Abstract
Background
It has been performed worldwidely to explore the potential of animals that might be a reservoir for community associated human infections of Clostridioides difficile. Several genetically undistinguished PCR ribotypes of C. difficile from animals and human have been reported, illustrating potential transmission of C. difficile between them. Pig and calf were considered as the main origins of C. difficile with predominant RT078 and RT033, respectively. As more investigations involved, great diversity of molecular types from pig and calf were reported in Europe, North American and Australia. However, there were quite limited research on C. difficile isolates from meat animals in China, leading to non-comprehensive understanding of molecular epidemiology of C. difficile in China.
Results
A total of 55 C. difficile were isolated from 953 animal stool samples, within which 51 strains were from newborn dairy calf less than 7 days in Shandong Province. These isolates were divided into 3 STs and 6 RTs, of which ST11/RT126 was predominant type, and responsible for majority antibiotic resistance isolates. All the isolates were resistant to at least one tested antibiotics, however, only two multidrug resistant (MDR) isolates were identified. Furthermore, erythromycin (ERY) and clindamycin (CLI) were the two main resistant antibiotics. None of the isolates were resistant to vancomycin (VAN), metronidazole (MTZ), tetracycline (TET), and rifampin (RIF).
Conclusions
In this study, we analyzed the prevalence, molecular characters and antibiotic resistance of C. difficile from calf, sheep, chicken, and pig in China. Some unique features were found here: first, RT126 not RT078 were the dominant type from baby calf, and none isolates were got from pig; second, on the whole, isolates from animals display relative lower resistant rate to these 11 tested antibiotics, compared with isolates from human in China in our previous report. Our study helps to deep understanding the situation of C. difficile from economic animals in China, and to further study the potential transmission of C. difficile between meat animals and human.
Keywords: Clostridioides difficile, Economic animal, Antibiotics resistance
Background
Clostridioides difficile is a spore-forming, Gram-positive, anaerobic bacillus found ubiquitously in the environment and the gastrointestinal tracts of humans and animals [1, 2]. C. difficile has emerged as the most common infectious pathogen of antibiotic-associated diarrhea (AAD), causing heavy disease economic burden [3]. Many studies have been performed to explore the potential of animals that might be a reservoir for community associated human infections of C. difficile around the world [4–6]. Pigs and calves were the most common meat animals for the isolation of C. difficile, in which PCR ribotype 078 was recognized to be frequently isolated from pigs and RT033 from cattles, however, there was extremely variations among countries [7–9]. Besides, there are other genetically indistinguishable strains which have been identified in human and animal isolates, such as, RT237, though to only be isolated from pigs in Australia, were reported to be found from C. difficile infection (CDI) patients [10]. This finding further supports the potential transmission of C. difficile from animals to human. In addition, high intestinal colonization percentages of up to 25% have been found in families and employees living and working on pig farms [11].
In China, until now, only one study focusing on C. difficile from animals has been published, however, in which calf was not included [12]. Here, we studied the prevalence, molecular characters and antibiotic resistance of C. difficile from calf, sheep, chicken, and pig. This study helps to deep understanding the situation of C. difficile from economic animals in China, and to further study the potential transmission of C. difficile between meat animals and human. In future, further studies on the genetic relationship between animals and human C. difficile strains are required to help better understanding its role in transmission of this pathogen.
Results
C. difficile isolates from economic animals in China
A total of 55 C. difficile strains was isolated from 953 fresh stool samples of economic animals, including sheep, cow, pig, and chicken, at a rate of 5.77%. Details were summarized in Table 1. Most of the C. difficile isolates were isolated from the feces of calves (aged < 7 days of age) with rate of 43.22% (51/118) in Shandong province (Table 1). Within the 51 isolates, 5 strains are from two different batch of N and ND with a rate of 4.24%, while the rest 46 isolates are from SN and YCVTN with a rate of 38.98% (Table 1). None C. difficile isolates was isolated from adult economic animals, except for 4 C. difficile isolates were obtained from 200 feces samples of adult sheep (Table 1).
Table 1.
Isolation of C. difficile from China economic animals
| Animal | Region | No. of specimens | Age group | No. of C. difficile isolates |
|---|---|---|---|---|
| Sheep | Yunnan | 200 | Adult | 4 |
| Cow | Yunnan | 200 | Adult | 0 |
| Shandong | 118 | Calf (< 7 days) | 51 | |
| Shandong | 57 | Calf (7 days~ 1 mouth) | 0 | |
| Pig | Yunnan | 200 | Adult | 0 |
| Shandong | 120 | Piglet (23 days~ 65 days) | 0 | |
| Chicken | Shandong | 58 | – | 0 |
| Total | 953 | 55 |
Molecular characters of C. difficile isolates from economic animals in China
All the 55 C. difficile isolates were positive for genes tcdA and tcdB (A+ B+), among which 92.73% (51/55) were also positive for the binary toxin genes (CDT+). And the rest 3 isolates from Yunnan adult sheep and 1 isolate from young calf in Shandong were CDT- (Table 2). Most of the isolates (50/51) from calve below 7-days-old were CDT+ (Table 2).
Table 2.
The molecular features and antibiotic susceptibility of 55 C. difficile isolates
| Source | NO. | STs | RTs | Toxin | No. of resistant isolates / Clinical breakpoints | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tcdA | tcdB | cdtA | cdtB | MXF | CLI | TET | ERY | LVX | CIP | CHL | MEM | VAN | MTZ | RIF | ||||
| ≥8 | ≥8 | ≥16 | ≥8 | ≥8 | ≥8 | ≥32 | ≥16 | ≥4 | ≥32 | ≥4 | ||||||||
| SN | 1 | 11 | ICDC028 (RT078) | + | + | + | + | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| SN | 21 | 11 | ICDC035 (RT126) | + | + | + | + | 0 | 14 | 0 | 21 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| SN | 7 | 11 | ICDC050 | + | + | + | + | 0 | 4 | 0 | 7 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| SN | 6 | 11 | ICDC052 | + | + | + | + | 0 | 1 | 0 | 6 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| N | 4 | 11 | ICDC035 (RT126) | + | + | + | + | 0 | 3 | 0 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| YCVTN | 1 | 11 | ICDC028 (RT078) | + | + | + | + | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
| YCVTN | 9 | 11 | ICDC035 (RT126) | + | + | + | + | 0 | 6 | 0 | 9 | 0 | 3 | 0 | 0 | 0 | 0 | 0 |
| YCVTN | 1 | 11 | ICDC050 | + | + | + | + | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| YNY | 1 | 11 | ICDC035 (RT126) | + | + | + | + | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ND | 1 | 3 | ICDC039 (RT220) | + | + | – | – | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| YNY | 3 | 468 | ICDC094 | + | + | – | – | 0 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 |
| Total | 55 | 0 | 31 | 0 | 51 | 0 | 15 | 1 | 1 | 0 | 0 | 0 | ||||||
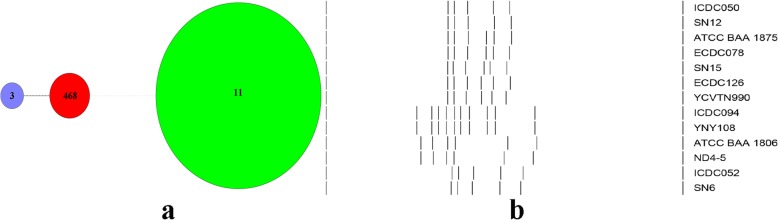
According to the multilocus sequence typing (MLST) [13], the total 55 C. difficile isolates belong to 3 MLST genotypes, in which sequence type (ST) 11 (51/55) was the most predominant one (92.73%), followed by ST468 and ST3 (Table 2). The new ST468 identified in our study, was only present in samples from sheep in Yunnan. All the C. difficile isolates in this study were divided into 2 clades as follows: clade1 (n = 4) and clade5 (n = 51) (Fig.1a).
Fig. 1.
The minimum spanning tree and PCR ribotypes (RTs) pattern of C. difficile. a Relationship of the 55 colonizing C. difficile strains by minimum spanning tree based on MLST data. Each circle corresponds to ST types, the number of which is indicated for the size of circles. The green color circle belongs to the clade5, the red and purple color circles belong to the clade1. The dotted lines between circles indicate the similarity between profiles (black, 3 alleles; gray, 7 alleles). b PCR ribotypes (RTs) banding patterns for C. difficile ICDC050, RT078 (ECDC078, ATCC BAA 1875), RT126 (ECDC126), ICDC094, RT220 (ATCC BAA 1806) and ICDC052 when visualised by BioNumerics version 7.6
In addition, all the 55 C. difficile isolates were divided into 6 different PCR ribotypes (RTs) according to the capillary electrophoresis based on QIAxcel [14] as follows: ICDC028 (n = 2), ICDC035 (n = 35), ICDC039 (n = 1), ICDC050 (n = 8), ICDC052 (n = 6) and ICDC094 (n = 3) (Fig.1b and Table 2). To be clear that ICDC028 in our study was the same as RT078, and ICDC035 was identical with RT126 (Fig.1b and Table 2).
Antibiotics resistance profile of these C. difficile strains
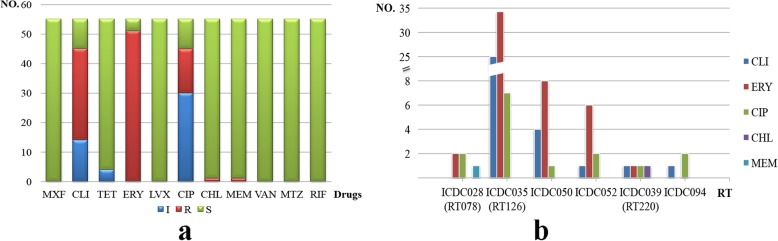
All the 55 C. difficile isolates were tested for their minimal inhibitory concentration (MIC) against 11 antimicrobial agents. As a result, they were all resistant to at least one antibiotics tested (Table 2 and Fig. 2a). Moreover, they all displayed higher resistant rate to macrolide-lincosamide-streptogramin B (MLSB), CLI and ERY, with a resistance rate at 53.36 and 92.73%, respectively (Table 2 and Fig. 2a). It is known that most isolates from human showing resistance to fluoroquinolones (FQs) in our previous study. Interestingly, all the isolates in this study are susceptible to moxifloxacin (MXF) and levofloxacin (LVX), except 15 isolates are resistant to ciprofloxacin (CIP) (Table 2 and Fig. 2a). There was only one each isolate resistant to chloramphenicol (CHL) and meropenem (MEM), respectivley (Table 2). None of the isolates were resistant to VAN, MTZ, TET, and RIF in this study (Table 2 and Fig. 2a). On the whole, isolates from animals display relative lower resistant rate to these 11 tested antibiotics, compared with isolates from human in China in our previous report [15].
Fig. 2.
The resistance characteristics and the relationship with RTs of the 55 C. difficile strains. a The resistance characteristics of all 55 C. difficile strains. The name of 11 tested drugs in the horizontal axis. For the vertical line, the numbers refer to number of strains involved in S (susceptible), I (intermediate) and R (resistant). b Different resistant number to 5 tested drugs among each RT group. CLI, ERY, CIP, CHL and MEM were included because at least one RT was resistant to them
It is indicated that RT126/ICDC035 was the main type in our study, and all the isolates with this type were resistant to at least one antibiotics (Table 2 and Fig. 2b). In addition, two isolates with RT078/ICDC028 were all resistant to both ERY and CIP, and one of them was also resistant to MEM (Table 2 and Fig. 2b). There were two isolates, ST3/RT220 and ST11/RT078, were confirmed as MDR, according to the definition. Furthermore, the MDR profile were CLI/CIP/ERY/CHL and ERY/CIP/MEM, respectively (Table 2 and Fig. 2b).
Discussion
Considering the potential zoonotic transmission of C. difficile from meat animal to humans, many studies have been performed to study the carriage of C. difficile in both economic and companion animals [11, 16–18]. Studies in North America reported the presence of C. difficile in food animals and meat with rates up to 42% [19]. Importantly, pig and calf were thought to be the predominant animals carrying C. difficile isolates. However, such kind of study is quite limited in China, although there are many molecular studies of clinical C. difficile in China, even in the Asia-Pacific area [20, 21]. Until now, there is only one report on C. difficile from pig, chicken and duck in China in 2019, but calf was not included [12]. Therefore, in this study, we determined to analyze the prevalence and genotypic characters of C. difficile from economic animals in China, including dairy calf, pig, sheep and chicken, which help to fully understand the situation of C. difficile from animals in China.
It is known that age is a key important factor affecting the isolation rate of C. difficile from animals, with a much higher prevalence in newborn than in adult animals [22]. Similarly, most of the C. difficile isolates (51/55) identified in this study, were from newborn dairy calf within 7 days. While this age effect in pigs and calves has been repeatedly reported in several studies [17, 23, 24], the reasons and the main sources for this high colonization in the first stage of life remain unknown. In a previously study from Belgium, the prevalence rate of C. difficile isolates from calves and adult cattle is 11.3 and 5.5%, respectively [25]. Although there was a higher probability of colonization in calves of less than 6 months in age than in cattle over 11 months of age [25], but none C. difficile isolates were identified from calves with age between 7 days to 1 month in our study. Furthermore, there were only 4 isolates from adult sheep, which is a little bit lower than that in Australia [5]. Most importantly, no C. difficile isolates were obtained from pig (which were thought to be the reservior of C. difficile), and chicken in our study. However, the carriage rate of that in Europe, North American and Australia is around 29.6% ~ 67.2% [26, 27]. Why no isolates are obtained from pig feces samples? It might be attributed to that the quality of feces samples collected, and the situation /location of the pig farm. In our next study, we may include more pig farms with distinct locations, and use the anaerobic swab or broth to carry the samples on site.
It is generally accepted that some PCR ribotypes seem to be more often associated with a particular animal host, such as RT078 in pigs [7–9], and RT033 in calves [28–30]. However, great diversity has been documented according to geographic locations. In Netherlands, RT012 was most prevalent in cattle in 2012, as expected, in pig samples RT078 predominated (77.8%), being the most reported type in pigs worldwide [19]. In observations from the USA and Canada, RT078 was also reported from calf samples [7, 31]. In addition, 4 RTs (127, 288, 033, and 126) with binary toxin-positive accounts for 70.3% (71/101) of isolates from a calf farm in Australia [32]. In this study, three STs (ST11, 3, and 468) and 6 RTs (RT126, ICDC050, ICDC052, ICDC094, RT078, and RT220) were identified. ST11 (51/55, 92.7%) and RT126 (35/55, 63.6%) were predominant molecular types. Besides, a new type, ST468 was found in our study. Different from our results, prevalence of RT126 from calf in Australia is about 2–6% [16, 32], and in a recent study from Germany is 4% [9]. Zidaric et al. [30] found that RT126 and RT078 predominated in calf on a single veal farm in Belgium. Interestingly, RT126 has also been found in 20% of pigs in Germany [26]. Prevalence of RT126 in humans is relatively low, accounting for 3% of infections in a 2008 European survey [33], however, RT126 is one of the most frequently isolated in humans in Spain [8]. There also are reports of increasing incidence in Taiwan [34], as well as small numbers recovered from patients in Kuwait [35] and Australia [36]. RT126 shares a very similar banding pattern with RT078, and these two RTs are often grouped together. Indeed, it is normally considered as a variant of 078 and has been reported in river water and different animal species in many countries [37, 38], indicating the possible zoonotic potential of this ribotype [6, 26]. In our study, there were 51 out of 55 isolates are tcdA, B gene and binary toxin gene positive, which are consistent with previous reports around the world that majority of the animal C. difficile isolates are toxigenic [8, 18].
The antibiotic susceptibility test displayed that none isolates were resistant to MXF, LVX, TET, RIF, VAN, and MTZ. While these isolates showed high resistant rate to ERY (92.73%), followed by CLI (53.36) and CIP (27.2%). The C. difficile isolates from economic animals in our study showed relative high resistance rate to ERY than other countries (45.5–52.9%) [12, 38]. In addition, the majority isolates resistant to ERY and CLI are RT126. The resistant rate to TET varies a lot among different studies. For example, C. difficile isolates from pig resistant to TET reached as high as 77.3% in a previous study in China, and similar 76.5% resistance rate in a Spain study [38]. Until now, almost none C. difficile isolates from meat animals are resistant to MTZ and VAN, including our study [15], which is a little bit different from drug resistant profile of human that susceptibility to MTZ is decreasing, and even resistant isolates were found [39]. Although all C. difficile isolates were resistant to at least one tested drug in our study, but only two MDR isolates were found, indicating lower MDR rate than that in human [40]. Furthermore, C. difficile isolates are resistant to less type of drugs compared with our previous study from human [15].
Conclusions
We explore the prevalence, molecular features, and antibiotic resistance of C. difficile from economic animals in China. This study helps to deep understanding the situation of C. difficile from economic animals in China, and to further study the potential transmission of C. difficile between meat animals and human.
Methods
Sample collection
During 2017 and 2018, we collected 953 fresh stool samples from economic animals from Shandong and Yunnan provinces, which including 200 sheep samples, 375 cow samples, 320 pig samples, and 58 chicken samples (Table 1). All animal’s stool samples were authorized and obtained from the farm and the relevant units.
In August 2017, we collected 200 samples from adult sheep (YNY), cows (YNN), pigs (YNZ) in Yunnan, all stool samples were cryopreserved after collection and delivered rapidly to the laboratory. The other stool samples were collected in Shandong province. In October 2017, we collected 79 from dairy calves (N, ND), 120 from pigs (PD), and 58 from chickens (CL) in Jinan. In addition, 50 stool samples (SN) and 46 stool samples (YCVTN) from dairy calves were randomly collected from the farm in Yucheng in 2017–12 and 2018–5. All fecal specimens were added directly to an enrichment broth (Cooked Meat Medium, Oxoid, UK) containing gentamicin (5 mg/L), cycloserine (250 mg/L) and cefoxitin (8 mg/L). Put them in the anaerobic bag (mitsubishi gas chemical company INC., GENBAG, BioMerieux, France), brought to the laboratory, and incubated in an anaerobic jar (Mart, NL) at 37 °Cfor 48 h.
Isolation of C. difficile
1 ml of enrichment broth was alcohol shocked with an equal volume of absolute ethanol for 1 h. A volume of 100 μl supernatant was plated directly on selective cycloserine-cefoxitin-fructose agar plates (CCFA, Oxoid, UK) with 5% egg yolk after ethanol shock treatment and incubated in an anaerobic jar at 37 °C for 48 h. Isolation of C. difficile was based on previously described methods, suspected colonies were further confirmed by the 16S rRNA gene [41]. All colonies were cultured on anaerobic medium in an anaerobic environment at 37 °C.
Multilocus sequence typing (MLST), toxin gene profile, and PCR-Ribotyping
The methods of MLST, toxin genes profile and PCR-Ribotyping was obtained according to previously reported [13, 41]. The primers and the amplification conditions used for PCR ribotyping have been described previously [42].
Antimicrobial susceptibility test
The minimum inhibitory concentrations (MICs) for 11 antimicrobial agents were determined by Etest strips (bioMérieux, France, and Liofilchem, Italy) on Brucella agar plates (Oxoid, Basingstoke, UK) containing 1 mg/L vitamine K1, 5 mg/L chlorhematin and 5% defibrinated sheep blood, according to the manufacturer’s instructions.
C. difficile isolates were tested for susceptibility to MXF, VAN, CLI, TET, ERY, RIF, LVX, CHL, MTZ, CIP and MEM using E-test strips (Biomerieux, France, and Liofilchem, Italy). The breakpoints for antimicrobial agents are shown in Table 2 and are based on recommendations of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucst.org) and the Clinical and Laboratory Standards Institute (CLSI) M11-A8 and M100-S28, were determined according to a previous study [40, 43, 44]. MDR were defined as resistance to at least three antimicrobial classes. C. difficile ATCC 700057 was included as a control in each experiment.
Acknowledgements
Not applicable.
Abbreviations
- PCR
Polymerase chain reaction
- RT
Ribotype
- MLST
Multilocus sequence typing
- MDR
Multidrug resistant
Authors’ contributions
YW and JXL designed the study. WGL, HL, ZJL and XZ collected and analyzed the data. YQL, QZ and WPG contributed samples collection and intellectual input. WZZ and YW drafted and wrote the manuscript. YQL, QZ and WPG revised the manuscript critically for intellectual content, all authors have contributed strains and participated at writing. All authors read and approved the final manuscript.
Funding
This study was supported by the National Sci-Tech Key Project (grant No. 2018ZX10733402), and by the State Key Laboratory for Infectious Disease Prevention and Control in China CDC (grant No. 2019SKLID317).
Availability of data and materials
The data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Ethics approval do not apply to our study, all institutes involved in this research are consent to participate.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of Clostridium difficile as Clostridioides difficile (hall and O'Toole 1935) Prévot 1938. Anaerobe. 2016;40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Knight DR, Elliott B, Chang BJ, Perkins TT, Riley TV. Diversity and evolution in the genome of Clostridium difficile. Clin Microbiol Rev. 2015;28(3):721–741. doi: 10.1128/CMR.00127-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janezic S, Zidaric V, Pardon B, Indra A, Kokotovic B, Blanco J, et al. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol. 2014;14(1):173. doi: 10.1186/1471-2180-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight DR, Riley TV. Prevalence of gastrointestinal Clostridium difficile carriage in Australian sheep and lambs. Appl Environ Microbiol. 2013;79(18):5689–5692. doi: 10.1128/AEM.01888-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai BY, Ko WC, Chen TH, Wu YC, Lan PH, Chen YH, et al. Zoonotic potential of the Clostridium difficile RT078 family in Taiwan. Anaerobe. 2016;41:125–130. doi: 10.1016/j.anaerobe.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Goorhuis A, Debast SB, van Leengoed LA, Harmanus C, Notermans DW, Bergwerff AA, et al. Clostridium difficile PCR Ribotype 078: an emerging strain in humans and in pigs? J Clin Microbiol. 2008;46(3):1157. doi: 10.1128/JCM.01536-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Álvarez-Pérez S, Blanco JL, Harmanus C, Kuijper E, García ME. Subtyping and antimicrobial susceptibility of Clostridium difficile PCR ribotype 078/126 isolates of human and animal origin. Vet Microbiol. 2017;199:15–22. doi: 10.1016/j.vetmic.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 9.Schneeberg A, Neubauer H, Schmoock G, Grossmann E, Seyboldt C. Presence of Clostridium difficile PCR ribotype clusters related to 033, 078 and 045 in diarrhoeic calves in Germany. J Med Microbiol. 2013;62(Pt 8):1190–1198. doi: 10.1099/jmm.0.056473-0. [DOI] [PubMed] [Google Scholar]
- 10.Mc Govern AM, Foster NF, Pereira LA, Knight DR, Elliott B, Chang BJ, et al. Human Clostridium difficile infection caused by a livestock-associated PCR ribotype 237 strain in Western Australia. JMM Case Rep. 2016;3(4):e005062. doi: 10.1099/jmmcr.0.005062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keessen EC, Harmanus C, Dohmen W, Kuijper EJ, Lipman LJ. Clostridium difficile infection associated with pig farms. Emerg Infect Dis. 2013;19(6):1032–1034. doi: 10.3201/eid1906.121645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang LJ, Yang L, Gu XX, Chen PX, Fu JL, Jiang HX. The first isolation of Clostridium difficile RT078/ST11 from pigs in China. PLoS One. 2019;14(2):e0212965. doi: 10.1371/journal.pone.0212965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao F, Li W, Gu W, Zhang W, Liu X, Fu X, et al. A retrospective study of community-acquired Clostridium difficile infection in Southwest China. Sci Rep. 2018;8(1):3992. doi: 10.1038/s41598-018-21762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zhang WZ, Li WG, Zhao HQ, Wu YH, Li H, et al. Standardization and application on ribotyping library of Clostridioides difficile in China. Chin J Epidemiol. 2019;40(12):1624–1628. doi: 10.3760/cma.j.issn.0254-6450.2019.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Li WG, Zhang WZ, Yu SB, Liu ZJ, Zhang X, et al. Antibiotic resistance of clinical isolates of Clostridioides difficile in China and its association with geographical regions and patient age. Anaerobe. 2019;60:102094. doi: 10.1016/j.anaerobe.2019.102094. [DOI] [PubMed] [Google Scholar]
- 16.Knight DR, Thean S, Putsathit P, Fenwick S, Riley TV. Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl Environ Microbiol. 2013;79:2630–2635. doi: 10.1128/AEM.03951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houser BA, Soehnlen MK, Wolfgang DR, Lysczek HR, Burns CM, Jayarao BM. Prevalence of Clostridium difficile toxin genes in the feces of veal calves and incidence of ground veal contamination. Foodborne Pathog Dis. 2012;9:32–36. doi: 10.1089/fpd.2011.0955. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez C, Taminiau B, Van Broeck J, Delmée M, Daube G. Clostridium difficile in food and animals: a comprehensive review. Adv Exp Med Biol. 2016;932:65–92. doi: 10.1007/5584_2016_27. [DOI] [PubMed] [Google Scholar]
- 19.Koene MG, Mevius D, Wagenaar JA, Harmanus C, Hensgens MP, Meetsma AM, et al. Clostridium difficile in Dutch animals: their presence, characteristics and similarities with human isolates. Clin Microbiol Infect. 2012;18(8):778–784. doi: 10.1111/j.1469-0691.2011.03651.x. [DOI] [PubMed] [Google Scholar]
- 20.Luo Y, Cheong E, Bian Q, Collins DA, Ye JL, Shin JH, et al. Different molecular characteristics and antimicrobial resistance profiles of Clostridium difficile in the Asia-Pacific region. Emerg Microbes Infect. 2019;8(1):1553–1562. doi: 10.1080/22221751.2019.1682472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins DA, Sohn KM, Wu Y, Ouchi K, Ishii Y, Elliott B, et al. Clostridioides difficile infection in the Asia-Pacific region. Emerg Microbes Infect. 2020;9(1):42–52. doi: 10.1080/22221751.2019.1702480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa MC, Stämpfli HR, Arroyo LG, Pearl DL, Weese JS. Epidemiology of Clostridium difficile on a veal farm: prevalence, molecular characterization and tetracycline resistance. Vet Microbiol. 2011;152:379–384. doi: 10.1016/j.vetmic.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Palacios A, Stämpfli HR, Duffield T, Peregrine AS, Trotz-Williams LA, Arroyo LG, et al. Clostridium difficile PCR ribotypes in calves, Canada. Emerg Infect Dis. 2006;12(11):1730–1736. doi: 10.3201/eid1211.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopman NE, Keessen EC, Harmanus C, Sanders IM, van Leegoed LA, Kuijper EJ, et al. Acquisition of Clostridium difficile by piglets. Vet Microbiol. 2011;149(1–2):186–192. doi: 10.1016/j.vetmic.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez C, Hakimi DE, Vanleyssem R, Taminiau B, Van Broeck J, Delmée M, et al. Clostridium difficile in beef cattle farms, farmers and their environment: assessing the spread of the bacterium. Vet Microbiol. 2017;210:183–187. doi: 10.1016/j.vetmic.2017.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Knight DR, Squire MM, Riley TV. Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes. Appl Environ Microbiol. 2015;81(1):119–123. doi: 10.1128/AEM.03032-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Susick EK, Putnam M, Bermudez DM, Thakur S. Longitudinal study comparing the dynamics of Clostridium difficile in conventional and antimicrobial free pigs at farm and slaughter. Vet Microbiol. 2012;157(1–2):172–178. doi: 10.1016/j.vetmic.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Romano V, Albanese F, Dumontet S, Krovacek K, Petrini O, Pasquale V. Prevalence and genotypic characterization of Clostridium difficile from ruminants in Switzerland. Zoonoses Public Health. 2012;59(8):545–548. doi: 10.1111/j.1863-2378.2012.01540.x. [DOI] [PubMed] [Google Scholar]
- 29.Pirs T, Ocepek M, Rupnik M. Isolation of Clostridium difficile from food animals in Slovenia. J Med Microbiol. 2008;57(6):790–792. doi: 10.1099/jmm.0.47669-0. [DOI] [PubMed] [Google Scholar]
- 30.Zidaric V, Pardon B, Dos Vultos T, Deprez P, Brouwer MS, Roberts AP, et al. Different antibiotic resistance and sporulation properties within multiclonal Clostridium difficile PCR ribotypes 078, 126, and 033 in a single calf farm. Appl Environ Microbiol. 2012;78(24):8515–8522. doi: 10.1128/AEM.02185-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keel K, Brazier JS, Post KW, Weese S, Songer JG. Prevalence of PCR ribotypes among Clostridium difficile isolates from pigs, calves, and other species. J Clin Microbiol. 2007;45(6):1963–1964. doi: 10.1128/JCM.00224-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight DR, Putsathit P, Elliott B, Riley TV. Contamination of Australian newborn calf carcasses at slaughter with Clostridium difficile. Clin Microbiol Infect. 2016;22(3):266.e1–266.e7. doi: 10.1016/j.cmi.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377(9759):63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 34.Hung YP, Lin HJ, Tsai BY, Liu HC, Liu HC, Lee JC, et al. Clostridium difficile ribotype 126 in southern Taiwan: a cluster of three symptomatic cases. Anaerobe. 2014;30:188–192. doi: 10.1016/j.anaerobe.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Jamal WY, Rotimi VO. Surveillance of antibiotic resistance among hospital- and community-acquired toxigenic Clostridium difficile isolates over 5-year period in Kuwait. PLoS One. 2016;11(8):e0161411. doi: 10.1371/journal.pone.0161411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight DR, Hart J, Gottardo NG, Eyre DW, Crook DW, Riley TV. Two cases of Clostridium difficile infection in unrelated oncology patients attributable to a single clone of C. difficile PCR ribotype 126. JMM Case Rep. 2015;2:e000043. doi: 10.1099/jmmcr.0.000043. [DOI] [Google Scholar]
- 37.Knetsch CW, Connor TR, Mutreja A, van Dorp SM, Sanders IM, Browne HP, et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 2014;19(45):20954. doi: 10.2807/1560-7917.ES2014.19.45.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrés-Lasheras S, Bolea R, Mainar-Jaime RC, Kuijper E, Sevilla E, Martín-Burriel I, et al. Presence of Clostridium difficile in pig faecal samples and wild animal species associated with pig farms. J Appl Microbiol. 2017;122(2):462–472. doi: 10.1111/jam.13343. [DOI] [PubMed] [Google Scholar]
- 39.Peláez T, Alcalá L, Alonso R, Rodríguez-Créixems M, García-Lechuz JM, Bouza E. Reassessment of Clostridium difficile susceptibility to metronidazole and vancomycin. Antimicrob Agents Chemother. 2002;46(6):1647–1650. doi: 10.1128/AAC.46.6.1647-1650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spigaglia P, Barbanti F, Mastrantonio P, European Study Group on Clostridium difficile (ESGCD) Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother. 2011;66(10):2227–2234. doi: 10.1093/jac/dkr292. [DOI] [PubMed] [Google Scholar]
- 41.Liu XS, Li WG, Zhang WZ, Wu Y, Lu JX. Molecular characterization of Clostridium difficile isolates in China from 2010 to 2015. Front Microbiol. 2018;9:845. doi: 10.3389/fmicb.2018.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stubbs SL, Brazier JS, O’Neill GL, Duerden BI. PCR targeted to the 16S-23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J Clin Microbiol. 1999;37(2):461–463. doi: 10.1128/JCM.37.2.461-463.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Y, Liu C, Li WG, Xu JL, Zhang WZ, Dai YF, et al. Independent microevolution mediated by mobile genetic elements of individual Clostridium difficile isolates from clade 4 revealed by whole-genome sequencing. mSystems. 2019;4(2):e00252–e00218. doi: 10.1128/mSystems.00252-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin D, Luo Y, Huang C, Cai J, Ye J, Zheng Y, et al. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J Clin Microbiol. 2017;55(3):801–810. doi: 10.1128/JCM.01898-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data generated and/or analyzed during the current study are available from the corresponding author on reasonable request.