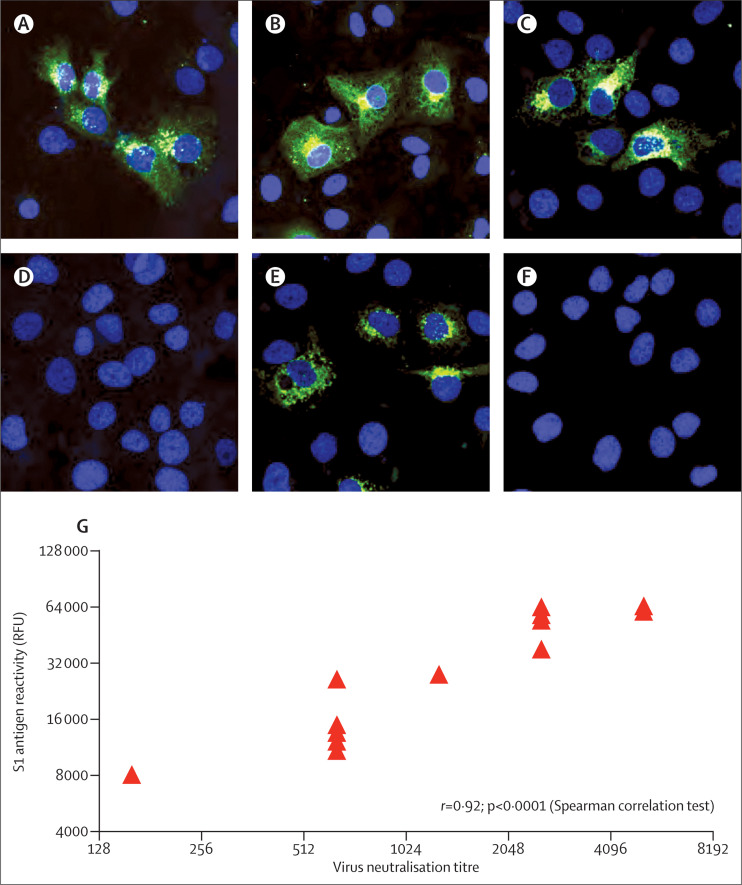
All camel sera were positive in the immunofluorescence assay when tested in a 1/200 dilution (figure 2 ). When serum samples from camels were tested at different dilutions on the microarray, we noted reactivity with MERS-CoV antigen, but not the SARS-CoV antigen. Reactivity to human coronavirus OC43 antigen, as a proxy to betacoronavirus, varied between negative (less than the cutoff value of 4000) and saturating signals (data not shown). Consistent with the array data, all serum samples had MERS-CoV neutralising capacity, with titres varying between 160 and 5120 (figure 2). We also noted a strong correlation between microarray titres and virus neutralising antibody titres (figure 2). The two camels with the highest MERS-CoV load in the swabs as shown by TaqMan assay (camels 5 and 7) had a relatively low serum neutralisation titre of 640.
Figure 2.
Middle East respiratory syndrome coronavirus (MERS-CoV) serological response in camels from Qatar
Immunofluorescence staining (green) of MERS-CoV infected and formalin fixed Huh7 cells (nuclei shown in blue) with serum samples from camel 5 (A), camel 7 (B), and camel 11 (C), negative camel control serum sample (D), a serum sample from a human case of MERS-CoV (E), and a serum sample from a healthy person (F). Fluorescent intensities (G) in relative fluorescence units (RFU) for MERS-CoV S1 antigen are shown for the 14 dromedary camels when tested at serum dilution 1/2560 versus virus neutralisation titres.
Discussion
We present, to our knowledge, the first virological confirmation of MERS-CoV in dromedary camels (panel ). We and others previously reported MERS-CoV neutralising antibodies in dromedary camels from the Canary Islands, Oman, and Egypt, suggesting circulation of MERS-CoV or a MERS-CoV-like virus in camels.13, 14 High prevalences and antibody titres were reported in the camels from Oman and Egypt suggesting widespread circulation. However, virological testing was unable to detect MERS-CoV viral sequences in camels, probably because only faecal and serum samples were analysed. In addition, shedding kinetics of MERS-CoV in camels (and human beings) are unknown, and therefore, interpretation of negative RT-PCR results from screening is difficult because positive and negative predictive values remain to be determined.22
Panel. Research in context.
Systematic review
We searched PubMed for articles published in English up to Dec 1, 2013, with the search terms “novel coronavirus EMC” or “MERS-CoV”. We identified 121 reports linked to the Middle East respiratory syndrome coronavirus (MERS-CoV). One report21 described the detection in one bat specimen of a single 181 bp fragment in a highly conserved area of the coronavirus genome, identical to the human MERS-CoV laboratory isolate EMC.
Interpretation
Our report describes the first detection of MERS-CoV in dromedary camels on a farm in Qatar that had been linked to human cases of the disease. Our study provides proof that MERS-CoV has infected dromedary camels in Qatar. Our results suggest the simultaneous occurrence of a MERS-CoV outbreak in people and camels. On the basis of the present data, we cannot conclude whether the people on the farm were infected by the camels or vice versa. Another possibility is that people and camels could have been infected from a third as yet unknown source. We recommend that detailed case histories be taken, including review of any animal exposures including animal products, and targeted (prospective) serosurveys to determine what risk factors—other than contact with an individual with the infection—are associated with human infection.
In the outbreak reported, we showed evidence for presence of the virus in six camels according to internationally recommended criteria of two independent RT-PCR targets. Furthermore, we used sequence confirmation as an additional test to increase the level of certainty—during an emerging disease outbreak such as this, options for validation of assays for clinical testing are limited.22 Discrepancies between assays can be explained by differences in detection limits, or by differences in specificity of the primer sets. In our study, N gene RT-PCR yielded four additional weak positive results that could not be confirmed by any other method, and an individual ORF1a RT-PCR identification was made for only one animal. These cases could have resulted from mispriming, or represent presence of levels of virus around the detection limit, as can be expected at the end of the infection cycle. Nevertheless, our results suggest a widespread and recent outbreak in this herd, coinciding with infection in two people.
Our data should not be taken as evidence for infection of people from the camels. Comparison of sequence data from the ORF1a gene and a concatenated 4·2 kb sequence from the human and camel viruses showed that these viruses are very similar, but distinct. Notably, samples from camels and people were analysed in different laboratories in the Netherlands and the UK. The data provided show proof that camels can be infected with MERS-CoV. The sequence difference between human and animal viruses from the farm, based on the available overlapping sequence, is so small that we cannot conclude whether the people on the farm were infected by the camels or vice versa. Another possibility is that people and camels were infected from a third as yet unknown source. Although additional sequencing might provide improved resolution, it probably will not provide conclusive evidence. The most important unknown is the exact timing of infections, both in the infected people and camels. If the virus entered the farm through either the people or the camels, the infections observed would probably have resulted from a chain of transmissions that introduced mutations.
Because we did not identify any seronegative animals that were PCR positive, and because all animals had MERS-CoV neutralising antibodies, our sampling probably took place relatively late in the outbreak on this farm. A more detailed analysis of the outbreak, including testing of additional animals and environmental samples is ongoing, as are attempts to obtain full MERS-CoV genomes of the human and animal specimens. We cannot exclude the possibility that other common livestock species including cattle, sheep, and goats, or other animals including wild species were involved in the spread of MERS-CoV. While confirmation of the source is awaited, we recommend that a detailed case history is taken of any cases of MERS-CoV, including review of any animal exposures (including animal products), and targeted (prospective) serosurveys to determine what risk factors are associated with human infection.
Acknowledgments
Acknowledgments
We thank WHO and the Food and Agriculture Organization of the United Nations for their generous help in organisation of the study, Theo Bestebroer for providing Middle East respiratory syndrome coronavirus specific primer sets, Berend Jan Bosch for providing antigens for the microarray, and Jeroen Cremer for technical support. Contributions to the study were funded through the European Union FP7 projects EMPERIE (contract number 223498; to BLH and ADMEO) and ANTIGONE (contract number 278976; to MPGK and ADMEO).
Contributors
BLH wrote the report, drew the figures, and generated, analysed, and interpreted data. SHSAD, MMA, and MPGK coordinated the study, wrote the report, and interpreted data. CBEMR wrote the report, drew the figures, did the literature search, and analysed and interpreted data. VSR drew the figures, and generated, analysed, and interpreted data. MG, RM, and AB generated and interpreted data. GJG generated, analysed, and interpreted data. MJ generated and analysed data. EF led the field investigation, obtained samples, and collected and interpreted data. AD supervised field outbreak management and collected and interpreted data. HG and FA supervised animal health field investigations. MA-T coordinated the study and interpreted data. SAA-M coordinated the study. HEAR led outbreak management and collected and interpreted data. AAK coordinated the study and interpreted data. ADMEO wrote the report and interpreted data.
Conflicts of interest
BLH, ADMEO, and VSR hold a pending patent for MERS-CoV. ADMEO is scientific adviser of Viroclinics Biosciences. All other authors declare that they have no conflicts of interest.
Contributor Information
Mohd M AlHajri, Email: malhajri1@sch.gov.qa.
Marion P G Koopmans, Email: m.koopmans@erasmusmc.nl.
References
- 1.The WHO MERS-CoV Research Group State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr. 2013 doi: 10.1371/currents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8. published online Nov 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Global Alert and Response (GAR): novel coronavirus infection—update 29 November 2013. http://www.who.int/csr/don/2013_11_29/en/index.html (accessed Nov 29, 2013).
- 3.Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2013;101C:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ithete NL, Stoffberg S, Corman VM. Close relative of human Middle East respiratory syndrome coronavirus in bat, South Africa. Emerg Infect Dis. 2013;19:1697–1699. doi: 10.3201/eid1910.130946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annan A, Baldwin HJ, Corman VM. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg Infect Dis. 2013;19:456–459. doi: 10.3201/eid1903.121503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau SK, Li KS, Tsang AK. Genetic characterization of betacoronavirus lineage C viruses in bats revealed marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: implications on the origin of the novel Middle East Respiratory Syndrome Coronavirus. J Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller MA, Raj VS, Muth D. Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines. MBio. 2012 doi: 10.1128/mBio.00515-12. published online Dec 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotten M, Watson SJ, Kellam P. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013 doi: 10.1016/S0140-6736(13)61887-5. published online Sept 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drosten C, Seilmaier M, Corman VM. Clinical features and virological analysis of a case of Middle East respiratory syndrome coronavirus infection. Lancet Infect Dis. 2013;13:745–751. doi: 10.1016/S1473-3099(13)70154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cauchemez S, Van Kerkhove MD, Riley S, Donelly CA, Fraser C, Ferguson NM. Transmission scenarios for Middle East respiratory syndrome coronavirus (MERS-CoV) and how to tell them apart. Euro Surveill. 2013;18:20503. [PMC free article] [PubMed] [Google Scholar]
- 11.Albarrak AM, Stephens GM, Hewson R, Memish ZA. Recovery from severe novel coronavirus infection. Saudi Med J. 2012;33:1265–1269. [PubMed] [Google Scholar]
- 12.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med. 2013;368:2487–2494. doi: 10.1056/NEJMoa1303729. [DOI] [PubMed] [Google Scholar]
- 13.Reusken CB, Haagmans BL, Muller MA. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perera R, Wang P, Gomaa M. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574. doi: 10.2807/1560-7917.es2013.18.36.20574. [DOI] [PubMed] [Google Scholar]
- 15.WHO Global Alert and Response (GAR): novel coronavirus infection—update 18 October 2013. http://www.who.int/csr/don/2013_10_18/en/index.html (accessed Nov 28, 2013).
- 16.WHO Global Alert and Response (GAR): novel coronavirus infection—update 29 October 2013. http://www.who.int/csr/don/2013_10_29a/en/index.html (accessed Nov 28, 2013).
- 17.Corman VM, Eckerle I, Bleicker T. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 18.Corman V, Muller M, Costabel U. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 19.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 20.Reusken C, Mou H, Godeke G. Specific serology for emerging human coronaviruses by protein microarray. Euro Surveill. 2013;18:20441. doi: 10.2807/1560-7917.es2013.18.14.20441. [DOI] [PubMed] [Google Scholar]
- 21.Memish ZA, Mishra N, Olival KJ. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis. 2013;19:1820–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Sousa R, Reusken C, Koopmans M. MERS coronavirus: data gaps for laboratory preparedness. J Clin Virol. 2013 doi: 10.1016/j.jcv.2013.10.030. published online Nov 7. [DOI] [PMC free article] [PubMed] [Google Scholar]